Abstract
DNA sequencing by synthesis (SBS) on a solid surface during polymerase reaction offers a paradigm to decipher DNA sequences. We report here the construction of such a DNA sequencing system using molecular engineering approaches. In this approach, four nucleotides (A, C, G, T) are modified as reversible terminators by attaching a cleavable fluorophore to the base and capping the 3′-OH group with a small chemically reversible moiety so that they are still recognized by DNA polymerase as substrates. We found that an allyl moiety can be used successfully as a linker to tether a fluorophore to 3′-O-allyl-modified nucleotides, forming chemically cleavable fluorescent nucleotide reversible terminators, 3′-O-allyl-dNTPs-allyl-fluorophore, for application in SBS. The fluorophore and the 3′-O-allyl group on a DNA extension product, which is generated by incorporating 3′-O-allyl-dNTPs-allyl-fluorophore in a polymerase reaction, are removed simultaneously in 30 s by Pd-catalyzed deallylation in aqueous buffer solution. This one-step dual-deallylation reaction thus allows the reinitiation of the polymerase reaction and increases the SBS efficiency. DNA templates consisting of homopolymer regions were accurately sequenced by using this class of fluorescent nucleotide analogues on a DNA chip and a four-color fluorescent scanner.
Keywords: DNA chip
DNA sequencing is driving genomics research and discovery. The completion of the Human Genome Project has set the stage for screening genetic mutations to identify disease genes on a genome-wide scale (1). Accurate high-throughput DNA sequencing methods are needed to explore the complete human genome sequence for applications in clinical medicine and health care. To overcome the limitations of the current electrophoresis-based sequencing technology (2–5), a variety of new DNA-sequencing methods have been investigated. Such approaches include sequencing by hybridization (6), mass spectrometry-based sequencing (7–9), sequence-specific detection of single-stranded DNA using engineered nanopores (10), and sequencing by ligation (11). More recently, DNA sequencing by synthesis (SBS) approaches such as pyrosequencing (12), sequencing of single DNA molecules (13), and polymerase colonies (14) have been widely explored.
The concept of DNA SBS was revealed in 1988 with an attempt to sequence DNA by detecting the pyrophosphate group that is generated when a nucleotide is incorporated in a DNA polymerase reaction (15). Pyrosequencing, which was developed based on this concept and an enzymatic cascade, has been explored for genome sequencing (16). However, there are inherent difficulties in this method for determining the number of incorporated nucleotides in homopolymeric regions of the template. Additionally, each of the four nucleotides needs to be added and detected separately, which increases the overall detection time. The accumulation of undegraded nucleotides and other components could also lower the accuracy of the method when sequencing a long DNA template. It is thus desirable to have a simple method to directly detect a reporter group attached to the nucleotide that is incorporated into a growing DNA strand in the polymerase reaction rather than relying on a complex enzymatic cascade. The SBS scheme based on fluorescence detection coupled with a chip format has the potential to markedly increase the throughput of DNA sequencing projects. Consequently, several groups have investigated such a system with an aim to construct an ultra high-throughput DNA sequencing method (17, 18). Thus far, no complete success of using such a system to unambiguously sequence DNA has been published.
Previous work in the literature exploring the SBS method is mostly focused on designing and synthesizing a cleavable chemical moiety that is linked to a fluorescent dye to cap the 3′-OH group of the nucleotides (19–21). The rationale is that, after the fluorophore is removed, the 3′-OH would be regenerated to allow subsequent nucleotide addition. However, no success has been reported for the incorporation of such a nucleotide with a cleavable fluorescent dye on the 3′ position by DNA polymerase into a growing DNA strand. The reason is that the 3′ position on the deoxyribose is very close to the amino acid residues in the active site of the polymerase, and the polymerase is therefore sensitive to modification in this area of the ribose ring, especially with a large fluorophore (22).
It is known that some modified DNA polymerases are highly tolerable for nucleotides with extensive modifications with bulky groups such as energy transfer dyes at the 5-position of the pyrimidines (T and C) and 7-position of purines (G and A) (23, 24). The ternary complexes of a rat DNA polymerase, a DNA template-primer, and dideoxycytidine triphosphate have been determined (22), which supports this fact. We thus reasoned that if a unique fluorescent dye is linked to the 5-position of the pyrimidines (T and C) and the 7-position of purines (G and A) via a cleavable linker, and a small chemical moiety is used to cap the 3′-OH group, the resulting nucleotide analogues should be able to incorporate into the growing DNA strand as terminators. Based on this rationale, we proposed an SBS approach using cleavable fluorescent nucleotide analogues as reversible terminators to sequence surface-immobilized DNA [supporting information (SI) Fig. 6] (25). In this approach, the nucleotides are modified at two specific locations so that they are still recognized by DNA polymerase as substrates: (i) a different fluorophore with a distinct fluorescent emission is linked to each of the four bases through a cleavable linker and (ii) the 3′-OH group is capped by a small chemically reversible moiety. DNA polymerase incorporates only a single nucleotide analogue complementary to the base on a DNA template covalently linked to a surface. After incorporation, the unique fluorescence emission is detected to identify the incorporated nucleotide and the fluorophore is subsequently removed. The 3′-OH group is then chemically regenerated, which allows the next cycle of the polymerase reaction to proceed. Because the large surface on a DNA chip can have a high density of different DNA templates spotted, each cycle can identify many bases in parallel, allowing the simultaneous sequencing of a large number of DNA molecules. We have previously established the feasibility of performing SBS on a chip using four photocleavable fluorescent nucleotide analogues (26) and discovered that an allyl group can be used as a cleavable linker to bridge a fluorophore to a nucleotide (27). We have also reported the design and synthesis of two photocleavable fluorescent nucleotides as reversible terminators for polymerase reaction (28, 29).
Our previous research efforts have firmly established the molecular level strategy to rationally modify the nucleotides by attaching a cleavable fluorescent dye to the base and capping the 3′-OH with a small chemically reversible moiety for SBS. This approach was recently adopted by Genomics Industry to potentially provide a platform for DNA sequencing (30). Here we report the design and synthesis of four chemically cleavable fluorescent nucleotide analogues as reversible terminators for SBS. Each of the nucleotide analogues contains a 3′-O-allyl group and a unique fluorophore with a distinct fluorescence emission at the base through a cleavable allyl linker. We first established that these nucleotide analogues are good substrates for DNA polymerase in a solution-phase DNA extension reaction and that the fluorophore and the 3′-O-allyl group can be removed with high efficiency in aqueous solution. We then performed SBS using these four chemically cleavable fluorescent nucleotide analogues as reversible terminators to identify ≈20 continuous bases of a DNA template immobilized on a chip. Accurate DNA sequences were obtained for DNA templates containing homopolymer sequences. The DNA template was immobilized on the surface of the chip that contains a PEG linker with 1,3-dipolar azide-alkyne cycloaddition chemistry. These results indicated that successful cleavable fluorescent nucleotide reversible terminators for four-color DNA sequencing by synthesis can be designed by attaching a cleavable fluorophore to the base and capping the 3′-OH with a small chemically reversible moiety so that they are still recognized by DNA polymerase as substrates. Further optimization of the approach will lead to even longer sequencing read lengths.
Results and Discussion
Design and Synthesis of Chemically Cleavable Fluorescent Nucleotide Analogues as Reversible Terminators for SBS.
To demonstrate the feasibility of carrying out de novo DNA sequencing by synthesis on a chip, four chemically cleavable fluorescent nucleotide analogues (3′-O-allyl-dCTP-allyl-bodipy-FL-510, 3′-O-allyl-dUTP-allyl-R6G, 3′-O-allyl-dATP-allyl-ROX and 3′-O-allyl-dGTP-allyl-bodipy-650/Cy5) (R6G, 6-carboxyrhodamine-6G; Rox, 6-carboxy-X-rhodamine; bodipy, 4,4-difluoro-4-bora-3α,4α-diaza-s-indacene; Fig. 1) were designed and synthesized as reversible terminators for DNA polymerase reaction. Modified DNA polymerases have been shown to be highly tolerant to nucleotide modifications with bulky groups at the 5-position of pyrimidines (C and U) and the 7-position of purines (A and G). Thus, we attached each unique fluorophore to the 5-position of C/U and the 7 position of A/G through an allyl carbamate linker. However, due to the close proximity of the 3′ position on the sugar ring of a nucleotide to the amino acid residues of the active site of the DNA polymerase, a relatively small allyl moiety was chosen as the 3′-OH reversible capping group. We have found that the fluorophore and the 3′-O-allyl group on a DNA extension product, which is generated by incorporation of the chemically cleavable fluorescent nucleotide analogues, are removed simultaneously in 30 s by Pd-catalyzed deallylation in aqueous solution. This one-step dual-deallylation reaction thus allows the re-initiation of the polymerase reaction. The detailed synthesis procedure and characterization of the four nucleotide analogues in Fig. 1 are described in SI Text.
Fig. 1.
Structures of 3′-O-allyl-dCTP-allyl-bodipy-FL-510 [λabs(max) = 502 nm; λem(max) = 510 nm], 3′-O-allyl-dUTP-allyl-R6G [λabs(max) = 525 nm; λem(max) = 550 nm], 3′-O-allyl-dATP-allyl-ROX [λabs(max) = 585 nm; λem(max) = 602 nm], and 3′-O-allyl-dGTP-allyl-bodipy-650 [λabs(max) = 630 nm; λem(max) = 650 nm].
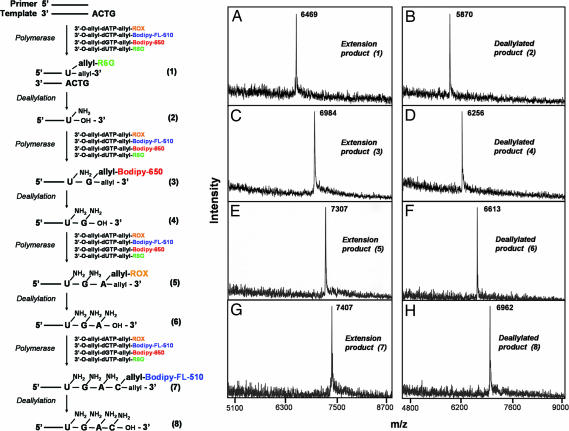
To verify that these fluorescent nucleotide analogues are incorporated accurately in a base-specific manner in a polymerase reaction, four continuous steps of DNA extension and deallylation were carried out in solution. This allows the isolation of the DNA product at each step for detailed molecular structure characterization by MALDI-TOF mass spectrometry (MS) as shown in Fig. 2. The first extension product 5′-U(allyl-R6G)-3′-O-allyl (1) was purified by HPLC and analyzed using MALDI-TOF MS (Fig. 2A). This product was then deallylated using a Pd-catalyzed deallylation mixture [1× Thermopol I reaction buffer/Na2PdCl4/P(PhSO3Na)3]. The active Pd catalyst is generated from Na2PdCl4 and a ligand P(PhSO3Na)3 (TPPTS) to mediate the deallylation reaction in DNA compatible aqueous condition to simultaneously cleave both the fluorophore and the 3′-O-allyl group (28). The deallylated product (2) was also analyzed by using MALDI-TOF MS (Fig. 2B). As can be seen in Fig. 2A, the MALDI-TOF MS spectrum consists of a distinct peak at m/z 6469 corresponding to the DNA extension product 5′-U(allyl-R6G)-3′-O-allyl (1), which confirms that the nucleotide analogue can be incorporated base specifically among pool of all four (A, C, G, T) by DNA polymerase into a growing DNA strand. Fig. 2B shows the deallylation result of the above DNA product. The peak at m/z 6469 has completely disappeared, whereas the peak corresponding to the dual deallylated product 5′–U (2) appears as the sole dominant peak at m/z 5870, which establishes that the Pd-catalyzed deallylation reaction completely cleaves both the fluorophore and the 3′-O-allyl group with high efficiency without damaging the DNA. The next extension reaction was carried out by using this deallylated DNA product with a free 3′-OH group regenerated as a primer along with four allyl modified fluorescent nucleotide mixture to yield an extension product 5′-UG(allyl-bodipy-650)-3′-O-allyl (3). As described above, the extension product 3 was analyzed by MALDI-TOF MS producing a dominant peak at m/z 6984 (Fig. 2C), and then deallylated for further MS analysis yielding a single peak at m/z 6256 (product 4) (Fig. 2D). The third extension reaction yielding 5′-UGA(allyl-ROX)-3′-O-allyl (5), the fourth extension reaction yielding 5′-UGAC(allyl-bodipy-FL-510)-3′-O-allyl (7), and their deallylation reactions to yield products 6 and 8 were similarly carried out and analyzed by MALDI-TOF MS as shown in Fig. 2 E–H. The chemical structures of the extension and cleavage products for each step are shown in SI Fig. 7. These results demonstrate that the above-synthesized four chemically cleavable fluorescent nucleotide analogues are successfully incorporated with high fidelity into the growing DNA strand in a polymerase reaction, and furthermore, both the fluorophore and the 3′-O-allyl group are efficiently removed by using a Pd-catalyzed deallylation reaction, which makes it feasible to use them for SBS on a chip.
Fig. 2.
The polymerase extension scheme (Left) and MALDI-TOF MS spectra of the four consecutive extension products and their deallylated products (Right). Primer extended with 3′-O-allyl-dUTP-allyl-R6G (1), and its deallylated product 2; Product 2 extended with 3′-O-allyl-dGTP-allyl-bodipy-650 (3), and its deallylated product 4; Product 4 extended with 3′-O-allyl-dATP-allyl-ROX (5), and its deallylated product 6; Product 6 extended with 3′-O-allyl-dCTP-allyl-bodipy-FL-510 (7), and its deallylated product (8). After 30 s of incubation with the palladium/TPPTS mixture at 70°C, deallylation is complete with both the fluorophores and the 3′-O-allyl groups cleaved from the extended DNA products.
Four-Color DNA Sequencing with Chemically Cleavable Fluorescent Nucleotide Analogues as Reversible Terminators on a DNA Chip.
The chemically cleavable fluorescent nucleotide analogues were then used in an SBS reaction to identify the sequence of the DNA template immobilized on a solid surface. A site-specific 1,3-dipolar cycloaddition coupling chemistry was used to covalently immobilize the alkyne-labeled self-priming DNA template on the azido-functionalized surface in the presence of a Cu(I) catalyst. The principal advantage offered by the use of a self-priming moiety as compared with using separate primers and templates is that the covalent linkage of the primer to the template in the self-priming moiety prevents any possible dissociation of the primer from the template during the process of SBS. To prevent nonspecific absorption of the unincorporated fluorescent nucleotides on the surface of the chip, a PEG linker is introduced between the DNA templates and the chip surface (SI Fig. 8). This approach was shown to produce very low background fluorescence after cleavage to remove the fluorophore as demonstrated by the DNA sequencing data described below.
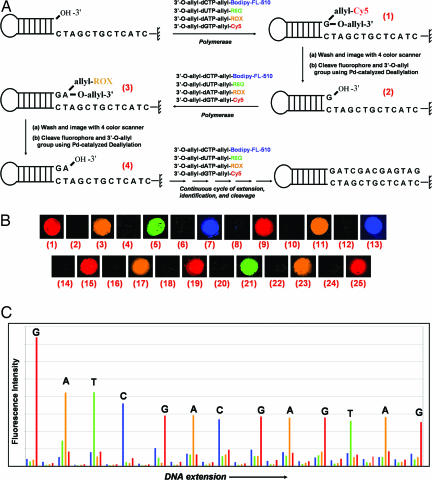
We first performed SBS on a chip-immobilized DNA template that has no homopolymer sequences using the four chemically cleavable fluorescent nucleotide reversible terminators (3′-O-allyl-dCTP-allyl-bodipy-FL-510, 3′-O-allyl-dUTP-allyl-R6G, 3′-O-allyl-dATP-allyl-ROX and 3′-O-allyl-dGTP-allyl-Cy5), and the results are shown in Fig. 3. The structure of the self-priming DNA moiety is shown schematically in Fig. 3A, with the first 13 nucleotide sequences immediately after the priming site. The de novo sequencing reaction on the chip was initiated by extending the self-priming DNA using a solution containing all four 3′-O-allyl-dNTPs-allyl-fluorophore, and a 9°N mutant DNA polymerase. To negate any lagging fluorescence signal that is caused by previously unextended priming strand, a synchronization step was added to reduce the amount of unextended priming strands after the extension with the fluorescent nucleotides. A synchronization reaction mixture consisting of all four 3′-O-allyl-dNTPs (Fig. 4), which have a higher polymerase incorporation efficiency due to the lack of a fluorophore compared with the bulkier 3′-O-allyl-dNTPs-allyl-fluorophore, was used along with the 9°N mutant DNA polymerase to extend any remaining priming strand that has a free 3′-OH group to synchronize the incorporation. The extension by 3′-O-allyl-dNTPs also enhances the enzymatic incorporation of the next nucleotide analogue, because after cleavage to remove the 3′-O-allyl group, the DNA product extended by 3′-O-allyl-dNTPs carry no modification groups. After washing, the extension of the primer by only the complementary fluorescent nucleotide was confirmed by observing a red signal (the emission from Cy5) in a four-color fluorescent scanner (Fig. 3B1). After detection of the fluorescent signal, the chip surface was immersed in a deallylation mixture [1× Thermolpol I reaction buffer/Na2PdCl4/P(PhSO3Na)3] and incubated for 5 min at 60°C to cleave both the fluorophore and 3′-O-allyl group simultaneously. The chip was then immediately immersed in a 3 M Tris·HCl buffer (pH 8.5) and incubated for 5 min at 60°C to remove the Pd complex. The surface was then washed, and a negligible residual fluorescent signal was detected to confirm cleavage of the fluorophore. This was followed by another extension reaction using 3′-O-allyl-dNTPs-allyl-fluorophore mixture to incorporate the next fluorescent nucleotide complementary to the subsequent base on the template. The entire process of incorporation, synchronization, detection, and cleavage was performed multiple times using the four chemically cleavable fluorescent nucleotide reversible terminators to identify 13 successive bases in the DNA template. The fluorescence image of the chip for each nucleotide addition is shown in Fig. 3B, and a plot of the fluorescence intensity vs. the progress of sequencing extension (raw four-color sequencing data) is shown in Fig. 3C. The DNA sequences are unambiguously identified from the four-color raw fluorescence data without any processing.
Fig. 3.
Four-color sequencing by synthesis data on a DNA chip. (A) Reaction scheme of SBS on a chip using four chemically cleavable fluorescent nucleotides. (B) The scanned four-color fluorescence images for each step of SBS on a chip: (1) incorporation of 3′-O-allyl-dGTP-allyl-Cy5; (2) cleavage of allyl-Cy5 and 3′-allyl group; (3) incorporation of 3′-O-allyl-dATP-allyl-ROX; (4) cleavage of allyl-ROX and 3′-allyl group; (5) incorporation of 3′-O-allyl-dUTP-allyl-R6G; (6) cleavage of allyl-R6G and 3′-allyl group; (7) incorporation of 3′-O-allyl-dCTP-allyl-bodipy-FL-510; (8) cleavage of allyl-bodipy-FL-510 and 3′-allyl group; images 9–25 are similarly produced. (C) A plot (four-color sequencing data) of raw fluorescence emission intensity at the four designated emission wavelength of the four chemically cleavable fluorescent nucleotides vs. the progress of sequencing extension.
Fig. 4.
Structures of 3′-O-allyl-dATP, 3′-O-allyl-dCTP, 3′-O-allyl-dGTP, and 3′-O-allyl-dTTP.
Comparison of Four-Color SBS with Pyrosequencing.
To further verify the advantage of SBS method using the four chemically cleavable fluorescent nucleotide reversible terminators, we carried out similar sequencing reaction as described above on a DNA template which contained two separate homopolymeric regions (stretch of 10 T's and five T's) as shown in Fig. 5A. These sequencing raw data were produced by adding all four fluorescent nucleotide reversible terminators together to the DNA template immobilized on the chip followed by synchronization reaction with four 3′-O-allyl-dNTPs, detecting the unique fluorescence emission for sequence determination, then cleaving the fluorophore and the 3′-O-allyl group in one step to continue the sequencing cycles. All 20 bases including the individual base (A, T, C, G), the 10 repeated A's and the five repeated A's are clearly identified. The small groups of peaks between the identified bases are fluorescent background from the DNA chip, which does not build up as the cycle continues. Fig. 5B shows the pyrosequencing data of the same DNA template containing the homopolymeric sequences. The first four individual bases are clearly identified. The two homopolymeric regions (10 A's) and (five A's) produce two large peaks, but it is very difficult to identify the exact sequence from the data.
Fig. 5.
Comparison of four-color sequencing by synthesis and pyrosequencing data. (A) Four-color DNA sequencing raw data with our sequencing by synthesis chemistry using a template containing two homopolymeric regions. The individual base (A, T, C, G), the 10 repeated A's, and the five repeated A's are clearly identified. The small groups of peaks between the identified bases are fluorescent background from the DNA chip, which does not build up as the cycle continues. (B) The pyrosequencing data of the same DNA template containing the homopolymeric regions (10 T's and five T's). The first four individual bases are clearly identified. The two homopolymeric regions (10 A's) and (five A's) produce two large peaks, which are very difficult to identify the exact sequence from the data.
Conclusion
We have synthesized and characterized four chemically cleavable fluorescent nucleotide analogues and used them to produce four-color de novo DNA sequencing data on a chip. In doing so, we have achieved two critical requirements for using SBS method to sequence DNA unambiguously. First, a strategy to use a chemically reversible moiety to cap the 3′-OH group of the nucleotide has been successfully implemented so that the nucleotide terminates the polymerase reaction to allow the identification of the incorporated nucleotide. In addition, these reversible terminators allow for the addition of all four nucleotides simultaneously in performing SBS. This ultimately reduces the number of cycles needed to complete the sequencing cycle, increases sequencing accuracy due to competition of the four nucleotides in the polymerase reaction, and enables accurate determination of homopolymeric regions of DNA template. Second, efficient removal of both the fluorophore and the 3′-OH capping group after the fluorescence signal detection have successfully been carried out, which increases the overall efficiency of SBS.
The key factor governing the sequencing read length of our four-color SBS approach is the step-wise yield that is determined by the nucleotide incorporation efficiency and the yield of the cleavage of the fluorophore and the 3′-OH capping group from the DNA extension products. This stepwise yield is ≈99% based on measurement of the DNA products in solution phase. The yield on the surface is difficult to measure precisely due to fluctuations in the fluorescence imaging using the current manual fluorescent scanner. The strong fluorescence signal even for the 20th base shown in Fig. 5 indicates that we should be able to extend the read length even further. In terms of read length, Sanger sequencing is still the gold standard with read length of >800 bp but limited in throughput and cost. The read length of pyrosequencing is ≈100 bp, but with high error rate due to difficulty in accurately determining the sequences of homopolymers. Our four-color SBS read length on a manual fluorescent scanner is currently at ≈20 bp with high accuracy. This read length is expected to increase when all of the extension, cleavage, and washing steps are automated. The DNA polymerases and fluorescent labeling used in the automated four-color Sanger sequencing method have undergone almost two decades of consistent incremental improvements after the basic fluorescent Sanger methods were established (2, 3). Following the same route, it is expected that the basic principle and strategy outlined in our four-color SBS method will stimulate further improvement of the sequencing by synthesis methodology with engineering of high-performance polymerases tailored for the cleavable fluorescent nucleotide terminators and testing alternative linkers and 3′-OH reversible capping moiety. It has been well established that, by using emulsion PCR on microbeads, millions of different DNA templates are immobilized on a surface of a chip (11, 16). These high-density DNA templates, coupled with our four-color SBS approach, will generate a high-throughput (>20 million bases per chip) and highly accurate platform for a variety of sequencing and digital gene expression analysis projects.
Materials and Methods
Synthesis of 3′-O-allyl-dNTPs-allyl-Fluorophore.
The detailed synthesis of the chemically cleavable fluorescent nucleotides 3′-O-allyl-dCTP-allyl-bodipy-FL-510, 3′-O-allyl-dUTP-allyl-R6G, 3′-O-allyl-dATP-allyl-ROX, 3′-O-allyl-dGTP-allyl-bodipy-650, and 3′-O-allyl-dGTP-allyl-Cy5 are described in SI Text.
Construction of a Chip with Immobilized Self-Priming DNA Template.
The DNA chip was constructed as shown in SI Fig. 8, and the detailed steps are described in SI Text.
Continuous DNA Polymerase Reaction Using Four Chemically Cleavable Fluorescent Nucleotides as Reversible Terminators in Solution.
We characterized the four nucleotide analogues 3′-O-allyl-dCTP-allyl-bodipy-FL-510, 3′-O-allyl-dUTP-allyl-R6G, 3′-O-allyl-dATP-allyl-ROX and 3′-O-allyl-dGTP-allyl-bodipy-650, by performing four continuous DNA-extension reactions sequentially using a primer (5′-AGAGGATCCAACCGAGAC-3′) and a synthetic DNA template (5′-GTGTACATCAACATCACCTACCACCATGTCAGTCTCG-GTTGGATCCTCTATTGTGTCCGG-3′) based on a portion of exon 7 of the human p53 gene. The detailed experimental procedures are described in SI Text.
Four-Color SBS Reaction on a Chip with Four Chemically Cleavable Fluorescent Nucleotides as Reversible Terminators.
Ten microliters of a solution consisting of 3′-O-allyl-dCTP-allyl-bodipy-FL-510 (3 pmol), 3′-O-allyl-dUTP-allyl-R6G (10 pmol), 3′-O-allyl-dATP-allyl-ROX (5 pmol), and 3′-O-allyl-dGTP-allyl-Cy5 (2 pmol), 1 unit of 9°N mutant DNA polymerase, and 1× Thermolpol II reaction buffer was spotted on the surface of the chip, where the self-primed DNA moiety was immobilized. The nucleotide analogue complementary to the DNA template was allowed to incorporate into the primer at 68°C for 10 min. To synchronize any unincorporated templates, an extension solution consisting of 30 pmol each of 3′-O-allyl-dCTP, 3′-O-allyl-dTTP, 3′-O-allyl-dATP and 3′-O-allyl-dGTP, 1 unit of 9°N mutant DNA polymerase, and 1X Thermolpol II reaction buffer was spotted on the same spot and incubated at 68°C for 10 min. After washing the chip with a SPSC buffer containing 0.1% Tween 20 for 5 min, the surface was rinsed with dH2O, dried briefly and then scanned with a four-color ScanArray Express scanner (PerkinElmer Life Sciences, Boston, MA) to detect the fluorescence signal. The four-color scanner is equipped with four lasers with excitation wavelengths of 488, 543, 594, and 633 nm and emission filters centered at 522, 570, 614, and 670 nm. For deallylation, the chip was immersed in a deallylation mixture [1× Thermolpol I reaction buffer/Na2PdCl4/P(PhSO3Na)3] and incubated for 5 min at 60°C. The chip was then immediately immersed in 3 M Tris·HCl buffer (pH 8.5) and incubated for 5 min at 60°C. Finally, the chip was rinsed with acetonitrile/dH2O (1:1 vol/vol) and dH2O. The chip surface was scanned again to compare the intensity of fluorescence after deallylation with the original fluorescence intensity. This process was followed by the next polymerase extension reaction using 3′-O-allyl-dNTPs-allyl-fluorophore and 3′-O-allyl-dNTPs, with the subsequent washing, fluorescence detection, and deallylation processes performed as described above. The same cycle was repeated multiple times using the four chemically cleavable fluorescent nucleotide mixture in polymerase extension reaction to obtain de novo DNA sequencing data on various different DNA templates.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants P50 HG002806 and R01 HG003582 and the Packard Fellowship for Science and Engineering.
Abbreviations
- SBS
sequencing by synthesis
- bodipy
4,4-difluoro-4-bora-3α,4α-diaza-s-indacene
- ROX
6-carboxy-X-rhodamine
- R6G
6-carboxyrhodamine-6G.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609513103/DC1.
References
- 1.Collins FS, Green ED, Guttmacher AE, Guyer MS. Nature. 2003;422:835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- 2.Smith LM, Sanders JZ, Kaiser RJ, Hughes P, Dodd C, Connell CR, Heiner C, Kent SB, Hood LE. Nature. 1986;321:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 3.Prober JM, Trainor GL, Dam RJ, Hobbs FW, Robertson CW, Zagursky RJ, Cocuzza AJ, Jensen MA, Baumeister K. Science. 1987;238:336–341. doi: 10.1126/science.2443975. [DOI] [PubMed] [Google Scholar]
- 4.Ju J, Ruan C, Fuller CW, Glazer AN, Mathies RA. Proc Natl Acad Sci USA. 1995;92:4347–4351. doi: 10.1073/pnas.92.10.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kan CW, Doherty EA, Barron AE. Electrophoresis. 2003;24:4161–4169. doi: 10.1002/elps.200305670. [DOI] [PubMed] [Google Scholar]
- 6.Drmanac S, Kita D, Labat I, Hauser B, Schmidt C, Burczak JD, Drmanac R. Nat Biotechnol. 1998;16:54–58. doi: 10.1038/nbt0198-54. [DOI] [PubMed] [Google Scholar]
- 7.Fu DJ, Tang K, Braun A, Reuter D, Darnhofer-Demar B, Little DP, O'Donnell MJ, Cantor CR, Koster H. Nat Biotechnol. 1998;16:381–384. doi: 10.1038/nbt0498-381. [DOI] [PubMed] [Google Scholar]
- 8.Roskey MT, Juhasz P, Smirnov IP, Takach EJ, Martin SA, Haff LA. Proc Natl Acad Sci USA. 1996;93:4724–4729. doi: 10.1073/pnas.93.10.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards JR, Itagaki Y, Ju J. Nucleic Acids Res. 2001;29:E104–E104. doi: 10.1093/nar/29.21.e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Proc Natl Acad Sci USA. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 12.Ronaghi M, Uhlen M, Nyren P. Science. 1998;281:363–365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- 13.Braslavsky I, Hebert B, Kartalov E, Quake SR. Proc Natl Acad Sci USA. 2003;100:3960–3964. doi: 10.1073/pnas.0230489100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitra RD, Shendure J, Olejnik J, Edyta Krzymanska O, Church GM. Anal Biochem. 2003;320:55–65. doi: 10.1016/s0003-2697(03)00291-4. [DOI] [PubMed] [Google Scholar]
- 15.Hyman ED. Anal Biochem. 1988;174:423–436. doi: 10.1016/0003-2697(88)90041-3. [DOI] [PubMed] [Google Scholar]
- 16.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen Y-J, Chen Z, et al. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheeseman PC. 5,302,509. US Patent. 1994
- 18.Metzker ML, Raghavachari R, Richards S, Jacutin SE, Civitello A, Burgess K, Gibbs RA. Nucleic Acids Res. 1994;22:4259–4267. doi: 10.1093/nar/22.20.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welch MB, Burgess K. Nucleosides Nucleotides. 1999;18:197–201. doi: 10.1080/15257779908043067. [DOI] [PubMed] [Google Scholar]
- 20.Lu G, Burgess K. Bioorg Med Chem Lett. 2006;16:3902–3905. doi: 10.1016/j.bmcl.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 21.Metzker ML. Genome Res. 2005;15:1767–1776. doi: 10.1101/gr.3770505. [DOI] [PubMed] [Google Scholar]
- 22.Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 23.Rosenblum BB, Lee LG, Spurgeon SL, Khan SH, Menchen SM, Heiner CR, Chen SM. Nucleic Acids Res. 1997;25:4500–4504. doi: 10.1093/nar/25.22.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Z, Chao J, Yu H, Waggoner AS. Nucleic Acids Res. 1994;22:3418–3422. doi: 10.1093/nar/22.16.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ju J, Li Z, Edwards J, Itagaki Y. 6,664,079. US Patent. 2003
- 26.Seo TS, Bai X, Kim DH, Meng Q, Shi S, Ruparel H, Li Z, Turro NJ, Ju J. Proc Natl Acad Sci USA. 2005;102:5926–5931. doi: 10.1073/pnas.0501965102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bi L, Kim DH, Ju J. J Am Chem Soc. 2006;128:2542–2543. doi: 10.1021/ja057136n. [DOI] [PubMed] [Google Scholar]
- 28.Ruparel H, Bi L, Li Z, Bai X, Kim DH, Turro NJ, Ju J. Proc Natl Acad Sci USA. 2005;102:5932–5937. doi: 10.1073/pnas.0501962102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng Q, Kim DH, Bai X, Bi L, Turro NJ, Ju J. J Org Chem. 2006;71:3248–3252. doi: 10.1021/jo060300k. [DOI] [PubMed] [Google Scholar]
- 30.Barnes C, Balasubramanian S, Liu X, Swerdlow H, Milton J. 7,057,026. US Patent. 2006
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.