Abstract
During the infection of plants, Agrobacterium tumefaciens introduces several Virulence proteins including VirE2, VirF, VirD5 and VirE3 into plant cells in addition to the T-DNA. Here, we report that double mutation of virF and virE3 leads to strongly diminished tumor formation on tobacco, tomato and sunflower. The VirE3 protein is translated from a polycistronic mRNA containing the virE1, virE2 and virE3 genes, in Agrobacterium. The VirE3 protein has nuclear localization sequences, which suggests that it is transported into the plant cell nucleus upon translocation. Indeed we show here that VirE3 interacts in vitro with importin-α and that a VirE3–GFP fusion protein is localized in the nucleus. VirE3 also interacts with two other proteins, viz. pCsn5, a component of the COP9 signalosome and pBrp, a plant specific general transcription factor belonging to the TFIIB family. We found that VirE3 is able to induce transcription in yeast when bound to DNA through the GAL4-BD. Our data indicate that the translocated effector protein VirE3 is transported into the nucleus and there it may interact with the transcription factor pBrp to induce the expression of genes needed for tumor development.
INTRODUCTION
Agrobacterium tumefaciens is the causative agent of the crown gall disease in plants. It provokes crown gall by transforming plant cells with a part of its Ti plasmid, the Transfer (T)-region or T-DNA, at infection sites. The T-DNA, which integrates in the plant genome contains genes encoding enzymes involved in the production and modification of plant hormones and in the production of tumor specific metabolites called opines. As a result plant cells are triggered to divide leading to crown gall tumor formation (1). The genes responsible for the processing and transfer of the T-DNA to plant cells are present in the Virulence region of the Ti plasmid and are called vir genes. These genes are spread over several different operons virA–virR (2), which together form a regulon. The vir genes are induced by phenolic compounds that are released by plants after wounding. The main inducer is a compound called acetosyringone. Some of the Vir proteins are responsible for the processing of the T-DNA, some others for transferring of the T-DNA to the plant cell and a third group is involved in the hijacking of the host cell metabolism in order to allow the integration and expression of the genes carried on the T-DNA. VirD1, VirD2 and VirC1 are responsible for the DNA processing reactions in the Ti plasmid leading to the formation of a single-stranded copy of the T-region (T-strand) that is introduced into plant cells. VirD2 generates nicks at the border repeats surrounding the T-DNA in the Ti plasmid and forms a covalent phosphodiester bond through its tyrosine 29 residue with the base at the 5′ end of the nick. Eventually, the ssDNA–VirD2 complex is transported into plant cells through the Type-IV Secretion System (TFSS) that is formed by the products of the genes of the virB operon (VirB1–VirB11) and the virD4 gene (3). VirE2 is the most abundant protein produced after induction of the vir genes. It binds in an unspecific and highly cooperative manner to single-stranded DNA. This protein probably coats the T-strand in the plant cell protecting it from the attack of host nucleases. VirD2 and VirE2 each have Nuclear Localization Signals (NLSs), which helps to target the T-strand to the plant cell nucleus. VirE1 is a chaperone protein of VirE2, which prevents the formation of VirE2 aggregates in the bacterium (4–6). The VirE2 protein is translocated into plant cells independently of the T-strand–VirD2 complex through the TFSS (7). Besides VirE2, a number of other Vir proteins are translocated into plant cells, including VirF, VirE3 and VirD5 (7–9).
The translocated virulence proteins are effector proteins, which interact with host proteins in the recipient (plant) cell to mediate the transformation of a normal plant cell into a crown gall tumor cell. The VirD2 protein, which is attached to the 5′ end of the T-strand, has a nuclear localization sequence (NLS) through which it interacts with importin-α (10,11). This guarantees the efficient nuclear uptake of the T-strand. Besides, VirD2 interacts with cyclophilins (12), which can function as chaperones and might keep VirD2 in an active conformation in the host cytoplasm. Recently, two other VirD2 interactors were described: one is the nuclear kinase CAK2Ms, a member of the conserved cyclin-dependent kinase-activating kinase family, which phosphorylates VirD2 in vitro and in vivo. The other is the TATA box-binding protein (TBP) (11). The CAK2-mediated phosphorylation may modulate VirD2 activity in vivo. Its interaction with TBP could lead the T-DNA to sites in the genome that are transcribed and where T-DNA seems preferentially integrated. The VirE2 protein also has an NLS, but it does not seem to interact with importin-α (10). Instead, it interacts with the proteins VIP1 and VIP2 (13). VIP1 is a nuclear protein that may facilitate the transport of VirE2 into the plant cell nucleus. VirF interacts through its F-box with plant homologs of the yeast Skp1 protein (14). F-box proteins and Skp1 proteins are subunits of a class of E3 ubiquitin ligases, called SCF complexes. These SCF complexes target specific proteins through the interaction with the F-box protein to ubiquitin-mediated proteolysis. It was recently published that VirF directs the targeted proteolysis of VIP1 and VirE2 (15).
Recently, it was found that two more Vir proteins are translocated from Agrobacterium into yeast and plant cells: VirE3 and VirD5 (8,9,16). VirE3 is conserved among all the Agrobacterium Ti plasmids studied so far. Even the Agrobacterium rhizogenes Ri-plasmid that lacks the genes virE1 and virE2 contains a copy of virE3, suggesting that the VirE3 protein itself plays an important role during transformation. Recently, it was published that VirE3 may mimic the function of VIP1 and assist in the transportation of VirE2 into the nucleus. Here, we describe the effects of virE3 deletion on virulence. Also, we have used the yeast two-hybrid system to find plant interactors to obtain further indications about its functions in plant transformation. We obtained indications that VirE3 may be involved in activation of genes in the plant cell nucleus during the early stages of transformation.
MATERIALS AND METHODS
Construction of a virF, virE3 and a double deletion mutant
The sacR/B gene cassette was subcloned as a 2.6 kb PstI fragment from pUM24 (17) into the PstI site of pMTL23 (18) resulting in pSDM3001. A 2.0 kb SacI fragment from plasmid pRAL7007 (19) containing the virF gene with flanking sequences was subcloned into the SacI site of pUC19 (20) resulting in pSDM3002. virF was deleted from pSDM3002 by EcoRV/EagI digestion and religation of the plasmid resulting in pSDM3003. The virF flanking sequences were then subcloned as a 1.5 kb SacI fragment from pSDM3003 into the SacI site of pSDM3001. Finally, the virF flanking sequences and the sacR/B gene cassette were cloned as a 4.1 kb BglI/BamHI fragment into the BamHI site of pBGS19 (21) resulting in pSDM3004.
The sacR/B gene cassette was cloned as a 2.6 kb PstI fragment from pUM24 (17) into the PstI site of pBGS8 (21) resulting in pSDM3005. The virE3 gene with flanking sequences from pTi15955 was subcloned as a 3.5 kb EcoRV/SacI fragment from pRAL3248 (19) into the EcoRV/SacI sites of pIC20R (22) (named pSDM3006). Subsequent deletion of the virE3 coding sequence from pSDM3006 by SmaI/StuI digestion and religation of the plasmid resulted in pSDM3007. The flanking sequences were then cloned as a 1.1 kb EcoRV/SacI fragment from pSDM3007 into the SacI/SmaI sites of pSDM3005 resulting in pSDM3008.
The Agrobacterium wild type strain LBA1010 harboring pTiB6 was electroporated with the non-replicative plasmids pSDM3004 and pSDM3008, respectively. Strains in which these plasmids had integrated into the Ti plasmid were grown on LC medium lacking NaCl and containing 6% sucrose (17) to select for virF and virE3 deletion mutants. In this way the virF deletion mutant LBA2560 and the virE3 deletion mutant LBA2564 were obtained. A double virF, virE3 deletion mutant was obtained after electroporation of mutant LBA2560 with plasmid pSDM3008 and by similarly applying the marker exchange-eviction mutagenesis method. The resulting virF, virE3 double mutant was named LBA2566. Deletion of the genes in the various mutants was confirmed by Southern blot analysis.
Tumor assay
A.tumefaciens cells from an overnight culture were washed three times with 0.9% (w/v) NaCl solution and diluted to an OD660 of ∼1.0. Stems of Nicotiana glauca, Nicotiana tabacum, Heliantus annuus, Lycopersicon esculentum, Kalanchoë tubiflora and Kalanchoë daigramontiana plants were wounded at three sites with a sterile toothpick and inoculated with 20 μl A.tumefaciens culture per wound site except for H.annuus which was inoculated with 15 μl bacterial culture.
RT–PCR
RNA isolation was carried out using the RNeasy kit of QIAGEN according to the instructions of the supplier.
Total RNA (0.5 μg) was annealed with 50 pmols of primer VirE3–6 (AACTGCAGGACCCGGGATGCGGTAATAC) to the 3′ end of the mRNA. The reaction was carried out in a final volume of 5 μl adjusting the volume with water. The samples were heated at 80°C for 3 min and then incubated at 60°C for 15 min. The RNA was reverse transcribed in 12 μl of 1× AMV–RT buffer, 0.2 μl of dNTP (25 mM), 1 μl of AMV–RT (Roche). Samples were incubated at 42°C for 1 h. A1 μl of the cDNA was then used for amplification with virE2 primers (VirE2/1 TTTGAACACACCGTCAAGCG) and VirE2/2 TCATGGATGTCACGCAACTC). The annealing temperature for these primers was 60°C and the number of cycles used 40. Similarly, amplification was done with virE3 primers VirE3-'NcoI (TACTCCATGGTGAGCACTACGAAGAAAA) and VirE3BglI (CCTGTAGATCTTTGCCGAAGGTA). The annealing temperature used with this set of primers was 64°C and the number of cycles 35.
Yeast two-hybrid
The Matchmaker yeast two-hybrid system (Clontech) was used to screen an Arabidopsis thaliana cDNA library fused to the GAL4-activation domain (pACT). The virE3 coding sequence (a SalI–PstI fragment) was fused to the GAL4 DNA-binding domain in pAS2-1 yielding plasmid pASE3. A BamHI–SmaI fragment from pASE3 containing virE3 except for the last 579 nt was cloned into pAS2-1 producing plasmid pASE3ΔC. The plasmid library was transformed into yeast strain PJ69-4A (23) harboring plasmid pASE3ΔC using the PEG-lithium acetate method (24) and screened for histidine and adenine auxotrophy. The screen was performed at 20°C. cDNA inserts from yeast cells growing in medium lacking histidine and adenine were amplified by PCR, using primers BC 304 (CTATTCGATGATGAAGATACC) and IN069 (TTGATTGGAGACTTGACC). PCR products were digested with HaeIII and separated on 1.5% agarose gels, leading to the identification of different groups of clones. The prey plasmids from each group of clones were transformed into Escherichia coli DH5α. The reproducibility and specificity of the interaction were tested by re-transformation of the prey plasmids into yeast strains containing a panel of different bait plasmids.
GST pull-down
Plasmid pASE3 was digested with PstI and overhang nucleotides removed using T4 DNA polymerase, after which it was digested with NcoI. The blunt end/NcoI fragment was isolated from gel. Similarly pGEX-KG (Amersham Pharmacia Biotech) was cut with SacI and overhang nucleotides removed with T4 DNA polymerase after which it was digested with NcoI. The blunt end/NcoI fragment obtained from pASE3 digestion was cloned into pGEX-KG digested as described above yielding plasmid pKGE3. The cDNAs of the importin-α genes present in the prey plasmids were cloned at the C-terminus of the His-tag (10× His) in pET-16H. E.coli BL21-DE3 cells were used for heterologous expression of the recombinant proteins. The GST–VirE3 fusion protein was purified from 4 ml of a culture expressing the fusion protein while GST protein was purified from 2 ml of expressing culture. Protein purification was done by binding to a gluthathione-Sepharose matrix (Amersham Pharmacia) essentially as described in (25). Four millilitres of E.coli cell lysate in 1 ml of binding buffer (50 mM Tris–HCl, pH 6.8; 100 mM NaCl, 10 mM CaCl2, 0.1% Triton X-100) expressing the His-tagged interactors, were incubated with the 100 μl of GST–VirE3 bound matrix or 50 μl of GST bound matrix for 2 h at 4°C. After four washes with binding buffer, 20 μl of all samples were loaded on a 12% SDS–polyacrylamide gel. Proteins were blotted on a PDVF membrane and the His-tagged interactors were detected using pentaHis-antibodies (QIAGEN).
Particle bombardment
The N-terminal part of the coding region of virE3 was amplified by PCR using primers VirE3-1B (TGAGATCTGCGTGAGCACTACGAAGAAAA) and VirE3-BglIIΔT (CCTGTAGATCTGCCGAAGGTAT). The amplified fragment was digested with BglII and cloned into the BglII site of pTH-BN in order to make a GFP–VirE3 fusion resulting in plasmid pTHBNE3. Deletion of NLS1 and NLS2 was done using primers ΔNLS1.1 (CTCCAGTCTGGTTTGGAGCGCCTCTTCTTC) and ΔNLS1.2 (GACTATCGCCAT ATCAGCAGCATAATCGGGGCTCTCCAGTCTGGTTTG) in order to delete NLS1 and primers ΔNLS2.1 (CGTTAATTCTTTTGGGTTGTCTACATTTCCCAGAATTTT ATAGCCTAT) and ΔNLS2.2 (CGTCACCGGGTTACGCTTCAGAACCGTACCGTG CTCACGCGTTAATTCTTTTGG) to delete NLS2 as descried by Fisher and Pei (26).
Gold suspensions were prepared as follows: 3 mg of 1.6 μm and 3 mg of 1.0 μm gold particles were washed once with 100% ethanol and twice with water. After the third wash, particles were resuspended in 100 μl of water. A 50 μl of the gold suspension were coated with 10 μg of plasmid DNA and bombarded at 1350 psi into the epidermis of onion scales using a Helios gene gun (PDS-1000/He, Bio-Rad) followed by incubation at 28°C in the dark for 24 h. The localization of GFP-VirE3 was determined by using confocal microscopy.
RESULTS
The protein encoded by virE3 plays a role in tumorigenesis
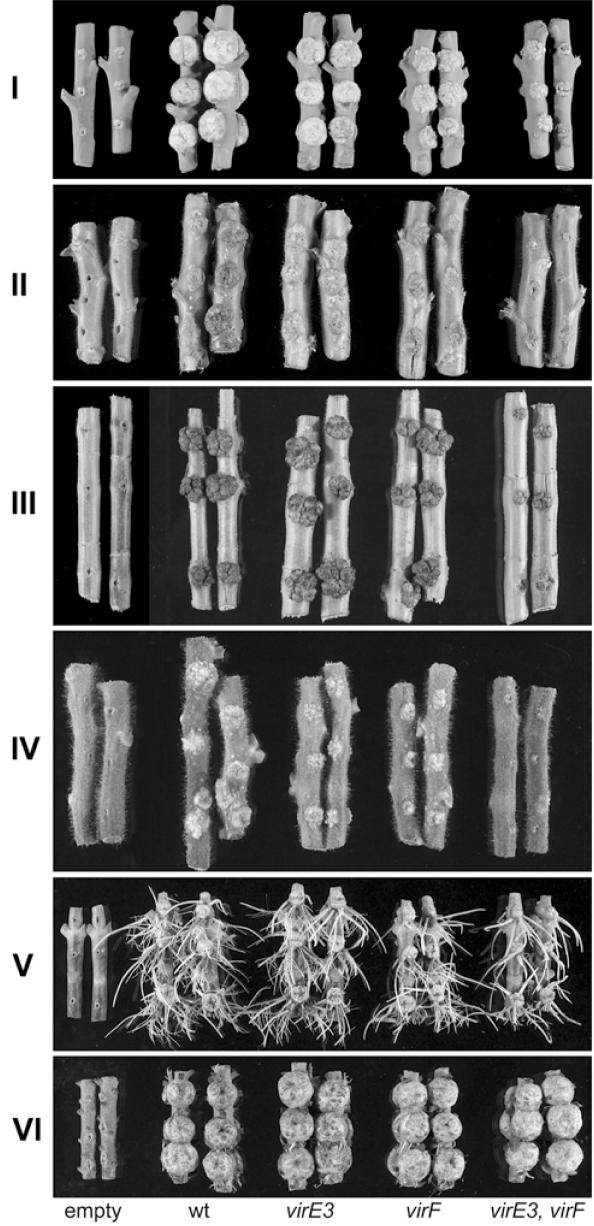
To study the role of VirE3 in tumorigenesis by A.tumefaciens, a virE3 deletion mutant, LBA2564, a virF deletion mutant LBA2560 and a virE3, virF double deletion mutant LBA2566 were constructed and compared to the wild-type (LBA1010) in virulence assays on N.glauca, N.tabacum, H.annuus, L.esculentum, K.daigramontiana and K.tubiflora. The results are shown in Figure 1. The virF, the virE3 and the double mutant had similar virulence on K.tubiflora and K.daigramontiana. The virF mutant gave smaller tumors on N.glauca, N.tabacum and L.esculentum, which is in line with earlier findings in our group (19, 27). Tumors induced by the virE3 deletion mutant were comparable in size to those induced by the wild-type, but the virE3 virF double mutant showed a much more severe decrease in tumor formation on the Nicotiana species, tomato and sunflower than the virF mutant, thus revealing that VirE3 contributes to virulence (Figure 1).
Figure 1.
Tumor formation on stems of various plant species, which were infected with (from left to right) the avirulent strain LBA288 (lacking the Ti plasmid), the wild-type octopine strain LBA1010, the virE3 deletion mutant LBA2564, the virF deletion mutant LBA 2560 or the virE3, virF double deletion mutant LBA2566. [(I) Nicotiana glauca, (II) Nicotiana tabacum, (III) Heliantus annuus, (IV) Lycopersicum esculentum, (V) Kalanchoë daigramontiana and (VI) Kalanchoë tubiflora].
Complementation experiments with the virE3 and virE3, virF double mutants (data not shown) showed that apart from virE3 and virF no other mutations are responsible for the attenuated virulence of the mutants.
virE3 is transcribed together with virE1 and virE2 in a polycistronic mRNA
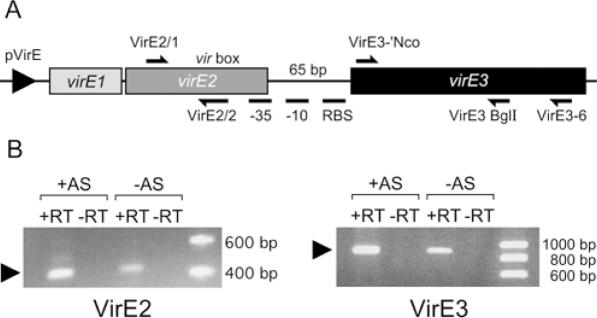
The virE3 gene is located downstream of virE2 with the start codon 65 bp after the stop codon of virE2. Previously, it was reported (28) that virE3 is inducible by acetosyringone. It is likely therefore, that activation of this gene depends on the VirA/VirG two-component regulatory system. The vir-box located in front of virE1 may regulate activation of virE3 transcription. However, close inspection of the nucleotide sequence upstream of virE3 revealed the presence of a putative ribosome binding site and −10 and −35 promoter sequences which partly overlap the 3′ end of the virE2 gene (Figure 2A). Furthermore, a putative vir-box is located upstream of these putative promoter sequences within the virE2 coding sequence. In order to find out whether one large polycistronic mRNA is produced containing virE1, virE2 and virE3 we performed RT–PCR experiments. Total RNA from wild-type strain LBA 1010 was isolated both from cells that were induced with acetosyringone and non-induced cells. The cDNA was synthesized from both samples using the primer VirE3-6 annealing in virE3. PCR were performed on the cDNA using two different sets of primers, one set annealing in virE3 (VirE3-1'NcoI; VirE3-BglII) and the other set annealing in virE2 (VirE2/1; VirE2/2). As shown in Figure 2B it is possible to detect mRNA containing virE2 when cDNA is synthesized with a primer annealing in virE3. This shows that virE2 and virE3 are present in one polycistronic mRNA. Therefore, we can conclude that virE3 is transcribed from the virE promoter in front of virE1 and that it belongs to the virE operon. PCR products of virE2 and virE3 were also detected, when synthesized cDNA represented the mRNA from non-induced cells. The medium used has a pH 5.5 and it has been reported that vir genes are already weakly expressed in media of low pH (29). As expected, the intensity of the bands is higher, when mRNA is used from cells induced by AS corroborating that the virE genes are induced by acetosyringone.
Figure 2.
(A) Schematic representation of virE operon. Position of primers used in RT-PCR and putative promoter and vir-box upstrem of virE3 are indicated. (B) Electrophoresis of the RT–PCR products. Total RNA from wild-type strain LBA 1010 was isolated both from cells that were induced with acetosyringone (+AS) and non-induced cells (−AS). The right panel labelled with VirE3 indicates that PCR was done with primers annealing in virE3 (VirE3 BglI and VirE3-'NcoI). The left panel labelled with VirE2 indicates that PCR was done using primers annealing in virE2 (VirE2/1 and VirE2/2). +RT = Samples treated with Reverse Transcriptase. −RT = Control samples without Reverse Transcriptase.
VirE3 activates transcription in yeast when bound to DNA
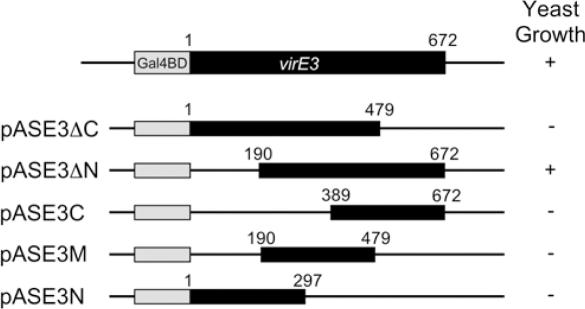
VirE3 was fused to the Gal4-DNA-binding domain (pASE3) for use in two-hybrid screens, but first tested in one-hybrid assays. The Gal4-VirE3 fusion protein turned out to activate the his3 and ade2 genes that in the yeast strain PJ69-49A are driven by Gal promoters, allowing yeast to grow in minus histidine or adenine medium. Deletion of the N-terminal part (amino acid 1–190) of VirE3 (pASE3ΔN) did not reduce the transcription-stimulating activity of the fusion protein. However, deletion of the C-terminal part (amino acid 479–672) of VirE3 (pASE3ΔC) led to the loss of the transcription activating activity. Also larger C-terminal (amino acid 297–672), N-terminal (amino acid 1–389) or combined C-terminal and N-terminal (amino acid 1–190 and 479–672) deletions led to a loss of transcriptional activation. These results suggest that VirE3, when bound to DNA, is itself able to promote gene transcription and that it can do this without its N-terminal 190 amino acid (Figure 3).
Figure 3.
Schematic representation of GAL4BD–VirE3 fusion proteins, which were tested in yeast one-hybrid assays for induction of yeast growth. Plus symbol indicates growth of yeast containing the fusion protein in medium lacking adenine. Minus symbol indicates no growth of yeast containing the fusion protein in medium lacking adenine.
VirE3 interacts with two A.thaliana importins-α, pCsn5-1 and pBrp (TFIIB-related protein)
We used the yeast two-hybrid system to identify plant interactors of VirE3, as this may give clues as to the function of this protein during tumorigenesis. A deleted version of VirE3 without the transcriptional activating activity was used as a bait (pASE3ΔC) and an Arabidopsis cDNA library was used as a prey. After performing all the control tests and back transformation into yeast, four different proteins were identified as positive interactors of VirE3. Two are members of the Arabidopsis importin-α family, At3g06720 (AtKapα) and At1g09270 (AtImpα-4), the third one is pCsn5-1 also called AJH1, a component of the COP9 signalosome (30), which is involved in protein degradation. The fourth interactor is pBrp, a novel plant specific transcription factor IIB (TFIIB)-related protein (31).
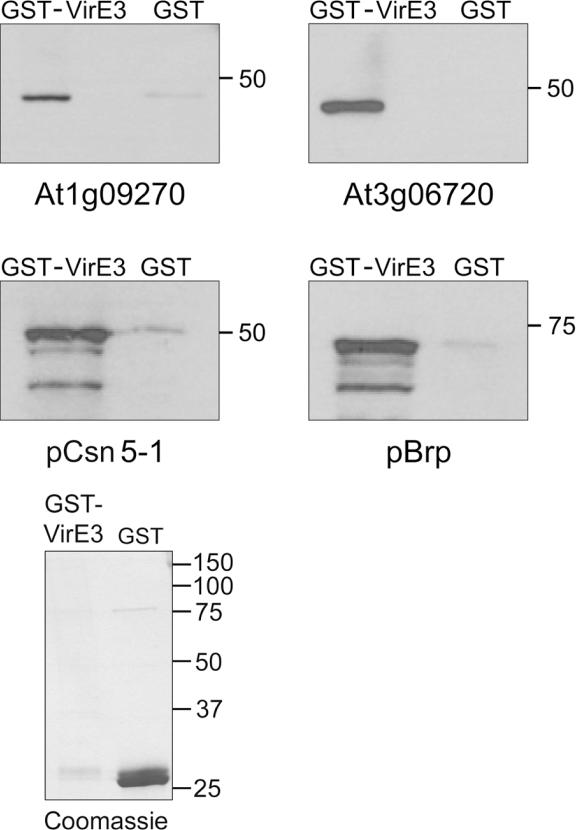
The specificity of the interactions between these proteins and VirE3 was further confirmed by GST pull-down assays. To this end, full-length VirE3 was cloned into the pGEX-KG vector (pKGE3) and the GST-tagged-VirE3 protein was expressed in BL21(DE3) cells. The interactors identified in the yeast two-hybrid system were His-tagged by cloning them into pET-16H. Beads bound to GST-VirE3 were mixed with supernatants of cells expressing His-tagged proteins. After removing the unbound protein, the presence of the His-tagged proteins on the beads was revealed by means of western blot analysis using antibodies against His-tag.
The importin-α At3g06720 is a 532 amino acids protein. The portion used in the pull-down assays was from the amino acid 166 till the end of the protein. For the importin-α At1g09270, a protein of 538 amino acids, a portion from amino acid 123 till the end of the protein was similarly used in pull-down assays. As shown in Figure 4 both fusion proteins interact with VirE3 in vitro. This interaction is specific for the VirE3 portion of the GST–VirE3 fusion protein because control GST retains no or at most only traces of His-importin-α proteins.
Figure 4.
In vitro interaction of VirE3 with two importin-α proteins (At1g09270, At3g06720), pCsn5-1 and pBrp. The interaction between VirE3 and these proteins was analyzed in GST pull-down assays. The interacting proteins were identified on a western blot using antibodies against the His-tag. Coomassie panel shows 10 μl of bounded bead to GST-VirE3 and GST isolated as described in Materials and Methods. Proteins level is shown to estimate amount of protein used in the assay.
The complete pCsn5-1 protein except the first 7 amino acids was used in pull-down assays. Figure 4 shows the in vitro interaction between VirE3 and pCsn5-1. A homologue to pCsn5-1 in mammals, Jab1, has been shown to interact with the GAL4-DNA-binding domain in yeast two-hybrid assays (32). Nevertheless in our pull-down experiments, where the GAL4-DNA-binding domain, is not present, the direct interaction of pCsn5-1 with VirE3 that was discovered in the yeast two-hybrid screen, is confirmed.
The TFIIB-related protein, pBrp, (At4g36650) was assayed in a pull-down as a full-length protein. The results shown in Figure 4 indicate that VirE3 and pBrp are direct interactors.
All proteins found as VirE3 interactors in the yeast two-hybrid clearly bind to VirE3 in pull-down assays suggesting a direct interaction between VirE3 and all these proteins.
VirE3 localizes in the nucleus of onion cells
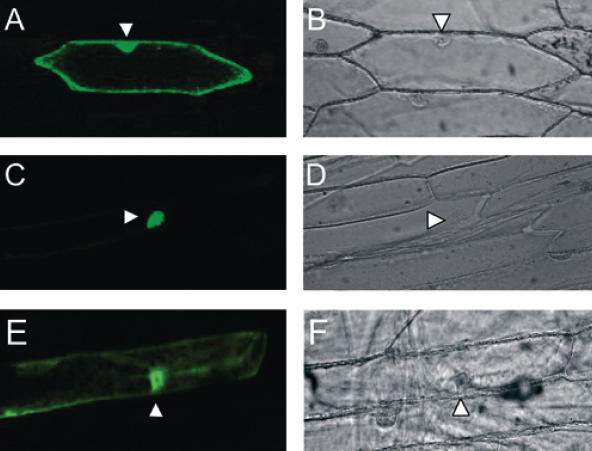
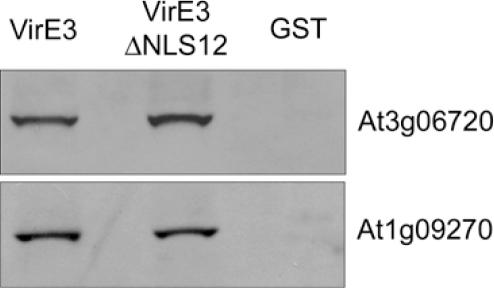
We have shown that VirE3 interacts with two importins-α. These proteins are part of the cell machinery involved in the translocation of proteins into the nucleus through the nuclear pore complex present in the double membrane of the nuclear envelope. Importins-α interact with proteins containing an NLS within their primary structure. The NLS consists of a cluster of basic amino acids (monopartite) or two clusters of basic amino acids separated by 10–12 amino acids (bipartite) (33,34). In VirE3 two putative bipartite NLSs can be found, both present in the N-terminal half of the protein between residues 40 and 53 (KRQTRLESPDRKRK)(NLS1) and between amino acids 80 and 101 (KRLRVDPKELTREHGRLRKTK)(NLS2). Therefore, we wished to analyze whether VirE3 localized to the nucleus of plant cells. To this end, we fused the first 297 amino acids of VirE3 to the C-terminus of GFP (GFP-VirE3N) and used particle bombardment to introduce the construct into onion epidermis cells. The results are shown in Figure 5, where it can be seen that the GFP-VirE3N fusion protein is located in the nucleus of the cells indeed (Figure 5C and D), whereas GFP alone (control) localizes both to the cytoplasm and nucleus (Figure 5A and B). The exclusion size of the nuclear pore is 40–60 kDa and the presence of small 27 kDa GFP protein in the nucleus is due to diffusion through the nuclear pore. These results demonstrate that VirE3 is specifically targeted to the nucleus of plant cells. Indeed when we introduced a construct encoding a GFP–VirE3N fusion protein from which both NLS1 and NLS2 had been deleted (GFP–VirE3NΔNLS1,2) in onion epidermis cells, green fluorescence was seen both in the nuclear area and in the cytoplasm (Figure 5E and F) as was seen for the GFP-control. The protein lacking NLS1 and NLS2 may still, though inefficiently, be able to find its way to the nucleus. Circumstantial evidence for this came from GST pull-down assays in which VirE3ΔNLS1,2 still was able to bind to the importin-α proteins Figure 6.
Figure 5.
Nuclear localization of VirE3 in onion epidermis cells. (A and B) GFP. (C and D) GFP-VirE3N. (E and F) GFP-VirE3NΔNLS1,2. (A, C and E) are fluorescent images. (B, D and F) visible light. Arrowhead indicates the position of the nuclei.
Figure 6.
In vitro interaction of VirE3 with both NLS1 and NLS2 deleted (VirE3ΔNLS1,2) and importin−α proteins (At3g06720, At1g09270). Both VirE3 and VirE3ΔNLS1,2 were expressed as GST fusion proteins. The interacting proteins were identified on a western blot using antibodies against the His-tag.
DISCUSSION
A.tumefaciens is able to transfer DNA into plant, yeast, fungal and human cells. Our study showed that Agrobacterium also transfers proteins to plants and yeast (7,16). Protein transfer is independent of DNA transfer although the same VirB/D4 TFSS is used. The effector proteins found to be transferred by Agrobacterium are VirE2, VirF, VirD5 and VirE3. The VirE2 protein coats the T-strand by cooperative binding and thus protects the T-strand from degradation in the plant cell and enables nuclear import. Unlike virE2 mutants, which are avirulent on most plants, virF mutants are attenuated in virulence on certain hosts only. VirF interacts with the plant protein Skp1, which forms part of so called SCF complexes that are involved in ubiquitination and proteolysis of target proteins. A third effector protein also translocated by Agrobacterium is VirE3. The virE3 gene is conserved among all the Agrobacterium Ti plasmids studied so far. Even the A.rhizogenes Ri-plasmid that lacks the virE1 and virE2 genes contains a copy of virE3, suggesting that this gene plays, by itself, an important role during transformation. Previously, it was reported that virE3 is not essential for tumorigenesis, as a transposon mutant in virE3 still is able to induce tumors on Kalanchoë leaves (28). However, here we show that VirE3 is in fact a host specific effector protein, as an Agrobacterium virE3 mutant has somewhat attenuated virulence and the virE3 mutation drastically diminishes the virulence of virF mutants on several plant species including L.esculentum, N.glauca and N.tabacum.
As shown previously by our group (16) VirE3 is translocated to plant cells. Here, we show that VirE3 interacts with several plant proteins. Two of these are importin-α proteins. These proteins are cytoplasmically localized and together with importin-β are involved in the translocation of proteins into the nucleus of eukaryotic cells. This nuclear import is dependent on a specific signal sequence, the NLS in the nuclear protein. In the cytoplasm, importin-α interacts on the one hand with the protein containing the NLS and on the other hand with importin-β. This complex importin-α/β/NLS-substrate migrates into the nucleus through the nuclear pore complex and once in the nucleus, the NLS-substrate is released and importin-α and -β recycled back into the cytoplasm. Structural analysis of importin-α has revealed three functional domains. One is an importin-β binding (IBB) domain, which is located in the N-terminus (35). The second is a hydrophobic central domain known as the armadillo (arm) repeat domain (36). A classical NLS binds to the arm repeat domain in importin-α (37). The third is a short acidic domain in the C-terminus. The function of this region is to export importin-α from the nucleus (38). The IBB domain contains an auto-inhibitory segment, which inhibits binding of importin-α to NLS-cargo protein. Importin-β binding to importin-α moves this auto-inhibitory segment away from the NLS-binding site in importin-α. This yields an importin-α/β heterodimer active for binding to an NLS-containing protein (39). The importin-α fragments isolated from the yeast two-hybrid library and the version used in our GST pull-down experiments lacked the IBB domain and consequently showed high affinity to NLS-containing proteins even in absence of importin-β.
The interaction with importin-α showed that VirE3 interacts directly with the importin machinery of the cell and no additional factors are needed to bridge VirE3 and the importin machinery, as is the case for VirE2, which needs VIP1 in order to interact with importin-α (40). At the same time, this interaction with importin-α suggested that VirE3 could be translocated into the nucleus of plant cells. We checked the localization of VirE3 in onion epidermis cells. Indeed, VirE3 was found in the nucleus, confirming that VirE3 produced (or introduced) in the cytoplasm is transported into the plant nucleus. During our studies this was also reported by Lacroix et al. (41)
Our data indicate that VirE3 interacts with two other proteins pCsn5-1 and pBrp. pCsn5-1 can be found both in the nucleus and cytoplasm. It can form part of the COP9 signalosome (CSN) or function as monomeric protein. It is not known whether VirE3 interacts with the monomeric form of pCsn5-1 or with CSN through pCsn5-1. The function of the CSN is not really clear, but it has been implicated in regulating proteins stability either leading to stabilization of a protein or reversely to proteolytic degradation (42). Therefore, the interaction with pCsn5-1 may lead to the degradation of VirE3 through the 26S proteasome or conversely increase its stability in the plant cell (43). Further experiments need to be performed in order to determine the significance of this pCsn5-1-VirE3 interaction for tumor development.
An intriguing result is the transcriptional activation activity that VirE3 shows in yeast. This transcription-stimulating activity lies in the C-terminal part of the protein, but no classical eukaryotic transcription activation domain can be found in VirE3 by sequence comparison. This activity suggests that VirE3 could act as a transcriptional activator or co-activator in the host. In order to have either of these functions VirE3 should interact with the plant transcriptional machinery. We have found that VirE3 interacts with pBrp, a plant specific member of the TFIIB family that binds to the TATA box (31). It is located at the cytosolic face of the plastid envelope, but it has been demonstrated that under certain conditions it is released from the plastids and accumulated in the nucleus. pBrp may be released from the plastids after Agrobacterium infection or it may interact with VirE3 in the cytoplasm and together be translocated into the nucleus, whereby VirE3 may act as a bridge between pBrp and the importin cell machinery. The general transcription factor TFIIB has been implicated as the direct target of many gene-specific transcriptional activators, leading to the proposal that certain activators stimulate transcription by TFIIB recruitment (44,45). Thus, certain genes could be induced by the pBrp–VirE3 interaction or just by pBrp brought into the nucleus by VirE3.
Among the genes that can be induced may not only be plant genes but also the genes harbored naturally in the T-DNA. In this sense, the results that we obtained with a double virE3, virF mutant could be explained. The VirF protein is thought to form a SCF complex that induces the specific proteolysis of VirE2 and thus promote the uncoating of the T-strand (15). The uncoated T-strand may then become double stranded and the genes on this dsT-DNA may become expressed even before integration of the T-DNA in the genome takes place. A double virE3, virF mutant would lack both activities and their infection ability would be greatly diminished.
Based on the data shown in this paper we suggest that the VirE3 protein is translocated by Agrobacterium into the plant cell cytoplasm and from there is transported into the nucleus. On its way to the nucleus it may assist in the nuclear transportation of pBrp in the same way as described by Lacroix et al. (41) for nuclear transportation of VirE2. In the nucleus it can activate transcription of genes through the interaction with the general transcription factor pBrp. The genes induced in this way might be needed for proper tumor development in specific hosts, whereas in the host in which virE3 is not needed for tumorigenesis, these genes are not required or are already sufficiently induced by other mechanisms.
This new concept of prokaryotic proteins functioning as transcriptional activators in eukaryotic organisms may also play a role in other interactions of pathogenic bacteria with plants (46), as the AvrBs3 protein, which is translocated into plants by the type III secretion channel of Xanthomonas campestris, similarly has an activation domain. However, a direct interaction between this prokaryotic translocated protein and the transcriptional machinery of the host cells has not yet been investigated.
Acknowledgments
We thank Peter Hock for preparing figures, Gerda Lamers for her help with the confocal microscopy, Amke den Dulk-Ras for her help with extracellular complementation experiments and Drs Sylvia de Pater and Bert van der Zaal for critical reading of the manuscript. The work was supported by the European Union through a Marie Curie fellowship to F.G., by the Stichting Binair Vector Systeem and by the Netherlands Organization for Scientific Research (NWO/ALW contract 834.02.006: State of the art equipment for proteome analysis). Funding to pay the Open Access publication charges for this article was provided by Leiden University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gelvin S.B. Agrobacterium-mediated plant transformation: the biology behind the ‘gene-jockeying’ tool. Microbiol. Mol. Biol. Rev. 2003;67:16–36. doi: 10.1128/MMBR.67.1.16-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu J., Oger P.M., Schrammeijer B., Hooykaas P.J.J., Farrand S.K., Winans S.C. The bases of crown gall tumorigenesis. J. Bacteriol. 2000;182:3885–3895. doi: 10.1128/jb.182.14.3885-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christie P.J. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transportes in eubacteria. J. Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng W., Chen L., Peng W.T., Liang X., Sekiguchi S., Gordon M.P., Comai L., Nester E.W. VirE1 is a specific molecular chaperone for the exported single-stranded-DNA-binding protein VirE2 in Agrobacterium. Mol. Microbiol. 1999;31:1795–1807. doi: 10.1046/j.1365-2958.1999.01316.x. [DOI] [PubMed] [Google Scholar]
- 5.Sundberg C.D., Ream W. The Agrobacterium tumefaciens chaperone-like protein, VirE1, interacts with VirE2 at domains required for single-stranded DNA binding and cooperative interaction. J. Bacteriol. 1999;181:6850–6855. doi: 10.1128/jb.181.21.6850-6855.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Z., Sagulenko E., Ding Z., Christie P.J. Activities of virE1 and the VirE1 secretion chaperone in export of the multifunctional VirE2 effector via an Agrobacterium Type IV secretion pathway. J. Bacteriol. 2001;183:3855–3865. doi: 10.1128/JB.183.13.3855-3865.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vergunst A.C., Schrammeijer B., den Dulk-Ras A., de Vlaam C.M.T., Regensburg-Tuink T.J.G., Hooykaas P.J.J. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–982. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- 8.Vergunst A.C., van Lier M.C., den Dulk-Ras A., Hooykaas P.J.J. Recognition of the Agrobacterium tumefaciens VirE2 translocation signal by the VirB/D4 transport system does not require VirE1. Plant Physiol. 2003;133:978–988. doi: 10.1104/pp.103.029223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vergunst A.C., van Lier M.C., den Dulk-Ras A., Stuve T.A., Ouwehand A., Hooykaas P.J.J. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc. Natl Acad. Sci. USA. 2005;102:832–837. doi: 10.1073/pnas.0406241102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballas N., Citovsky V. Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc. Natl Acad. Sci. USA. 1997;94:10723–10728. doi: 10.1073/pnas.94.20.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bakó L., Umeda M., Tiburcio A.F., Schell J., Koncz C. The VirD2 pilot protein of Agrobacterium-transferred DNA interacts with the TATA box-binding protein and nuclear protein kinase in plants. Proc. Natl Acad. Sci. USA. 2003;100:10108–10113. doi: 10.1073/pnas.1733208100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng W., Chen L., Wood D.W., Metcalfe T., Liang X., Gordon M.P., Comai L., Nester E.W. Agrobacterium VirD2 protein interacts with plant host cyclophilins. Proc. Natl Acad. Sci. USA. 1998;95:7040–7045. doi: 10.1073/pnas.95.12.7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tzfira T., Citovsky V. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 2001;20:3596–3607. doi: 10.1093/emboj/20.13.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schrammeijer B., Risseeuw E., Pansegrau W., Regensburg-Tuink T.J.G., Crosby W.L., Hooykaas P.J.J. Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homologs of the yeast Skp1 protein. Curr. Biol. 2001;11:258–262. doi: 10.1016/s0960-9822(01)00069-0. [DOI] [PubMed] [Google Scholar]
- 15.Tzfira T., Vaidya M., Citovsky V. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature. 2004;431:87–92. doi: 10.1038/nature02857. [DOI] [PubMed] [Google Scholar]
- 16.Schrammeijer B., den Dulk-Ras A., Vergunst A.C., Jurado Jácome E., Hooykaas P.J.J. Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3. Nucleic Acids Res. 2003;31:860–868. doi: 10.1093/nar/gkg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ried J.L., Collmer A. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene. 1987;57:239–246. doi: 10.1016/0378-1119(87)90127-2. [DOI] [PubMed] [Google Scholar]
- 18.Chambers S.P., Prior S.E., Barstow D.A., Minton N.P. The pMTL nic-cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene. 1988;68:139–149. doi: 10.1016/0378-1119(88)90606-3. [DOI] [PubMed] [Google Scholar]
- 19.Melchers L.S., Maroney M.J., den Dulk-Ras A., Thompson D.V., van Vuuren H.A., Schilperoort R.A., Hooykaas P.J.J. Octopine and nopaline strains of Agrobacterium tumefaciens differ in virulence; molecular characterization of the virF locus. Plant Mol. Biol. 1990;14:249–259. doi: 10.1007/BF00018565. [DOI] [PubMed] [Google Scholar]
- 20.Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 21.Spratt B.G., Hedge P.J., te Heesen S., Edelman A., Broomer-Smith J.K. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 22.Marsh J.L., Erfle M., Wykes E.J. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene. 1984;32:481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- 23.James P., Halladay J., Craig E.A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gietz D., St-Jean A., Woods R.A., Schiestl R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;25:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christensen S.K., Dagenais N., Chory J., Weigel D. Regulation of auxin response by the protein kinase PINOID. Cell. 2000;100:469–478. doi: 10.1016/s0092-8674(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 26.Fisher C.L., Pei G.K. Modification of a PCR-based site directed mutagenesis method. Biotechniques. 1997;23:570–574. doi: 10.2144/97234bm01. [DOI] [PubMed] [Google Scholar]
- 27.Hooykaas P.J.J., Hofker M., den Dulk-Ras A., Schilperoort R.A. A comparison of virulence determinants in an octopine Ti plasmid, a nopaline Ti plasmid, and an Ri plasmid by complementation analysis of Agrobacterium tumefaciens mutants. Plasmid. 1984;11:195–205. doi: 10.1016/0147-619x(84)90026-x. [DOI] [PubMed] [Google Scholar]
- 28.Kalogeraki V.S., Zhu J., Stryker J.L., Winans S.C. The right end of the vir region of an octopine-type Ti plasmid contains four new members of the vir regulon that are not essential for pathogenesis. J. Bacteriol. 2000;182:1774–1778. doi: 10.1128/jb.182.6.1774-1778.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang C.H., Winans S.C. Functional roles assigned to the periplasmic, linker, and receiver domains of the Agrobacterium tumefaciens VirA protein. J. Bacteriol. 1992;174:7033–7039. doi: 10.1128/jb.174.21.7033-7039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwok S.F., Solano R., Tsuge T., Chamoviz D.A., Ecker J.R., Matsui M., Deng X-W. Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell. 1998;10:1779–1790. doi: 10.1105/tpc.10.11.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagrange T., Hakimi M-A., Pontier D., Courtois F., Alcaraz J.P., Grunwald D., Lam E., Lerbs-Marhe S. Transcription factor IIB (TFIIB)-related protein (pBrp), a plant-specific member of the TFIIB-related protein family. Mol. Cell. Biol. 2003;23:3274–3286. doi: 10.1128/MCB.23.9.3274-3286.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nordgård O., Dahle Ø., Andersen T.Ø., Gabrielsen O.S. Jab1/CSN5 interacts with the Gal4 DNA binding domain: A note of caution about two-hybrid interactions. Biochimie. 2001;83:969–971. doi: 10.1016/s0300-9084(01)01329-3. [DOI] [PubMed] [Google Scholar]
- 33.Kalderon D., Roberts B.L., Richardson W.D., Smith A.E. A short amino acid sequence able to specify nuclear localization. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 34.Robbins J., Dilworth S.M., Laskey R.A., Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 35.Görlich D., Henklein P., Laskey R.A., Hartmann E. A 41 amino acid motif in importin-α confers binding to importin-β and hence transit into the nucleus. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- 36.Yano R., Oakes M.L., Tabb M.M., Nomura M. Yeast Srp1p has homology to armadillo/plakoglobin/β-catenin and participates in apparently multiple nuclear functions including the maintenance of the nucleolar structure. Proc. Natl Acad. Sci. USA. 1994;91:6880–6884. doi: 10.1073/pnas.91.15.6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fontes M.R., The T., Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α. J. Mol. Biol. 2000;197:1183–1194. doi: 10.1006/jmbi.2000.3642. [DOI] [PubMed] [Google Scholar]
- 38.Kutay U., Bischoff F.R., Kostka S., Kraft R., Görlich D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 39.Kobe B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin α. Nat. Struct. Biol. 1999;6:388–397. doi: 10.1038/7625. [DOI] [PubMed] [Google Scholar]
- 40.Citovsky V., Kapelnikov A., Oliel S., Zakai N., Rojas M.R., Lucas W.J., Gilbertson R.L., Tzfira T., Loyter Abraham. Protein interactions involved in nuclear import of Agrobacterium VirE2 protein in vivo and in vitro. J. Biol. Chem. 2004;279:29528–29533. doi: 10.1074/jbc.M403159200. [DOI] [PubMed] [Google Scholar]
- 41.Lacroix B., Vaidya M., Tzfira T., Citovsky V. The VirE3 protein of Agrobacterium mimics a host cell function required for plant genetic transformation. EMBO J. 2005;24:428–437. doi: 10.1038/sj.emboj.7600524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolf D.A., Zhou C., Wee S. The COP9 signalosome: an assembly and maintenance platform for cullin ubiquitin ligases? Nature Cell Biol. 2003;5:1029–1033. doi: 10.1038/ncb1203-1029. [DOI] [PubMed] [Google Scholar]
- 43.Bech-Otschir D., Seeger M., Dubiel W. The COP9 Signalosome: at the interface between signal transduction and ubiquitin-dependent proteolysis. J. Cell Sci. 2002;115:467–473. doi: 10.1242/jcs.115.3.467. [DOI] [PubMed] [Google Scholar]
- 44.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cosma M.P. Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell. 2002;7:227–236. doi: 10.1016/s1097-2765(02)00604-4. [DOI] [PubMed] [Google Scholar]
- 46.Yang B., Zhu W., Johnson L.B., White F.F. The virulence factor AvrXa7 of Xanthomonas oryzae pv. oryzae is a type III secretion parthway-dependent nuclear-localized double-stranded DNA-binding protein. Proc. Natl Acad. Sci. USA. 2000;97:9807–9812. doi: 10.1073/pnas.170286897. [DOI] [PMC free article] [PubMed] [Google Scholar]