Abstract
The cyclin-dependent kinase inhibitor 1A (CDKN1A), also known as p21 (WAF1/CIP1) modulates cell cycle, apoptosis, senescence and differentiation via specific protein–protein interactions with the cyclins, cyclin-dependent kinase (Cdk), and many others. Expression of the p21 gene is mainly regulated at the transcriptional level. By conducting both ligation-mediated PCR (LMPCR) and chromatin immunoprecipitation (ChIP) in vivo, we identified a functional target site for the transcription factor, nuclear factor I (NFI), in the basal promoter from the p21 gene. Transfection of recombinant constructs bearing mutations in the p21 NFI site demonstrated that NFI acts as a repressor of p21 gene expression in various types of cultured cells. Inhibition of NFI in human skin fibroblasts through RNAi considerably increased p21 promoter activity suggesting that NFI is a key repressor of p21 transcription. Over-expression of each of the four NFI isoforms in HCT116 cells established that each of them contribute to various extend to the repression of the p21 gene. Most of all, over-expression of NFI-B in doxorubicin, growth-arrested HCT116 increased the proportion of cells in the S-phase of the cell cycle whereas NFI-A and NFI-X reduced it, thereby establishing a role for NFI in the cell cycle dependent expression of p21.
INTRODUCTION
p21 (CDKN1A) belongs to a family of cell-cycle dependent kinase inhibitors. p21 modulates various processes such as cell growth (1), differentiation (2) and apoptosis (3). DNA-damaging agents such as UV-irradiations or carcinogens induce the transcriptional activation of p21 via a p53-dependent pathway. Phosphorylated p53 binds to regulatory elements present in distal regions of the p21 promoter and transactivates p21 transcription via physical and functional interactions with the ubiquitous transcription factor Sp1 bound to the proximal region (4,5). Strong p21 accumulation lead to suppression of cdk activity, allowing the accumulation of hypophosphorylated Rb, inhibition of E2F-dependent transcriptional processes, and cell cycle arrest in G1 (6–8). By interacting directly with PCNA, an auxiliary factor for DNA polymerases δ and ɛ, p21 also prevents DNA synthesis and regulates DNA methylation without alteration of the PCNA-dependent nucleotide-excision repair (9,10). Some reports also suggest a p53-independent activation of p21 following UV-irradiation (11,12) but the mechanisms of induction remained poorly understood. Inducers of differentiation such as steroid hormones (13), transforming growth factor-β (TGF-β), phorbol ester or phosphatase inhibitors (14), interferon γ (15), progesterone (16), nerve growth factor (17) and platelet-derived growth factor (18) enhance p21 gene expression in many different cell systems. As for p53, the majority of these modulators affect p21 gene transcription via Sp1 and Sp3 interactions at the proximal promoter. Novel alternate p21 transcripts that are upregulated as a result of DNA damage-induced p53 activation and are dependent on p53 for their basal or induced expression were recently discovered (19,20).
In proliferating cells, p21 is expressed at a basal level in a constitutive and cell cycle dependant way (21). Under these conditions, most of the p21 proteins are components of the cyclin/cdk active complex (22). Therefore, it is likely that p21, when expressed at a moderate level, can act as an anchor protein as well as an assembly factor for cyclin D-cdk4/cdk6 thereby promoting their mutual interactions and consequently cell cycle progression, which is in complete contrast to its function as a cdk inhibitor (23). However, there seems to be a lack of understanding on the regulatory mechanisms involved in this basal expression of the p21 gene. In this study, we provided evidence that NFI is a major contributor of p21 gene expression as it could repress its transcription by interacting with the p21 proximal promoter in vivo.
MATERIALS AND METHODS
Cell culture
Human skin fibroblasts (HSFs) were isolated from skin biopsies of healthy male donors and primary cultured in DMEM (Gibco BRL, Burlington, ON, Canada) supplemented with 10% fetal bovine serum (FBS, Gibco BRL). Human epitheloid carcinoma HeLa cells (ATCC CCL 2) were grown in DMEM supplemented with 10% FBS. Rat pituitary GH4C1 cells were grown in Ham's F10 medium (Sigma-Aldrich, Oakville, ON, Canada) supplemented with 10% FBS. The human colorectal cancer cell line HCT116, kindly donated by Dr Chantal Guillemette (Oncology and Molecular Endocrinology Research Center, CHUL, Québec, Canada), was grown in McCoy's medium (Invitrogen cat# 16600) supplemented with 10% FBS. All cells were grown under 5% CO2 at 37°C and gentamycin was added to all media at a final concentration of 15 μg/ml.
Cell cycle analysis
HCT116 cells (2.5 × 105 cells/well) were plated into 6-wells tissue culture plates (Sarstedt, Montréal, QC, Canada) in complete McCoy's medium and then growth arrested by the addition of 500 nM doxorubicin (Sigma-Aldrich). Approximately 48 h later, cells were collected, fixed with 70% cold ethanol, and stored at −20°C overnight. Fixed cells were washed with PBS and incubated with PBS containing RNase (100 μg/ml) and PI (10 μg/ml) at room temperature in the dark for 30 min. The DNA content of the cells were analyzed using flow cytometry and acquisition of data for 105 events were performed using an Epics XL flow cytometer (Beckman Coulter, Miami, FL). The distribution of HCT116 cells within each phase of the cell cycle was analyzed from dual parameters histograms using an Epics® Elite flow cytometry workstation version 4.5 software.
In vivo footprinting
HSFs were seeded into 150 mm culture plates at a density of 50% cells/plate and grown as above for 3 days. Living cells and purified DNA (referred as in vivo and in vitro, respectively) were treated with one of the following probing agents: 0.02% dimethylsulphate (DMS; Sigma-Aldrich Canada), ultraviolet C (UVC; 1500 J/m2) irradiation (G15T8 germicidal lamp; Philips, Montreal, QC, Canada) and DNase I (8.75 g/μl; Worthington Biochemical, Lakewood, NJ, USA) as previously described (24,25). Strand break frequencies were estimated on an alkaline agarose gel. LMPCR was carried out as previously described (24,25) using two primer sets (1A: 5′-TCTCTCACCTCCTCTGA-3′, position +97 to +81; 1B: 5′-CTGAGTGCCTCGGTGCCTCGGCG-3′, position +85 to +62; and 2A: 5′-CCAGA TTTGTGGCTCAC-3′, position −280 to −264; 2B: 5′CTCACTTCGGGGGAAATGTGTCCAGCG-3′; position −268 to −241), allowing the analysis of the proximal promoter of p21 from nt −220 to +40. Briefly, gene-specific primers were annealed to the genomic fragments of varying sizes and then extended using cloned Pfu exo-DNA polymerase (Stratagene, LaJolla, CA) to produce double-strand blunt ends. An asymmetric double-strand linker (L25: 5′-GCGGTGACCCGGGAGATCTG-AATTC-3′ and oligo L11: 5′-GAATTCAGATC-3′) was then ligated to the phosphorylated terminal end of each fragment, providing a common sequence on the 5′ end of all fragments. Using Pfu exo-DNA polymerase, a linker-specific primer was used for a single round of linear amplification, followed by PCR amplification using the appropriate primer sets in combination with the linker primer. All primer extensions and PCR amplifications were carried out on T gradient thermocycler from Biometra (Montreal Biotech, Inc. Kirkland, Canada) as described (24,25). The PCR-amplified fragments were phenol/chloroform extracted, ethanol precipitated and subjected to electrophoresis on 8% polyacrylamide, 7 M urea gels alongside a Maxam and Gilbert sequencing ladder, followed by electrotransfer to nylon membranes (Roche Diagnostics Corp., Laval, Canada), hybridization to a 32P-labeled gene-specific probe and visualization by autoradiography on Kodak films (Amersham Biosciences, Baie d'Urfé, Canada).
Plasmid constructs
The p21 promoter fragment spanning region −192 to +36 (p21–192) relative to the mRNA start site was produced by KpnI/BglII digestion of the plasmid p21 (0–2300)-Luc containing the entire p21 promoter (kindly provided by Dr Claude Labrie, Oncology and Molecular Endocrinology Research Center, CHUL Research Center, Québec, Canada). Synthetic oligomers were used to produce two distinct linkers allowing the ligation of the fragment upstream of the chloramphenicol acetyltransferase (CAT) reporter gene in the HindIII/XbaI-linearized vector pCATbasic (Promega, Madison, WI). The plasmid lacking the region −192 to −125 (p21–124) was obtained through double-digestion of the p21–192 construct with the KpnI and BstX1 restriction enzymes. The p21–192 mNFI construct that bear a mutated NFI binding site was produced by the PCR, using p21–192 as a template and the synthetic oligomers p21-NFImA/B (5′-GGACCGGCTGGCCTGCTAAAACTCGATTAGGCTCAGCTG-GCTCC-3′)/(5′-GGAGCCAGCTGAGCCTAATCGAGTTTTAGCAGGCCACCGGTCC-3′). PCR amplifications were performed using the QuickChange® Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's specifications. The DNA insert from each recombinant plasmid was sequenced by chain-termination dideoxy sequencing (26) to confirm the mutations. The pCH-NFI-A1.1, pCH-NFI-B, pCH-NFI-C and pCH-NFI-X expression plasmids that encode high levels of the NFI-A, -B, -C and -X NFI isoforms, as well as the empty vector pCH that was used to derive the NFI expressing plasmids have been kindly provided by Dr Richard M. Gronostajski (Department of Biochemistry, SUNY at Buffalo, Buffalo, NY, USA).
Protein extracts and electrophoretic mobility shift assays
Nuclear extracts from primary cultured HSFs deprived or not of serum for 48–72 h, or from rat pituitary GH4C1 and HeLa cells were prepared as described previously (27,28) and kept frozen at −80°C until use. The NFI-L-enriched rat liver carboxymethyl (CM)-Sepharose fraction has been described previously (29). The protein concentration was determined for each nuclear extract by the Bradford procedure and further validated by Coomassie blue staining of SDS–polyacrylamide fractionated nuclear proteins. Double-stranded synthetic oligonucleotides corresponding to: (i) the p21 promoter region located from −167 to −127 and bearing the target region for NFI (p21.1: 5′-GCCTGCTGGAACTCGGCCAGGCTCAGCTGGCTCCGCGCGG-3′), (ii) the high-affinity binding site for CTF/NFI (5′-GATCTTATTTTGGATTGAAGCCAAT-ATGAG-3′) or (iii) the 80 bp HindIII–SmaI fragment from p21 to 192 were 5′ 32P-end-labeled as described (30) and used as probes in the electrophoretic mobility shift assays (EMSAs). Approximately 3 × 104 c.p.m. labeled DNA was incubated for 5 min at room temperature with nuclear extracts in the presence of poly(dI:dC) (Amersham) and 50 mM KCl in buffer D (10 mM HEPES, 10% v/v glycerol, 0.1 mM EDTA, 0.25 mM phenylmethylsulfonyl fluoride). DNA–protein complexes were separated by electrophoresis on native 4 or 6% polyacrylamide gels run in Tris/glycine buffer at 4°C, as described previously (30).
Competition experiments in EMSA were conducted as described above except that up to 500-fold molar excesses of the following unlabeled double-stranded oligonucleotides were added to the reaction mixture as unlabeled competitors: p21.1, or derivatives of p21.1 that bear mutations (bold, small letters) into either the 5′ (in p21.1m5′: 5′-GCCTGCTaaAACTCGGCCAGGCTCAGCTGGCTCCGCGCTGG-3′) or the 3′ NFI half-site (in p21.1m3′: 5′-GCCTGCTGGAACTCGattAGGCTCAGCTGGCTCCGCGCTGG-3′), or mutated into both half-sites (in p21.1m5′3′: 5′-GCCTGCTaaAACT-CGattAGGCTCAGCTGGCTCCGCGCTGG-3′), p21.2 (5′-GCCTGCTGGAACTCGGCCAGGC-TC-3′) and p21.3 (5′-CAGCTGGCTCCGC-GCTGG-3′) which correspond to each half of the p21.1 sequence, CTF/NFI, AP1 (5′-GATCCCCG-CGTTGAGTCATTCGCCTC-3′) and Sp1 (5′-GATCATATCTGCGGGGCGGG-GCAGACACAG-3′). EMSA supershift experiments were conducted as above except that 3 μl of either a pre-immune serum, or a polyclonal antibody that can recognize all isoforms of NFI (sc-5567, Santa Cruz Biotechnology, Santa Cruz, CA, USA) was incubated with the proteins for 5 min prior to the addition of the probe.
Methylation interference
The 83 bp SmaI–HindIII fragment from the p21–192 plasmid that covers p21 promoter sequences from −110 to −192 was 5′ end-labeled at the −110 position (top strand) and partially methylated with DMS as described (31). Approximately 3 × 105 c.p.m. methylated, labeled probe was incubated with 100 μg crude nuclear proteins from HeLa cells in the presence of 1 μg poly(dI:dC) in buffer D and DNA–protein complexes were separated by electrophoresis through a 6% native polyacrylamide gel in Tris-glycine buffer. Following electrophoresis, the DNA–protein complexes were visualized by autoradiography and isolated by electroelution as described (32). The isolated labeled DNA was then treated with piperidine (33) and further analyzed on a 6% sequencing gel.
DNase I in vitro footprinting
The 228 bp XbaI/KpnI DNA fragment spanning the p21 promoter sequence from position −192 to +36 was 5′ end-labeled and used as a probe in DNase I footprinting. DNase I digestion was performed in DNase I buffer (34) by incubating 4 × 104 c.p.m. labeled probe with 75 μg crude nuclear proteins from HSF cells. Further analysis of the digested products onto polyacrylamide sequencing gels was done as described previously (35).
SDS–PAGE and Western blot
One volume of sample buffer (6 M urea, 63 mM Tris [pH 6.8], 10% [vol/vol] glycerol, 1% SDS, 0.00125% [wt/vol] bromophenol blue and 300 mM β-mercaptoethanol) was added to 15 μg proteins before they were size fractionated on a 10 or 12% SDS–polyacrylamide minigel and electro-transferred onto a nitrocellulose membrane o/n at 4°C. The blot was then washed once (5 min) with TS buffer (150 mM NaCl and 10 mM Tris–HCl [pH 7.4]) and once (30 min) in TSM buffer (TS buffer plus 5% [wt/vol] fat-free powdered milk and 0.1% Tween-20). Then, a 1:5000 dilution of the NFI antibody in TSM buffer was added to the membrane and incubation proceeded for at least 90 min at 22°C. The blot was then washed four times in TSM buffer and incubated an additional 90 min at 22°C in a 1:5000 dilution of a peroxidase-conjugated goat anti-rabbit immunoglobulin G (Jackson Immuno-research, Biocan Scientific, Mississauga, ON, Canada). The membrane was successively washed in TSM (twice, 5 min each) and TS (twice, 5 min each) before immunoreactive complexes were revealed using western blot chemiluminescence reagents (Renaissance, NEN Dupont, Boston, MA) and autoradiographed.
Transient transfections and CAT assay
Primary cultured HSFs and the established tissue culture cell lines HeLa and GH4C1 were all transiently transfected with the p21-CAT recombinant constructs (p21–2300, p21–192 and its mutated derivatives p21–192 mNFI, p21–124) by the calcium phosphate precipitation method as described (36), using 15 μg of the test plasmid and 5 μg of the human growth hormone (hGH) gene-encoding plasmid pXGH5 (37). CAT activities were measured as described previously (38) and normalized to hGH secreted into the culture medium, which was assayed using a radioimmunoassay kit (Medicorp, Montréal, Québec, Canada). Each single value is expressed as 100 × (% CAT in 4h)/100 μg protein/ng hGH. To be considered significant, each individual value needed to be at least 3-fold the value of the background level caused by the reaction buffer used (usually corresponding to 0.15% chloramphenicol conversion). To compare the groups, Student's t-test was performed. Differences were considered to be statistically significant at P < 0.05. All data are expressed as mean ± SD.
Chromatin immunoprecipitation assays
The ChIP analyses were conducted as previously described (39). Briefly, HSFs were grown on 150-mm tissue culture dishes to ∼75% confluence in complete DMEM, or in FBS-depleted DMEM for periods of time ranging from 48 to 72 h. Cells were then cross-linked with 1% formaldehyde for 10 min, harvested, and chromatin immunoprecipitated with 1 μg polyclonal antibodies directed against the Sp1 (sc-59; Santa Cruz Biotechnology), Sp3 (sc-644; Santa Cruz Biotechnology), NFI and E2F1 (sc-193×; Santa Cruz Biotechnology) transcription factors (Santa Cruz Biotechnology, Inc. Santa Cruz, CA). The resulting DNA was analyzed by PCR using a pair of primers (p21–279U: 5′CAGATTTGTGGCTC-ACTTCGTGG-3′ and p21+223L: 5′AGAAACA-CCTGTGAACGCAGCAC-3′) spanning the entire p21 gene promoter area from nt –279 to +223. As a negative control, each ChIP sample was also subjected to PCR using primers (p21–2587U: 5′AATTCCTCTGAAAGCTGACTGCC3′ and p21–2132L: 5′AGGTTTACCTGGGGTCTTTA-GA-3′) specific to a region located ∼2 kbp upstream from the p21 promoter.
RNAi assays
Silencer™ negative control (id #4611, #4613, #4615), and Silencer™ pre-designed siRNA duplexes against NFI-A (id #115686, #115687, #144076), NFI-B (id #115688, #115689, #115690), NFI-C (id #215174, #215173) and NFI-X (id #115298, #115297, #3296) were purchased from Ambion, Inc. (Austin, TX, USA) and used according to manufacturer's specifications. Briefly, 2.5 μg of all three siRNAs directed against NFI-A mRNA were combined and transfected in triplicate into HSFs cultured to sub-confluence onto 39-mm tissue culture dishes (1 × 106 cells per dish at start) by the calcium phosphate precipitation method along with 10 μg of the appropriate plasmid construct (p21–124, p21–192, p21–192-mNFI, p21–2300 and p21–2300-mNFI). Cells were harvested 48 h following the addition of fresh medium and processed as mentioned above for the CAT assay.
RESULTS
In vivo footprinting of the p21 gene promoter by LMPCR
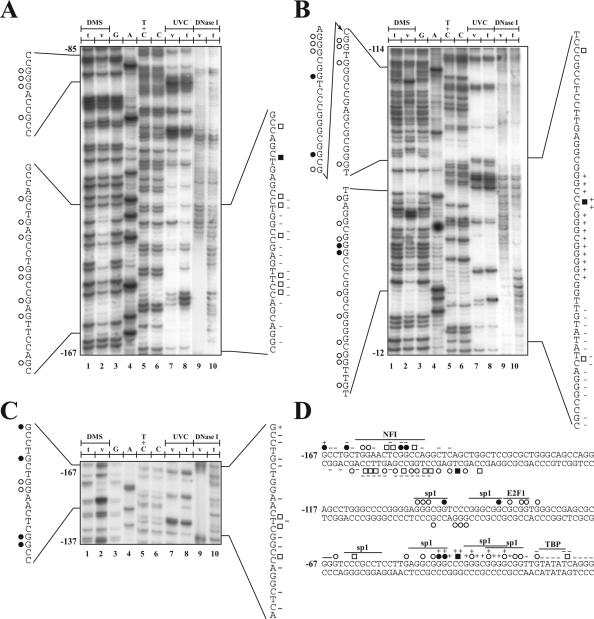
In an attempt to identify the transcription factors that regulate p21 gene transcription, we performed in vivo footprinting analyses of the proximal promoter region from the p21 gene in normal, proliferating primary cultures of HSFs by exploiting the LMPCR procedure (24). Footprint analyses were performed on the p21 promoter region covering both strands from position −220 to +40 relative to the p21 mRNA start site. Many protected and hyper-reactive sites were detected along the area from the p21 promoter that bear a putative NFI target site, on both the top and bottom strands. As shown on Figure 1A to 1C (and also summarized in Figure 1D), the comparison of the band intensity from the in vivo and in vitro treatments revealed many protected and hypersensitive nucleotides. Consistent with studies that reported binding of TBP to the TATA-box of other active gene promoters in different human cells (40,41), we also observed protected residues along the TATA-box of the p21 gene (from nt −28 to −14) as shown on Figure 1B (and summarized on Figure 1D). The DNase I protection extended many base pair beyond the 3′ end of the TATA-box that is typical of the association of other members of the holoenzyme with TBP. In addition, four of the six putative Sp1 binding sites reported to be required for p21 gene expression under basal and inducible conditions [(4,42); see discussion for details] were indeed protected from methylation by DMS. Two additional p21 promoter regions that revealed DNA–protein interactions were also identified by LMPCR. The first corresponds to the DNA target site for the transcription factor E2F1 (−84 to −80) that has already been shown to up-regulate transcription of the p21 gene [(43,44); see discussion for details]. The second protected area extends from position −161 to −138 and encompasses a potential binding site for NFI (−161 to −149). This large DNA–protein interaction interface also extended 10 bp beyond the 3′ end of the NFI site, from −148 to −138. However, search using computer databases for the identification of transcription factors target sites could not suggest any known putative transcription factor that could interact with this specific DNA segment from the p21 promoter.
Figure 1.
Genomic footprinting of the human p21 gene promoter. The region shown was analyzed with primer set 1, to reveal the bottom strand sequence from nt −167 to −85 (A), and the primer set 2, to reveal the upper strand sequence from nt −114 to −12 (B) and nt −167 to −137 (C) relative to the transcription initiation site. Lane 1, 8 and 10: LMPCR of naked DNA purified from primary cultures of HSFs treated in vitro (t) with DMS (lane 1), UVC (lane 8) or DNase I (lane 10). Lanes 2, 7 and 9: LMPCR of DNA purified from HSFs treated in vivo (v) with DMS (lane 2), UVC (lane 7) or DNase I (lane 9) prior to DNA purification. Lanes 3–6: Maxam-Gilbert sequencing. DMS protected and hypersensitive guanines are indicated by opened and closed circles, respectively, on each side of the autoradiograms, whereas UVC protected and hypersensitive sites are indicated by opened and closed squares. The DNase I protected and hypersensitive sites are indicated by − and +, respectively. (D) Summary of the in vivo DMS, UVC and DnaseI footprints identified along the −187 to −136 human p21 gene promoter. The position of the consensus sequence for the specified transcription factors is also indicated above the sequence.
Proteins from the NFI family bind the −161/−149 sequence of the p21 promoter
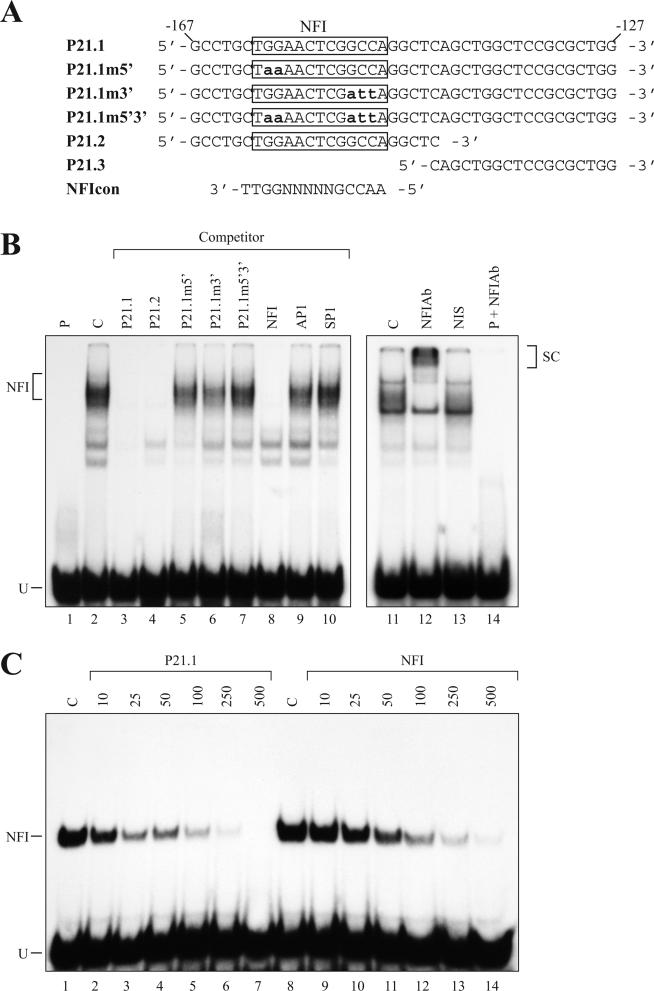
Both competition and supershift experiments in EMSA were conducted to demonstrate whether NFI possesses the ability to bind the −161/−149 region from the p21 gene (Figure 2). Incubation of a labeled probe (p21.1) bearing the entire footprinted region from positions −167 to −127 (Figure 2A), with nuclear proteins from primary cultured HSFs (Figure 2B) yielded a diffuse pattern of DNA–protein complexes (lane 2) identical to that reported previously for NFI (45–47). The formation of these DNA–protein complexes was almost completely abolished by the addition of a 150-fold molar excess of an unlabeled competitor oligonucleotide bearing the consensus sequence for CTF/NFI (lane 8). Formation of these complexes was also prevented when either the p21.1 (lane 3) or the p21.2 double-stranded oligonucleotides (lane 4), both of which has the NFI-binding site from the p21 promoter (Figure 2A), were used as competitors (Figure 2B). However, the p21.3 oligonucleotide, which is deleted from the NFI binding site (Figure 2A), was unable to compete for the formation of the p21.1/protein complexes (data not shown). This result suggests that the −127 to −144 p21 sequence, which bear the protected residues identified immediately 3′ from the NFI site in the p21 promoter, is not required for NFI binding and is likely recognized by a yet unknown nuclear protein. Derivatives of the p21.1 oligonucleotide that bear mutations either in the 5′ (in p21.1m5′) or the 3′ (in p21.1m3′) half-site of NFI could still compete, although not as efficiently as the wild-type p21.1 oligomer, for NFI binding to the labeled probe (28% reduction with p21.1m5′ (lane 5) and 43% with p21.1m3′ (lane 6) as evaluated using Biorad Quantity One software) indicating that NFI possesses the ability to bind to each single half-site from the p21 NFI site in vitro. The oligomer bearing both mutated sites (p21.1m5′3′) was unable to compete for the formation of this complex in EMSA (lane 7). Competitors bearing the target sequences for the unrelated transcription factors AP1 (lane 9) and Sp1 (lane 10) were unable to compete for the formation of the shifted complexes yielded by the HSFs nuclear extract, a further evidence that the complexes detected indeed result from the recognition of the p21.1 labeled probe by NFI.
Figure 2.
EMSA analysis of nuclear proteins from primary cultured HSFs interacting with the p21 NFI target site. (A) DNA sequence of the double-stranded oligonucleotides used as probes or competitors in the EMSA experiments. The consensus sequence for NFI is also indicated for comparison purpose (NFIcon). (B) The 5′ end-labeled p21.1 oligonucleotide was incubated with nuclear proteins (5 μg) from HSFs either alone (C; lane 2) or with a 150-fold molar excess of various unlabeled competitor oligonucleotides (p21.1, p21.1m5′, p21.1m3′, p21.1m5′3′, p21.2, NFI, AP1 and Sp1; lanes 3 to 10). Formation of DNA–protein complexes was then monitored by EMSA on a 6% native polyacrylamide gel. The position of multiple DNA–protein complexes corresponding to the recognition of the labeled probe by human NFI proteins is indicated (NFI). The p21.1 probe was also incubated with 5 μg HSFs nuclear proteins either alone (C; lane 11) or in the presence of either 2 μl of a polyclonal antibody directed against NFI (lane 12) or 2 μl of a mouse non-immune serum (NIS; lane 13). Formation of DNA–protein complexes was then monitored by EMSA as above. As an additional negative control, the probe was also incubated with the NFI Ab in the absence of nuclear proteins (lane 14). SC; supershifted complex resulting from the recognition of the NFI-p21 labeled probe complex by the NFI Ab. P: labeled probe with no added protein (lane 1). U: unbound fraction of the probe. (C) The p21.1 labeled probe used in B was incubated with 5 μg nuclear proteins from HSFs either alone (lanes 1 and 8), or in the presence of increasing concentrations (5- to 500-fold molar excesses) of unlabeled, double-stranded competitors bearing the sequence of either the p21 NFI site (p21.1) or that of the high-affinity, NFI prototypical target site (NFI). Formation of DNA–protein complexes was then monitored by EMSA as above. The position of the NFI complex is indicated along with that of the free probe (U).
To validate the identity of NFI as the transcription factor binding the p21.1 labeled probe, supershift experiments were conducted using a polyclonal Ab directed against an N-terminal epitope shared by all members of the NFI family. The NFI Ab recognized at least one of the proteins binding the p21.1 probe and led to the formation of a larger supershifted complex (SC in Figure 2B, lane 12). This new complex did not result from the non-specific recognition of the labeled probe by the NFI Ab or by an unrelated serum protein as neither condition (Figure 2B, lanes 13 and 14) could yield the supershifted complex seen when the NFI Ab is incubated with the p21.1 probe in the presence of nuclear proteins (Figure 2B, lane 12). Altogether, these results suggest that NFI binds the p21 promoter through its consensus sequence located from positions −161 to −149 in primary cultured HSFs.
The NFI target site from the p21 promoter differs from the prototypical sequence by 3 nucleotides over the entire 15 bp that constitutes this site (Figure 2A). Recently, Roulet et al. (48) conducted a very elegant study in which they functionally validated a binding site predictor that could predict the affinity of a degenerated NFI site relative to that of the prototypical NFI sequence. According to their mathematical model, NFI is expected to bind the p21 NFI target site with an affinity of over 91% that normally yielded when it binds the high-affinity NFI prototypical (consensus) sequence (which, in this case, is considered to correspond to an affinity of 100%). We therefore assessed the affinity of NFI toward both the p21 and the prototypical NFI target sites by incubating crude nuclear proteins from HSFs with the p21.1 labeled probe in the presence of varying concentrations (5- to 500-fold molar excesses) of either unlabeled p21.1 or the NFI consensus oligomer. As shown on Figure 2C, the unlabeled p21.1 oligomer was as efficient as the prototypical NFI-bearing oligonucleotide in competing for the formation of the NFI complex in EMSA. This result provided evidence that NFI binds to the p21 degenerated NFI site with an affinity undistinguishable from that obtained with the prototypical NFI site.
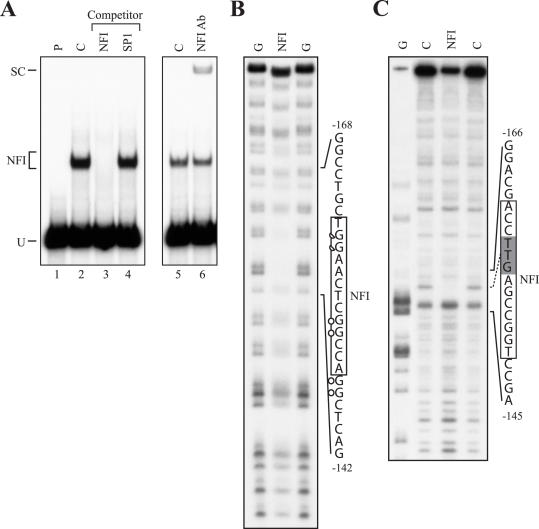
Recognition of DNA target sites by transcription factors might on occasion differ depending on whether the labeled probe used in the EMSA is synthetic (as the double-stranded oligonucleotide p21.1 used above) or derived from the gene's regulatory sequence. To further validate the results from the EMSAs conducted using the p21.1 oligonucleotide, EMSA experiments were repeated by using an 83 bp genomic fragment obtained from the SmaI–HindIII digestion of the p21–192 plasmid and covering the p21 promoter sequence from positions −110 to −192 as labeled probe. For this specific purpose, and to increase the sensitivity of the assay, we used a carboxymethyl (CM)-Sepharose enriched preparation of rat liver NFI-L (29) as the source of NFI protein. As shown on Figure 3A (lane 2), NFI-L indeed bound to the −110/−192 p21 labeled probe very efficiently. Specificity for the formation of the NFI complex was further demonstrated by both competition experiments in EMSA (only the NFI but not the unrelated Sp1 competitor oligonucleotide could compete for the formation of the shifted complex; Figure 3A, lanes 3 and 4, respectively) and supershift experiments [Figure 3A; addition of the NFI Ab (lane 6) supershifted the NFI complex from its normal position on the gel (NFI) to that corresponding to a complex with low electrophoretic mobility in gel (SC)].
Figure 3.
In vitro DMS and DNase I footprinting of NFI binding to the p21 promoter. (A) The 5′ end-labeled 83 bp SmaI–HindIII fragment from the p21–192 plasmid that covers p21 promoter sequences from −110 to −192 was incubated with 2 μg nuclear proteins from a CM-Sepharose-enriched preparation of rat liver NFI in the presence of either no (C; lanes 2 and 5) or a 150-fold molar excess of unlabeled competitor oligomers (NFI or Sp1; lanes 3 and 4, respectively) Formation of DNA–protein complexes was then monitored by EMSA on a 4% native polyacrylamide gel. The position of the NFI/p21–192 DNA–protein complex is indicated (NFI). The −110/−192 labeled probe was also incubated with 2 μl of the NFI Ab to monitor the formation of the NFI/NFIAb/p21 protein–protein–DNA supershifted complex (SC in lane 6). P: labeled probe with no added protein (lane 1). U: unbound fraction of the labeled probe. (B) The labeled probe used in A was methylated with DMS and incubated with CM-Sepharose-enriched NFI before separation of the DNA–protein complex by EMSA. Both the labeled DNA from the NFI complex (NFI in panel A) and the unbound fraction of the probe (U in panel A) were isolated and further treated with piperidine before being analyzed on a 8% polyacrylamide sequencing gel. The DNA sequence from the p21 promoter that includes the protected G residues (full and half circles correspond to fully and partly protected G residues, respectively) is indicated along with its positioning relative to the p21 mRNA start site. The p21 NFI target site is also indicated (box). (C) A 228 bp DNA fragment spanning p21 promoter sequences from position −192 to +36 was 5′ end-labeled and incubated with 75 μg CM-Sepharose-enriched NFI before being digested with DNase I. The position of the NFI site protected from digestion by DNase I is shown (shaded box) along with that of the p21 NFI site identified by in vivo footprinting (full line box). G: Maxam and Gilbert ‘G’ sequencing ladder; C: labeled DNA digested by DNase I in the absence of proteins.
We next performed both DMS methylation interference assays and DNase I footprinting in vitro in order to more precisely define the DNA target site bound by NFI along the −110 to −192 p21 promoter fragment. As shown in Figure 3B, methylated G residues that interfere with binding of NFI to its target site in the p21 promoter in vitro also perfectly mapped within the site protected by NFI and defined through the use of the in vivo footprinting procedures (see Figure 1). In vitro DNase I footprinting identified only a short stretch of protected sequence (∼3 bp) that is contained within the NFI site identified in vitro by DMS footprinting and in vivo by LMPCR. The lack of DNase I break points in the binding site of NFI, as revealed by the absence of bands on the autoradiogram, did not allow the determination of the entire sequence protected by the protein. This in vitro result contrasts with the in vivo footprinting data for this particular p21 promoter region as a larger protection could be observed (extending over ∼14 bp; see Figure 1). We therefore conclude that NFI indeed possesses the ability to bind efficiently to the p21 basal promoter both in vivo and in vitro by interacting with a target site located between positions −161 and −149.
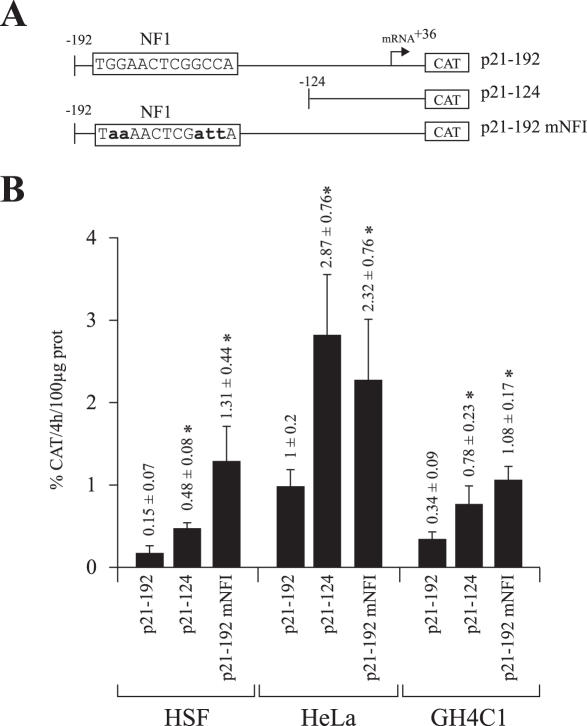
NFI functions as a repressor of p21 promoter activity and triggers G2-arrested cells into the S-phase of the cell cycle
In order to assess the regulatory influence exerted by the p21 NFI site, DNA fragments spanning the p21 basal promoter and extending from 5′ position −192 up to either 3′ positions +36 (which include the NFI site) or –124 to +36 (in which the NFI site has been deleted) was inserted upstream the CAT reporter gene (Figure 4A) and transiently transfected into either primary cultured HSFs or tissue culture cells (HeLa and GH4C1). Deletion of the entire NFI site identified by in vivo footprinting (in p21–124), yielded a significant increase in the activity of the CAT reporter gene upon transfection of the three different types of cells used in this study (3.1-, 2.9- and 2.3-fold increase in CAT activity relative to the level directed by the p21–192 plasmid in HSFs, HeLa and GH4C1 cells, respectively; Figure 4B). In order to prevent undesirable regulatory effects that might have resulted from the deletion of sequences nearby the p21 NFI site in the p21–124 construct, the NFI site from the parental plasmid p21–192 was also altered through site-directed mutagenesis. The five residues identified through both DMS methylation interference and LMPCR footprinting as the most important for NFI binding out of the 15 bp that constitute the p21 NFI site were changed for As and Ts (the wild-type NFI site 5′-CTGGAACTCGGCCAG-3′ was changed to 5′-CTaaAACTCGattAG-3′) to create the NFI-mutated construct p21–192 mNFI. Consistent with the data from the deletion analysis, mutation of the p21 NFI site in p21–192 mNFI also yielded a substantial increase in CAT activity relative to the unmutated, wild-type NFI-bearing construct p21–192 upon transfection of all cell types (8.5-, 2.3- and 3.1-fold increase in HSFs, HeLa and GH4C1 cells, respectively), further validating the results from the deletional analysis (Figure 4B).
Figure 4.
Transient transfection analysis. (A) Schematic representation of the plasmids used. The NFI target site from the p21 promoter is shown. Numbers indicate positions relative to the p21 mRNA start site. (B) The plasmids shown in A were transfected into HSFs, as well as in the tissue culture cell lines GH4C1 and HeLa. Cells were harvested and CAT activities determined and normalized to secreted hGH. *CAT activities that are statistically different from those obtained with the p21–192 construct (P < 0.05; paired samples, t-test).
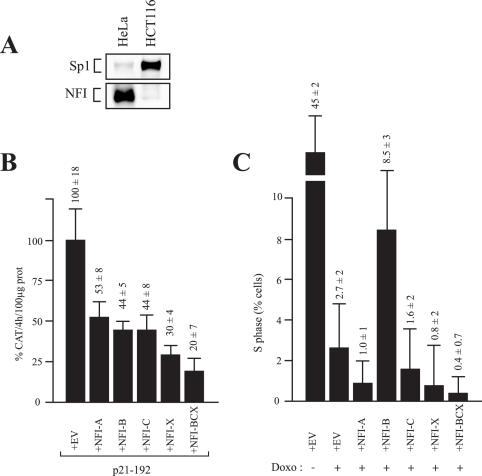
To further demonstrate the negative regulatory influence of NFI on the p21 promoter, p21–192 was co-transfected along with expression plasmids encoding high level of expression of each of the NFI isoforms into HCT116 cells. This cell line proved to be particularly interesting as it expresses very little NFI protein in Western blot analyses, unlike Sp1, which is abundantly expressed in these cells (Figure 5A). Because HCT116 cells can be easily growth arrested in the G2-phase of the cell cycle by the addition of doxorubucin, p21–192 was therefore transfected either alone or in the presence of each of the NFI expression plasmid (NFI-A, -B, -C and -X) in growth-arrested HCT116 cells. Part of the transfected HCT116 cells were used for the measurement of the CAT activity while the remaining cells were used to monitor cell cycle progression of the doxo-growth-arrested cells by FACS analyses. Transfection of HCT116 cells with the NFI expression plasmids revealed that all four NFI isoforms could reduce the activity of the p21–192 construct (up to 3.5-fold repression with NFI-X; Figure 5B). Furthermore, co-transfection of p21–192 along with the combination of NFI-B, -C and -X brought down p21 promoter function further to a 5-fold repression. Culturing HCT116 cells in the presence of doxorubicin indeed arrested the cells primarily into the G2-phase of the cell cycle, with ∼3% of the cells in the S-phase (compare 1st and 2nd columns on Figure 5C) but did not affect the negative regulatory influence of the NFI isoforms on the activity of the p21 promoter (results not presented). Interestingly, while over-expressing NFI-A, -C and -X substantially reduced the proportion of the cells committed into growth arrest (down to 1% with NFI-A), over-expression of NFI-B considerably increased those in the S-phase (up to ∼9%). Transfection of doxorubicin-treated HCT116 cells with the combination of the NFI-B, -C and -X dramatically reduced the cells remaining in S-phase from 2.7 ± 2.0 to 0.4 ± 0.7.
Figure 5.
Influence of NFI over-expression on p21 promoter activity in growth-arrested HCT116 cells. (A) Crude nuclear extracts were prepared from both HCT116 and HeLa cells (used as a control) and examined for expression of Sp1 and NFI in Western blot analyses. (B and C) The p21–192 recombinant construct was co-transfected with the empty vector pCH (+EV), or with expression plasmids encoding each of the NFI isoforms, either individually (+NFI-A, -B, -C and -X) or in combination (+NFI-BCX), in doxorubicin, growth-arrested HCT116 cells. Cells were collected 48 h later and used either for the measurement of the CAT activity (panel B) or to determine the proportion of the cells engaged in the S-phase of the cell cycle by FACS analyses (panel C).
Serum starvation alters the NFI-mediated repression of p21 gene transcription
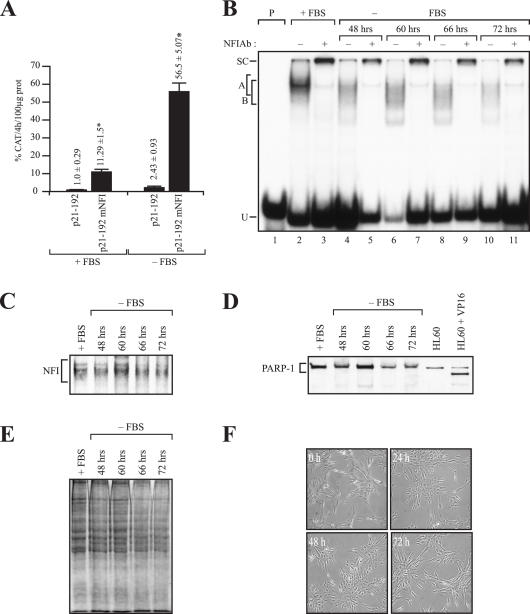
P21 being cell cycle regulated, we investigated the NFI influence on p21 gene transcription upon transfection of both the wild-type p21–192 construct or its NFI-mutated derivative p21–192 mNFI into primary HSFs grown in serum-free medium, a condition under which p21 is known to be highly expressed (49–51). As expected, basal p21 promoter activity increased considerably upon serum starvation (about 3-fold for p21–192 and 4-fold for p21–192 mNFI; Figure 6A). Under the experimental conditions used to conduct this transfection, which differ slightly from those used above in Figure 4 (see Experimental Procedures), mutation of the NFI site yielded an 11-fold increase in CAT activity when HSFs were maintained in complete medium. However, p21 promoter activity increased further up to 17-fold in serum-starved HSFs. This increase in p21 promoter activity when cells are maintained under serum-free condition correlated with a significant decrease in NFI binding in EMSA (Figure 6B) when fibroblasts are deprived of serum for various periods of time (24, 48 and 72 h), despite that no alteration in the absolute amount of the NFI protein is observed in Western blot (Figure 6C). Indeed, while extracts from HSFs grown in the presence of serum yielded a very clear and intense NFI-labeled probe DNA–protein complex in the EMSA (Figure 6B, lane 2), culturing them under serum starvation for various periods of time (48, 60, 66 and 72 h) resulted in a more diffused, strongly reduced NFI signal (Figure 6B, compare lanes 4, 6, 8 and 10 with lane 2). This reduction in NFI binding could not be accounted for from the degradation of NFI as no significant quantitative nor qualitative changes are observed in the total amount of NFI as revealed by Western blot (Figure 6C). Besides, maintaining HSFs under serum-free condition up till 72 h did not committed the cells into apoptosis as poly(ADP-ribose) polymerase-1 (hPARP-1), a 113-kDa DNA repair enzyme whose cleavage by caspase-3 into degradation products of 89- and 22-kDa (of which the larger is efficiently recognized by the C-2–10 mAb) is recognized as an early marker of apoptosis (52,53), remained entirely intact during serum starvation (Figure 6D). This is further validated by the fact that proteins in the high molecular range showed no evidence of degradation upon Coomassie blue staining on gel (Figure 6E). In addition, HSFs exhibited a normal cell morphology under phase-contrast microscopy with no sign of apoptosis, even when the cells were maintained for 72 h without serum (Figure 6F).
Figure 6.
Influence of serum starvation on NFI binding and p21 promoter function in vitro. (A) The p21–192 construct and its NFI-mutated derivative p21–192 mNFI were transfected into HSF grown either in complete (+FBS) or serum-free DMEM (−FBS) for 72 h. Cells were then harvested and CAT activities determined and normalized to secreted hGH. Asterisks indicate CAT activities that are statistically different from those obtained with the p21–192 construct (P < 0.05; paired samples, t-test). (B) A double-stranded oligonucleotide bearing the high-affinity binding site for human CTF/NFI was incubated with 5 μg proteins from HSFs grown for various periods of time post-transfection (48, 60, 66 and 72 h) in complete (+FBS) or serum-deprived DMEM (−FBS). Formation of DNA–protein complexes was then monitored by EMSA on a 6% native polyacrylamide gel. The position of multiple DNA–protein complexes corresponding to the recognition of the labeled probe by human NFI is indicated (A and B). Extracts from each condition were also incubated either alone (−; lanes 2, 4, 6, 8 and 10) or with 2 μl of the NFI Ab (+; lanes 3, 5, 7, 9 and 11). P: labeled probe with no added protein (lane 1); U: unbound fraction of the labeled probe. (C) Western blot analysis of NFI in nuclear extracts prepared from HSFs cultured in complete DMEM (+FBS) or FBS-free DMEM (−FBS) for 48–72 h. (D) Western blot analysis of PARP-1 expression in HSFs cultured in complete DMEM (+FBS) or FBS-free DMEM (−FBS) for 48–72 h. Nuclear extracts from HL60 cells cultured in the absence and presence of the apoptosis inducer VP16 were also loaded in lanes 6 and 7 as negative and positive controls, respectively. (E) 15 μg nuclear proteins from each of the extracts used above were loaded on a 10% SDS–polyacrylamide gel and stained with Coomassie blue for comparative purpose. (F) Phase-contrast images of HSFs cultured in complete DMEM (+FBS) or FBS-free DMEM (−FBS) for 48–72 h. Magnification, ×200.
NFI binds the p21 promoter in vivo in dividing but not serum-starved HSFs
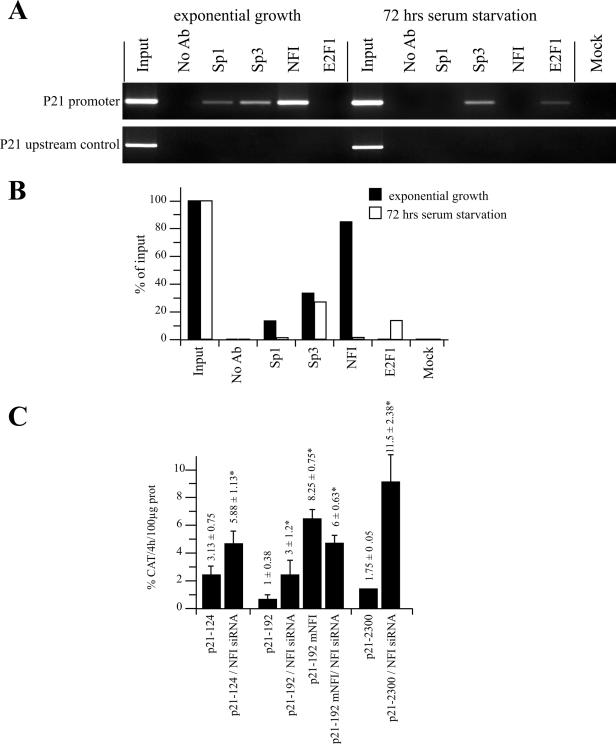
The binding of NFI to the p21 gene promoter area was further examined in vivo in HSFs by ChIP assays. As a control, ChIP was also conducted on other transcription factors, such as Sp1, Sp3 and E2F1, also reported to bind the p21 basal promoter. Primers that spanned the entire p21 promoter from −279 down to +223 relative to the p21 mRNA start site were selected for the assay. Antibodies against Sp1, Sp3 and NFI all enriched the p21 promoter sequences in HSFs grown in complete, serum-containing medium, indicating that this genomic area is bound in vivo by these transcription factors (Figure 7A). In contrast, only the Sp3 and E2F1 antibodies enriched the p21 promoter in HSFs maintained under serum starvation for 72 h, relative to the ‘no antibody’ control sample. These results suggest that Sp1, Sp3 and NFI are bound to cis-acting elements present on the p21 promoter in the in vivo chromatin configuration of HSF cells during the dividing phase of the cell cycle (basal level of p21). Upon serum starvation (where high levels of p21 expression are observed), binding of both Sp1 and NFI were completely abrogated. Interestingly, while the level of Sp3 binding to the p21 promoter remained unaffected by serum starvation, that of E2F1, initially undetectable in actively growing cells, was found to bind weakly in serum-deprived HSFs. The significant occupancy of NFI in relation to the ‘input’ (Figure 7B), suggest a critical role for this transcription factor on the p21 gene regulation. These results demonstrate that NFI binds the p21 promoter area in vivo in actively growing, but not serum-starved, growth-arrested HSFs.
Figure 7.
Chromatin immunoprecipitation of NFI and RNAi assays in HSFs. (A) ChIP assays were performed on HSFs in an exponential state of growth or after 72 h of serum starvation. Chromatin was isolated and immunoprecipited with antibodies directed against the transcription factors Sp1, Sp3, NFI and E2F1. PCR of the p21 gene promoter was then carried out on the ChIP samples along with a ‘no antibody’ control (No Ab) that contains chromatin but no antibody, an ‘input’ sample corresponding to 0.2% of the total input chromatin, and a ‘mock’ sample that does not contain chromatin. PCR amplification of a gene segment located ∼2000 bp upstream from the p21 promoter was also conducted on the same sample as a negative control for all immunoprecipitates. (B) Graphical representation of the amount of specific PCR products expressed as the percentage of antibody binding versus the amount of PCR product obtained using a standardized aliquot of input chromatin. The signal in the no-antibody lane corresponds to the non-specific binding background and was subtracted from each sample. (C) RNAi was performed using a combination of siRNA complementary to the NFI-A, -B, -C and -X transcripts. The p21 promoter-bearing recombinant constructs p21–124, p21–192 and p21–2300, and the derivative from p21–192 that comprise a mutated NFI site (p21–192 mNFI) were transfected together with either the siRNA silencer negative control or with the combination of NFI siRNA duplexes (siNFI) into subconfluent HeLa cells. Cells were harvested and CAT activities determined. *CAT activities that are statistically different from those obtained with the p21 promoter constructs transfected solely with the siRNA silencer negative control (P < 0.05; paired samples, t-test).
Suppression of NFI expression by RNAi alleviates p21 repression
Evidence that NFI functionally represses p21 promoter activity was further examined through the suppression of the endogenous NFI transcript by RNAi (Figure 7C). Because HeLa cells are much easier to culture and are transfected with a higher efficiency, they were selected over HSFs for conducting the RNAi assays. Recombinant constructs bearing various lengths from the p21 promoter (up to 5′ positions −124, −192 and −2300) were co-transfected with and without a combined pool of siRNAs directed against NFI-A, -B, -C and -X. As expected, the P21–124 construct lacking the NFI binding site yielded a promoter activity higher than the NFI-bearing constructs p21–192 and p21–2300, which are both under the negative regulatory influence of NFI. Co-transfection of these constructs along with the NFI siRNAs substantially released repression by NFI for both p21–192 (3-fold increase) and p21–2300 (near 7-fold increase). Again, mutating the NFI binding site in p21–192 (p21–192 mNFI) strongly increased CAT activity in HeLa cells (by ∼8-fold). However, while inhibition of endogenous NFI through RNAi considerably increased the activity of the unmutated, wild-type p21–192 construct (3-fold increase) it had no such influence when the NFI site is mutated in p21–192 mNFI (which yielded a weak 27% reduction in p21 promoter activity).
DISCUSSION
Cyclin-dependent kinase inhibitor p21 plays a key function in cell cycle arrest at the G1/S checkpoint in response to DNA damage, and is involved in the assembly of active cyclin-kinase complexes especially cyclin D-Cdk4/6. Transcription of the p21 gene relies on the control of numerous regulators of which many still have to be identified. In the present study, we precisely mapped, through both in vivo (LMPCR) and in vitro (DMS and DNase I footprinting) approaches, a target site for the transcription factor NFI in the basal promoter of the human p21 gene that proved to function as a powerful repressor of p21 gene transcription in both non-damaged and proliferating primary cultures of human fibroblasts and established tissue culture cells.
To our knowledge, no evidence regarding a regulatory function exerted by NFI on the transcription of the p21 gene has ever been published until now. We demonstrated that members of the NFI family could bind a target site on the p21 basal promoter located between positions −161 and −149, and considerably repress its transcription in a variety of cells. Although each of the four NFI isoforms were found to contribute to this repression, NFI-X turned out to be the most effective. Intriguingly, each member from this family exerted very specific influences on the growth properties of the cells by either restricting or promoting progression of the cells into the S-phase from the cell cycle. Indeed, over-expression of NFI-A, -C and -X reduced the number of cells remaining in the S-phase in HTC116 cells that have been growth arrested with doxorubicin, whereas over-expressing the NFI-B isoform triggered more cells to enter the S-phase, thereby establishing the biological significance for the regulatory influence of NFI on cell cycle progression. Consistent with these results, Luciakova et al. (54) have recently demonstrated that mutation of the NFI sites present in the promoter of the adenine nucleotide translocase-2 gene (ANT2) totally annihilated the growth-arrest properties of NFI in NIH3T3 cells. Most interestingly, they attributed these growth-arrest properties of NFI to the NFI-A, -C and -X isoforms but not NFI-B as the former were all found to repress expression of an ANT2-driven reporter gene construct, whereas NFI-B activated it. Members from the NFI family can either repress or activate their target genes depending on the cell type or promoter context. This means that although all NFI isoforms were found to repress to varying degrees the transcription directed by the p21 promoter in HCT116 cells, yet they may activate transcription of other genes in the same cell that may or may not be related to cell growth. For instance, NFI has been shown to activate expression of p53 (55) and gadd153 (56), both of which play key roles in growth arrest of damaged or environmentally stressed cells. Paradoxically, NF1-X1 was identified as one of three genes (together with c-myc and MDM2) that prevent TGFβ-induced growth arrest (57). Therefore, while all NFI isoforms clearly repress p21 gene expression in HCT116 cells, NFI-B might as well regulate expression of additional, cell-growth related genes that are not under the influence of the other NFI isoforms. This differential property of NFI-B may rely on its ability to recruit either co-repressors or co-activators that are distinct from those recruited by other NFI proteins. For instance, of all the NFI isoforms, only NFI-B has been found to activate whey acidic protein (WAP) gene expression in cooperation with STAT5A and the glucocorticoid receptor (GR) in JEG-3 cells (58). Most interestingly, NFI-B deficient mice have been shown to die early after birth and display severe lung hypoplasia (59), lung development being arrested at the late pseudoglandular stage. These results suggest that NFI-B is most likely required for normal lung cell proliferation in order to ensure proper lung development. It is noteworthy that the in vivo footprint identified in the present study extends 10 bp beyond the 3′ end of the NFI consensus site (from −148 to −138). A search conducted using databases for the identification of transcription factor target sites failed to identify any known nuclear protein that may interact with the −148/−138 p21 sequence. The presence of additional co-factors that can bind DNA in vivo but not in vitro, as well as structural events, such as DNA folding, could explain the detection of the protected residues located downstream from the −161 to −149 NFI site. Indeed, the RNAi experiments conducted in this study (Figure 7C) suggested that other DNA binding proteins might contribute to the appropriate transcription of the p21 gene by interacting somewhere along the −124/−148 segment from the p21 promoter as the p21–124 construct did not mimic levels achieved with the p21–192 that bears mutations in the −161/−149 NFI site. Besides, reducing the endogenous level of NFI through RNAi is also likely to considerably alter the transcription of many genes in the transfected cells. Some of them may as well encode transcription factors other than NFI that also participate to the cascade of regulatory events required to ensure appropriate transcription of the p21 gene. Further works will be required in order to determine the precise identity of such cis-acting transcriptional regulators and whether their regulatory influences are mediated through their interaction with the −148/−138 area from the p21 promoter.
The NFI family of transcription factors is made-up of four protein subtypes (NFI-A, -B, -C and -X) (60), each encoded by a different gene, that can form either homo- or heterodimers. The large diversity of this family of proteins, for which 19 isoforms have been described to date [reviewed in (61)], rely on the fact that each individual NFI mRNA transcript is subjected to alternative splicing (33,62). Members from the NFI family have been shown to function either as repressors (35,46,56) or activators (63,64) of gene transcription. Through interactions with weak binding sites, NFI may also regulate gene expression by its ability to either cooperate or compete with many transcription factors (35,65). Considering the negative influence exerted by NFI on p21 gene transcription, we propose that NFI may act in cooperation with other nearby co-activators or transcription factors to restrict the expression of the p21 gene throughout the cell cycle progression while DNA is undamaged. NFI was reported to interact with and antagonize Sp1, which result in the down-regulation of the platelet-derived growth factor (PDGF)-A gene expression (65). Interestingly, in vivo footprinting analysis of the p21 promoter also revealed DNA–protein interactions at four of the six putative consensus binding sites reported for Sp1 (4). Besides NFI and Sp1, these in vivo analyses also identified protections at target sites for nuclear proteins, such as E2F1 (Figure 1) that have already been reported to bind the p21 proximal promoter. Members from the E2F family of transcription factors play a crucial role in the transactivation of G1/S transition specific genes (66). Previous studies already demonstrated the involvement of E2F1 in the p53-independent activation of p21 (43,44).
Results from the ChIP analyses revealed variations in the pattern of p21 promoter occupancy by the transcription factors Sp1, NFI and E2F1 between actively growing and serum-starved, quiescent HSFs. Indeed, whereas both Sp1 and NFI but not E2F1 substantially bind the p21 promoter in proliferating cells, HSFs grown under serum deprivation completely lost p21 promoter recognition by both Sp1 and NFI but gained binding of E2F1. The relative occupancy of the p21 promoter by NFI in vivo clearly exceeded that of both Sp1 and Sp3 in proliferating HSFs, thereby favoring repression rather than activation of p21 transcription. On the contrary, the lack of any NFI binding in serum-starved cells combined to the recognized positive influence of both Sp3 and E2F1 as both factors were shown to bind the p21 promoter under this condition, is consistent with the increased expression of p21 reported when cells progress toward growth arrest (44,67).
Interestingly the presence of Sp3, whose binding to the p21 promoter does not change with alterations in the proliferative state of the cells, raised the interesting possibility that most of the Sp1 target sites identified in the p21 promoter might became occupied by Sp3 under serum deprivation as both transcription factors have been reported to bind the same GC-rich target sites (67). It is noteworthy that Sp1/Sp3 share extensive structural and sequence homology (68), and often cooperate in activating gene transcription (69–71). The presence of Sp3 in HSFs grown with or without serum suggests that p21 is constitutively activated by this transcription factor and that fine tuning of p21 transcription is ensured by subtle variations in the ability of nuclear repressors such as NFI to interact with its corresponding target site in the p21 promoter. Our results are consistent with those published by other groups who demonstrated that while basal transcription of p21 is regulated by both Sp1 and Sp3 in cultured primary mouse keratinocytes, only Sp3 contributes to the calcium-induced p21 promoter activity (67).
The identification of each of the nuclear proteins yielding the new protected sites identified in this study as well as the precise regulatory function they play will be required to assess the precise mechanisms that modulate p21-dependent regulation of cell cycle progression. We must now look upon NFI as a new and central player in the molecular mechanisms regulating p21 expression during cell cycle progression. The relationship between NFI and other transcription factors such as p53, Sp1, Sp3 and E2F1 that also bind the p21 promoter will be of a particular interest and should prove to be particularly fascinating regarding p21 gene regulation.
Acknowledgments
The authors are grateful to Drs R. Stephen Lloyd and Tim R. O'Connor for supplying T4 endonuclease V and photolyase, respectively and to Dr R.M. Gronostajski for the NFI expression plasmids. This work was supported by grants from the NSERC (grant OGP0138624) to S.L.G. and the National Cancer Institute of Canada (NCIC) (with funds from the Canadian Cancer Society and the Terry Fox Run) to R.D. F.V. holds a Doctoral Research Award from the Canadian Institute for Health Research (CIHR). R.D. and S.L.G. are research scholars (Senior and National levels, respectively) from the ‘Fonds de la Recherche en Santé du Québec (FRSQ). R.D. holds a Canada Research Chair in ‘Genetics, Mutagenesis and Cancer’. Funding to pay the Open Access publication charges for this article was provided by the NSERC grant OGP0138624 to S.L.G.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gartel A.L., Serfas M.S., Tyner A.L. p21—negative regulator of the cell cycle. Proc. Soc. Exp. Biol. Med. 1996;213:138–149. doi: 10.3181/00379727-213-44046. [DOI] [PubMed] [Google Scholar]
- 2.Wu H., Wade M., Krall L., Grisham J., Xiong Y., Van Dyke T. Targeted in vivo expression of the cyclin-dependent kinase inhibitor p21 halts hepatocyte cell-cycle progression, postnatal liver development and regeneration. Genes Dev. 1996;10:245–260. doi: 10.1101/gad.10.3.245. [DOI] [PubMed] [Google Scholar]
- 3.Ostrovsky O., Bengal E. The mitogen-activated protein kinase cascade promotes myoblast cell survival by stabilizing the cyclin-dependent kinase inhibitor, p21WAF1 protein. J. Biol. Chem. 2003;278:21221–21231. doi: 10.1074/jbc.M211357200. [DOI] [PubMed] [Google Scholar]
- 4.Koutsodontis G., Moustakas A., Kardassis D. The role of Sp1 family members, the proximal GC-rich motifs, and the upstream enhancer region in the regulation of the human cell cycle inhibitor p21WAF-1/Cip1 gene promoter. Biochemistry. 2002;41:12771–12784. doi: 10.1021/bi026141q. [DOI] [PubMed] [Google Scholar]
- 5.Pardali K., Kurisaki A., Moren A., ten Dijke P., Kardassis D., Moustakas A. Role of Smad proteins and transcription factor Sp1 in p21(Waf1/Cip1) regulation by transforming growth factor-beta. J. Biol. Chem. 2000;275:29244–29256. doi: 10.1074/jbc.M909467199. [DOI] [PubMed] [Google Scholar]
- 6.Afshari C.A., Nichols M.A., Xiong Y., Mudryj M. A role for a p21-E2F interaction during senescence arrest of normal human fibroblasts. Cell Growth Differ. 1996;7:979–988. [PubMed] [Google Scholar]
- 7.Delavaine L., La Thangue N.B. Control of E2F activity by p21Waf1/Cip1. Oncogene. 1999;18:5381–5392. doi: 10.1038/sj.onc.1202923. [DOI] [PubMed] [Google Scholar]
- 8.Linke S.P., Harris M.P., Neugebauer S.E., Clarkin K.C., Shepard H.M., Maneval D.C., Wahl G.M. p53-mediated accumulation of hypophosphorylated pRb after the G1 restriction point fails to halt cell cycle progression. Oncogene. 1997;15:337–345. doi: 10.1038/sj.onc.1201200. [DOI] [PubMed] [Google Scholar]
- 9.Ando T., Kawabe T., Ohara H., Ducommun B., Itoh M., Okamoto T. Involvement of the interaction between p21 and proliferating cell nuclear antigen for the maintenance of G2/M arrest after DNA damage. J. Biol. Chem. 2001;276:42971–42977. doi: 10.1074/jbc.M106460200. [DOI] [PubMed] [Google Scholar]
- 10.Cayrol C., Knibiehler M., Ducommun B. p21 binding to PCNA causes G1 and G2 cell cycle arrest in p53-deficient cells. Oncogene. 1998;16:311–320. doi: 10.1038/sj.onc.1201543. [DOI] [PubMed] [Google Scholar]
- 11.Haapajarvi T., Kivinen L., Heiskanen A., des Bordes C., Datto M.B., Wang X.F., Laiho M. UV radiation is a transcriptional inducer of p21(Cip1/Waf1) cyclin-kinase inhibitor in a p53-independent manner. Exp. Cell Res. 1999;248:272–279. doi: 10.1006/excr.1999.4403. [DOI] [PubMed] [Google Scholar]
- 12.Loignon M., Fetni R., Gordon A.J., Drobetsky E.A. A p53-independent pathway for induction of p21waf1cip1 and concomitant G1 arrest in UV-irradiated human skin fibroblasts. Cancer Res. 1997;57:3390–3394. [PubMed] [Google Scholar]
- 13.Liu M., Iavarone A., Freedman L.P. Transcriptional activation of the human p21(WAF1/CIP1) gene by retinoic acid receptor. Correlation with retinoid induction of U937 cell differentiation. J. Biol. Chem. 1996;271:31723–31728. doi: 10.1074/jbc.271.49.31723. [DOI] [PubMed] [Google Scholar]
- 14.Biggs J.R., Kudlow J.E., Kraft A.S. The role of the transcription factor Sp1 in regulating the expression of the WAF1/CIP1 gene in U937 leukemic cells. J. Biol. Chem. 1996;271:901–906. doi: 10.1074/jbc.271.2.901. [DOI] [PubMed] [Google Scholar]
- 15.Chin Y.E., Kitagawa M., Su W.C., You Z.H., Iwamoto Y., Fu X.Y. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science. 1996;272:719–722. doi: 10.1126/science.272.5262.719. [DOI] [PubMed] [Google Scholar]
- 16.Owen G.I., Richer J.K., Tung L., Takimoto G., Horwitz K.B. Progesterone regulates transcription of the p21(WAF1) cyclin- dependent kinase inhibitor gene through Sp1 and CBP/p300. J. Biol. Chem. 1998;273:10696–10701. doi: 10.1074/jbc.273.17.10696. [DOI] [PubMed] [Google Scholar]
- 17.Yan G.Z., Ziff E.B. Nerve growth factor induces transcription of the p21 WAF1/CIP1 and cyclin D1 genes in PC12 cells by activating the Sp1 transcription factor. J. Neurosci. 1997;17:6122–6132. doi: 10.1523/JNEUROSCI.17-16-06122.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michieli P., Chedid M., Lin D., Pierce J.H., Mercer W.E., Givol D. Induction of WAF1/CIP1 by a p53-independent pathway. Cancer Res. 1994;54:3391–3395. [PubMed] [Google Scholar]
- 19.Gartel A.L., Radhakrishnan S.K., Serfas M.S., Kwon Y.H., Tyner A.L. A novel p21WAF1/CIP1 transcript is highly dependent on p53 for its basal expression in mouse tissues. Oncogene. 2004;23:8154–8157. doi: 10.1038/sj.onc.1207820. [DOI] [PubMed] [Google Scholar]
- 20.Radhakrishnan S.K., Gierut J., Gartel A.L. Multiple alternate p21 transcripts are regulated by p53 in human cells. Oncogene. 2006;25:1812–1815. doi: 10.1038/sj.onc.1209195. [DOI] [PubMed] [Google Scholar]
- 21.Li Y., Jenkins C.W., Nichols M.A., Xiong Y. Cell cycle expression and p53 regulation of the cyclin-dependent kinase inhibitor p21. Oncogene. 1994;9:2261–2268. [PubMed] [Google Scholar]
- 22.Besson A., Yong V.W. Involvement of p21(Waf1/Cip1) in protein kinase C alpha-induced cell cycle progression. Mol. Cell Biol. 2000;20:4580–4590. doi: 10.1128/mcb.20.13.4580-4590.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaBaer J., Garrett M.D., Stevenson L.F., Slingerland J.M., Sandhu C., Chou H.S., Fattaey A., Harlow E. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 24.Drouin R., Therrien J.-P., Angers M., Ouellet S. In vivo DNA analysis. In: Moss T., editor. DNA-Protein Interactions: Principles and Protocols. 2nd edn. Totowa, NJ: Humana Press; 2001. pp. 175–219. [DOI] [PubMed] [Google Scholar]
- 25.Rouget R., Vigneault F., Codio C., Rochette C., Paradis I., Drouin R., Simard L.R. Characterization of the survival motor neuron (SMN) promoter provides evidence for complex combinatorial regulation in undifferentiated and differentiated P19 cells. Biochem. J. 2005;385:433–443. doi: 10.1042/BJ20041024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergeron M.J., Leclerc S., Laniel M.A., Poirier G.G., Guerin S.L. Transcriptional regulation of the rat poly(ADP-ribose) polymerase gene by Sp1. Eur. J. Biochem. 1997;250:342–353. doi: 10.1111/j.1432-1033.1997.0342a.x. [DOI] [PubMed] [Google Scholar]
- 28.Roy R.J., Gosselin P., Guerin S.L. A short protocol for micro-purification of nuclear proteins from whole animal tissue. Biotechniques. 1991;11:770–777. [PubMed] [Google Scholar]
- 29.Roy R.J., Guerin S.L. The 30-kDa rat liver transcription factor nuclear factor 1 binds the rat growth-hormone proximal silencer. Eur. J. Biochem. 1994;219:799–806. doi: 10.1111/j.1432-1033.1994.tb18560.x. [DOI] [PubMed] [Google Scholar]
- 30.Laniel M.A., Poirier G.G., Guerin S.L. Nuclear factor 1 interferes with Sp1 binding through a composite element on the rat poly(ADP-ribose) polymerase promoter to modulate its activity in vitro. J. Biol. Chem. 2001;276:20766–20773. doi: 10.1074/jbc.M010360200. [DOI] [PubMed] [Google Scholar]
- 31.Baldwin A.S.J. Methylation and Uracil Interference Assays for Analysis of Protein-DNA Interactions. In: Struhl K., editor. Current Protocols in Molecular Biology. New York: Wiley; 1989. pp. pp. 12.18–12.10. [DOI] [PubMed] [Google Scholar]
- 32.Harvey M., Brisson I., Guerin S.L. A simple apparatus for fast and inexpensive recovery of DNA from polyacrylamide gels. Biotechniques. 1993;14:942–948. [PubMed] [Google Scholar]
- 33.Marin M., Karis A., Visser P., Grosveld F., Philipsen S. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell. 1997;89:619–628. doi: 10.1016/s0092-8674(00)80243-3. [DOI] [PubMed] [Google Scholar]
- 34.Robidoux S., Gosselin P., Harvey M., Leclerc S., Guerin S.L. Transcription of the mouse secretory protease inhibitor p12 gene is activated by the developmentally regulated positive transcription factor Sp1. Mol. Cell. Biol. 1992;12:3796–3806. doi: 10.1128/mcb.12.9.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laniel M.A., Poirier G.G., Guerin S.L. Nuclear factor 1 interferes with Sp1 binding through a composite element on the rat poly(ADP-ribose) polymerase promoter to modulate its activity in vitro. J. Biol. Chem. 2001;276:20766–20773. doi: 10.1074/jbc.M010360200. [DOI] [PubMed] [Google Scholar]
- 36.Graham F.L., van der Eb A.J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 37.Selden R.F., Howie K.B., Rowe M.E., Goodman H.M., Moore D.D. Human growth hormone as a reporter gene in regulation studies employing transient gene expression. Mol. Cell. Biol. 1986;6:3173–3179. doi: 10.1128/mcb.6.9.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pothier F., Ouellet M., Julien J.P., Guerin S.L. An improved CAT assay for promoter analysis in either transgenic mice or tissue culture cells. DNA Cell Biol. 1992;11:83–90. doi: 10.1089/dna.1992.11.83. [DOI] [PubMed] [Google Scholar]
- 39.Oberley M.J., Farnham P.J. Probing chromatin immunoprecipitates with CpG-island microarrays to identify genomic sites occupied by DNA-binding proteins. Methods Enzymol. 2003;371:577–596. doi: 10.1016/S0076-6879(03)71043-X. [DOI] [PubMed] [Google Scholar]
- 40.Temple M.D., Cairns M.J., Kim A., Murray V. Protein-DNA footprinting of the human epsilon-globin promoter in human intact cells using nitrogen mustard analogues and other DNA-damaging agents. Biochim. Biophys. Acta. 1999;1445:245–256. doi: 10.1016/s0167-4781(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 41.Zhao X.T., You Z.S., Cheng P., Wang Y. Investigation of protein-DNA interactions in Enhancer II and core promoter of HBV by in vivo footprinting. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 1999;31:489–493. [PubMed] [Google Scholar]
- 42.Koutsodontis G., Tentes I., Papakosta P., Moustakas A., Kardassis D. Sp1 plays a critical role in the transcriptional activation of the human cyclin-dependent kinase inhibitor p21(WAF1/Cip1) gene by the p53 tumor suppressor protein. J. Biol. Chem. 2001;276:29116–29125. doi: 10.1074/jbc.M104130200. [DOI] [PubMed] [Google Scholar]
- 43.Gartel A.L., Najmabadi F., Goufman E., Tyner A.L. A role for E2F1 in Ras activation of p21(WAF1/CIP1) transcription. Oncogene. 2000;19:961–964. doi: 10.1038/sj.onc.1203411. [DOI] [PubMed] [Google Scholar]
- 44.Hiyama H., Iavarone A., Reeves S.A. Regulation of the cdk inhibitor p21 gene during cell cycle progression is under the control of the transcription factor E2F. Oncogene. 1998;16:1513–1523. doi: 10.1038/sj.onc.1201667. [DOI] [PubMed] [Google Scholar]
- 45.Eskild W., Simard J., Hansson V., Guerin S.L. Binding of a member of the NF1 family of transcription factors to two distinct cis-acting elements in the promoter and 5′-flanking region of the human cellular retinol binding protein 1 gene. Mol. Endocrinol. 1994;8:732–745. doi: 10.1210/mend.8.6.7935489. [DOI] [PubMed] [Google Scholar]
- 46.Steffensen K.R., Holter E., Tobin K.A., Leclerc S., Gustafsson J.A., Guerin S.L., Eskild W. Members of the nuclear factor 1 family reduce the transcriptional potential of the nuclear receptor LXRalpha promoter. Biochem. Biophys. Res. Commun. 2001;289:1262–1267. doi: 10.1006/bbrc.2001.6078. [DOI] [PubMed] [Google Scholar]
- 47.Wickenheisser J.K., Nelson-DeGrave V.L., Quinn P.G., McAllister J.M. Increased cytochrome P450 17alpha-hydroxylase promoter function in theca cells isolated from patients with polycystic ovary syndrome involves nuclear factor-1. Mol. Endocrinol. 2004;18:588–605. doi: 10.1210/me.2003-0090. [DOI] [PubMed] [Google Scholar]
- 48.Roulet E., Bucher P., Schneider R., Wingender E., Dusserre Y., Werner T., Mermod N. Experimental analysis and computer prediction of CTF/NFI transcription factor DNA binding sites. J. Mol. Biol. 2000;297:833–848. doi: 10.1006/jmbi.2000.3614. [DOI] [PubMed] [Google Scholar]
- 49.Chang B.D., Watanabe K., Broude E.V., Fang J., Poole J.C., Kalinichenko T.V., Roninson I.B. Effects of p21Waf1/Cip1/Sdi1 on cellular gene expression: implications for carcinogenesis, senescence, and age-related diseases. Proc. Natl Acad. Sci. USA. 2000;97:4291–4296. doi: 10.1073/pnas.97.8.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Funato N., Ohtani K., Ohyama K., Kuroda T., Nakamura M. Common regulation of growth arrest and differentiation of osteoblasts by helix-loop-helix factors. Mol. Cell. Biol. 2001;21:7416–7428. doi: 10.1128/MCB.21.21.7416-7428.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang Z.Y., Wu Y., Hedrick N., Gutmann D.H. T-cadherin-mediated cell growth regulation involves G2 phase arrest and requires p21(CIP1/WAF1) expression. Mol. Cell. Biol. 2003;23:566–578. doi: 10.1128/MCB.23.2.566-578.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaufmann S.H., Desnoyers S., Ottaviano Y., Davidson N.E., Poirier G.G. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53:3976–3985. [PubMed] [Google Scholar]
- 53.Lazebnik Y.A., Kaufmann S.H., Desnoyers S., Poirier G.G., Earnshaw W.C. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature. 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- 54.Luciakova K., Barath P., Poliakova D., Persson A., Nelson B.D. Repression of the human adenine nucleotide translocase-2 gene in growth-arrested human diploid cells: the role of nuclear factor-1. J. Biol. Chem. 2003;278:30624–30633. doi: 10.1074/jbc.M303530200. [DOI] [PubMed] [Google Scholar]
- 55.Furlong E.E., Keon N.K., Thornton F.D., Rein T., Martin F. Expression of a 74-kDa nuclear factor 1 (NF1) protein is induced in mouse mammary gland involution. Involution-enhanced occupation of a twin NF1 binding element in the testosterone-repressed prostate message-2/clusterin promoter. J. Biol. Chem. 1996;271:29688–29697. doi: 10.1074/jbc.271.47.29688. [DOI] [PubMed] [Google Scholar]
- 56.Nakamura M., Okura T., Kitami Y., Hiwada K. Nuclear factor 1 is a negative regulator of gadd153 gene expression in vascular smooth muscle cells. Hypertension. 2001;37:419–424. doi: 10.1161/01.hyp.37.2.419. [DOI] [PubMed] [Google Scholar]
- 57.Sun P., Dong P., Dai K., Hannon G.J., Beach D. p53-independent role of MDM2 in TGF-beta1 resistance. Science. 1998;282:2270–2272. doi: 10.1126/science.282.5397.2270. [DOI] [PubMed] [Google Scholar]
- 58.Mukhopadhyay S.S., Wyszomierski S.L., Gronostajski R.M., Rosen J.M. Differential interactions of specific nuclear factor I isoforms with the glucocorticoid receptor and STAT5 in the cooperative regulation of WAP gene transcription. Mol. Cell. Biol. 2001;21:6859–6869. doi: 10.1128/MCB.21.20.6859-6869.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grunder A., Ebel T.T., Mallo M., Schwarzkopf G., Shimizu T., Sippel A.E., Schrewe H. Nuclear factor I-B (Nfib) deficient mice have severe lung hypoplasia. Mech. Dev. 2002;112:69–77. doi: 10.1016/s0925-4773(01)00640-2. [DOI] [PubMed] [Google Scholar]
- 60.Gronostajski R.M. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249:31–45. doi: 10.1016/s0378-1119(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 61.Kane R., Murtagh J., Finlay D., Marti A., Jaggi R., Blatchford D., Wilde C., Martin F. Transcription factor NFIC undergoes N-glycosylation during early mammary gland involution. J. Biol. Chem. 2002;277:25893–25903. doi: 10.1074/jbc.M202469200. [DOI] [PubMed] [Google Scholar]
- 62.Osada S., Matsubara T., Daimon S., Terazu Y., Xu M., Nishihara T., Imagawa M. Expression,D.NA-binding specificity and transcriptional regulation of nuclear factor 1 family proteins from rat. Biochem. J. 1999;342(Pt 1):189–198. [PMC free article] [PubMed] [Google Scholar]
- 63.Gao B., Jiang L., Kunos G. Transcriptional regulation of alpha(1b) adrenergic receptors (alpha(1b)AR) by nuclear factor 1 (NF1): a decline in the concentration of NF1 correlates with the downregulation of alpha(1b)AR gene expression in regenerating liver. Mol. Cell. Biol. 1996;16:5997–6008. doi: 10.1128/mcb.16.11.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clark R.E., Jr, Miskimins W.K., Miskimins R. Cyclic AMP inducibility of the myelin basic protein gene promoter requires the NF1 site. Int. J. Dev. Neurosci. 2002;20:103–111. doi: 10.1016/s0736-5748(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 65.Rafty L.A., Santiago F.S., Khachigian L.M. NF1/X represses PDGF A-chain transcription by interacting with Sp1 and antagonizing Sp1 occupancy of the promoter. EMBO J. 2002;21:334–343. doi: 10.1093/emboj/21.3.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Araki K., Nakajima Y., Eto K., Ikeda M.A. Distinct recruitment of E2F family members to specific E2F-binding sites mediates activation and repression of the E2F1 promoter. Oncogene. 2003;22:7632–7641. doi: 10.1038/sj.onc.1206840. [DOI] [PubMed] [Google Scholar]
- 67.Prowse D.M., Bolgan L., Molnar A., Dotto G.P. Involvement of the Sp3 transcription factor in induction of p21Cip1/WAF1 in keratinocyte differentiation. J. Biol. Chem. 1997;272:1308–1314. doi: 10.1074/jbc.272.2.1308. [DOI] [PubMed] [Google Scholar]
- 68.Kennett S.B., Udvadia A.J., Horowitz J.M. Sp3 encodes multiple proteins that differ in their capacity to stimulate or repress transcription. Nucleic Acids Res. 1997;25:3110–3117. doi: 10.1093/nar/25.15.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu N., Heuchel R., Barczyk M., Zhang W.M., Gullberg D. Tandem Sp1/Sp3 sites together with an Ets-1 site cooperate to mediate alpha11 integrin chain expression in mesenchymal cells. Matrix Biol. 2006;25:118–129. doi: 10.1016/j.matbio.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 70.Tsuda M., Watanabe T., Seki T., Kimura T., Sawa H., Minami A., Akagi T., Isobe K., Nagashima K., Tanaka S. Induction of p21(WAF1/CIP1) by human synovial sarcoma-associated chimeric oncoprotein SYT-SSX1. Oncogene. 2005;24:7984–7990. doi: 10.1038/sj.onc.1208942. [DOI] [PubMed] [Google Scholar]
- 71.Li L., He S., Sun J.M., Davie J.R. Gene regulation by Sp1 and Sp3. Biochem. Cell Biol. 2004;82:460–471. doi: 10.1139/o04-045. [DOI] [PubMed] [Google Scholar]