Abstract
Avian plumage traits are the targets of both natural and sexual selection. Consequently, genetic changes resulting in plumage variation among closely related taxa might represent important evolutionary events. The molecular basis of such differences, however, is unknown in most cases. Sequence variation in the melanocortin-1 receptor gene (MC1R) is associated with melanistic phenotypes in many vertebrate taxa, including several avian species. The blue-crowned manakin (Lepidothrix coronata), a widespread, sexually dichromatic passerine, exhibits striking geographic variation in male plumage colour across its range in southern Central America and western Amazonia. Northern males are black with brilliant blue crowns whereas southern males are green with lighter blue crowns. We sequenced 810 bp of the MC1R coding region in 23 individuals spanning the range of male plumage variation. The only variable sites we detected among L. coronata sequences were four synonymous substitutions, none of which were strictly associated with either plumage type. Similarly, comparative analyses showed that L. coronata sequences were monomorphic at the three amino acid sites hypothesized to be functionally important in other birds. These results demonstrate that genes other than MC1R underlie melanic plumage polymorphism in blue-crowned manakins.
Keywords: ecological genetics, Lepidothrix, melanocortin-1 receptor, melanism, plumage polymorphism, sexual selection
1. Introduction
The diversity of avian plumage colours and patterns has fascinated naturalists for centuries. As the targets of both natural and sexual selection, plumage traits can have direct effects on individual fitness, and thus, evolutionary changes in plumage traits may represent important mechanisms in avian speciation (Price 1998). Plumage traits often vary considerably among closely related species, suggesting rapid evolution of plumage differences, which is consistent with their importance as reproductive isolating mechanisms (Omland & Lanyon 2000). Despite the importance of plumage traits in avian speciation, the molecular genetic and developmental pathways responsible for plumage differences among closely related species are entirely unknown in almost all cases.
Elucidating the molecular genetic control of adaptive phenotypic traits, such as many avian plumage characters, is a central goal of evolutionary biology. Only recently, however, it has become possible to explore the basis of such traits in non-model organisms. In this regard, the melanocortin-1 receptor gene (MC1R) is of great interest due to its role in melanistic plumage and pelage patterns (Kijas et al. 1998; Theron et al. 2001; Andersson 2003; Eizirik et al. 2003; Hoekstra & Nachman 2003; Doucet et al. 2004; Mundy et al. 2004; Hoekstra et al. 2004; Rosenblum et al. 2004; Mundy 2005). Variation at MC1R underlies melanic phenotypes in avian, mammalian and reptilian taxa. In birds, MC1R variation is associated with striking melanic plumage polymorphism in bananaquits (Coereba flaveola; Theron et al. 2001), snow geese (Anser caerulescens) (Mundy et al. 2004), Arctic skuas (Stercorarius parasiticus; Mundy et al. 2004), white-winged fairy-wrens (Malurus leucopterus; Doucet et al. 2004) and domestic chickens (Gallus gallus; Takeuchi et al. 1996; Andersson 2003). The taxonomic breadth of this survey suggests that MC1R variation may underlie plumage polymorphism in many avian taxa. Indeed, we are aware of only a single published case in which MC1R variation was not associated with plumage polymorphism. MacDougall-Shackleton et al. (2003) showed that MC1R variation does not underlie unmelanized plumage patterns in Phylloscopus warblers. This result, however, may have been expected since plumage patterns of many Phylloscopus warblers involve complex patterns of unmelanized wing bars, crown stripes and rump patches on a greenish grey background (Mundy 2005), rather than the typical pattern of polymorphism consisting of one predominately dark phenotype and one light phenotype exhibited by bananaquits, skuas, snow geese and other vertebrate taxa (e.g. rock pocket mice (Chaetodipus intermedius)—Hoekstra & Nachman 2003). Here, we show that MC1R variation does not underlie plumage polymorphism in the blue-crowned manakin (Lepidothrix coronata), a species that exhibits a light–dark geographic pattern of plumage variation.
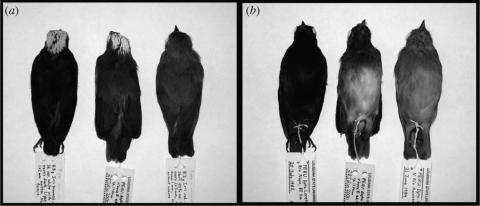
Lepidothrix coronata is a widespread neotropical suboscine passerine, with a striking degree of geographic variation in male plumage colour across its range (Snow 2004; figure 1). In the northern portion of its range (Costa Rica south to northern Peru) males are predominately black with a blue crown patch. In the southern portion of the range (southern Peru to northern Bolivia), males are predominately green with a blue crown patch (figure 1). A broad zone of intergradation between these two plumage groups exists in east-central Peru and western Brazil (Haffer 1970). In this zone, male plumage is generally greenish black, with the degree of green or black plumage in introgressed birds depending on the proximity to the parental populations (Haffer 1970). Males on the southern end of the zone of intergradation are greener than those on the northern end. Throughout the entire range, females are entirely green and lack the blue crown patch.
Figure 1.
Variation in L. coronata male plumage colour. From left to right in both frames is a black male (LSUMZ 110508), a green male (LSUMZ 133193), and a female (LSUMZ 120035). The male specimens correspond, respectively, to tissue samples B-3105 and B-40543 sequenced in this study, and have the same MC1R genotype (see table 2). (a) Dorsal view. (b) Ventral view.
Male plumage is not concordant with mtDNA phylogeographic patterns (Cheviron et al. 2005). Phylogeographic analyses revealed a monophyletic mtDNA haplotype clade in central Peru that encompassed a large proportion of the spectrum of plumage variation, consisting of individuals with pure black, pure green and intermediate plumage (Cheviron et al. 2005). Conversely, populations on the north bank of the Amazon river are identical in plumage colour to those on the south bank of the Amazon, yet despite this similarity in plumage, individuals from these populations fall into relatively distantly related haplotype clades (Cheviron et al. 2005). These results suggest that the microevolutionary forces shaping geographic variation in male plumage may differ from those shaping phylogeographic mtDNA patterns. Since L. coronata is a polygynous, lekking species (Snow 2004), male plumage traits, and the genes underlying them, may be under strong sexual selection, possibly explaining incongruence between mtDNA phylogeographic patterns and geographic variation in male plumage (Brumfield et al. 2001).
In this study, we examined sequence variation in the coding region of MC1R to test the hypothesis that MC1R variation is associated with variation in male plumage colour in L. coronata.
2. Material and methods
(a) Taxon sampling
We sampled 23 individuals from across the range of L. coronata (table 1), with most samples coming from the central Amazonian portion of the range that encompasses the zone of intergradation between the green and black plumage groups. North and south of this zone of intergradation, male plumage colour is relatively invariant. We included representatives of all the phylogeographic mtDNA clades identified by Cheviron et al. (2005), as well as all of the major plumage types. To determine polarity of substitutions, we sequenced MC1R from the congeneric white-crowned manakin (L. serena).
Table 1.
Sampling localities and voucher numbers of specimens used to examine MC1R sequence variation in L. coronata. (Geographic coordinates were obtained either from specimen tags or museum catalogues.)
| locality | species | tissue source | tissue voucher numbers |
|---|---|---|---|
| Costa Rica: Puntarenas; Rio Copey, 4 km E Jaco, 9.62°, −84.63° | L. coronata | LSUMZ | B-16077 |
| Panama: Pamama; E Panama Canal, 9.33°, −79.92° | L. coronata | LSUMZ | B-28461 |
| Venezuela: Amazonas; San Carlos de Rio Negro, −2.08°, −66.48° | L. coronata | AMNH | RWD17053 |
| Peru: San Martin; 20 km NE Tarapoto on road to Yurimaguas, −6.41°, −75.66° | L. coronata | LSUMZ | B-5394 |
| L. coronata | LSUMZ | B-5487 | |
| Peru: Loreto; 1.5 km S Libertad, S bank of Rio Napo, 80 km N Iquitos, −3.03°, −72.75° | L. coronata | LSUMZ | B-3105 |
| Peru: Loreto; S bank of Rio Amazonas, ca 10 km SSW mouth of Rio Napo, −3.42°, −72.58° | L. coronata | LSUMZ | B-4657 |
| Peru: Loreto; lower Rio Napo region, ca 90 km N Iquitos, −3.91°, −73.08° | L. coronata | LSUMZ | B-4545 |
| Peru: Loreto; 86 km SE Juanjui, −7.58°, −75.93° | L. coronata | LSUMZ | B-40543 |
| Peru: Ucayali; SE slope of Cerro Tahuayo, −8.13°, −74.04° | L. coronata | LSUMZ | B-10492 |
| Peru: Loreto; 79 km WNW Contamana, −7.13°, −75.68° | L. coronata | LSUMZ | B-27578 |
| L. coronata | LSUMZ | B-27832 | |
| Peru: Madre de Dios; Moskitania, 13.4 km NNW Atalaya, −13.82°, −71.63° | L. coronata | FMNH | B-433689 |
| L. coronata | FMNH | B-433691 | |
| L. coronata | FMNH | B-433692 | |
| L. coronata | FMNH | B-433693 | |
| L. coronata | FMNH | B-433694 | |
| Brazil: Rondônia; ca 50 km NW Jaciparana, w bank Rio Maderia, −9.25°, −64.40° | L. coronata | LSUMZ | B-31333 |
| Bolivia: La Paz Department; Rio Beni, ca 20 km by river N Puerto Linares, −12.56°, −67.20° | L. coronata | LSUMZ | B-901 |
| Bolivia: Pando Department; Nicolás Suarez, 12 km by road S Cobija, −11.18°, −69.03° | L. coronata | LSUMZ | B-9125 |
| L. coronata | LSUMZ | B-9177 | |
| L. coronata | LSUMZ | B-9269 | |
| L. coronata | LSUMZ | B-9306 | |
| Brazil: Amazonas Department; ca 80 km N Manaus | L. serena | LSUMZ | B-20306 |
(b) Amplification and sequencing of MC1R
Total genomic DNA was extracted from approximately 25 mg of pectoral muscle tissue using QIAGEN DNeasy extraction kits (QIAGEN, Valencia, California). We amplified an 810 bp fragment of the avian MC1R gene using the following primers modified from Mundy et al. (2004): lcorMSHR9—5′CTGGCTCCGGAAGGCRTAGAT 3′ and lcorMSHR72—5′AYGCCAGYGAGGGCAACCA3′. This fragment, which corresponds to bases 70–880 of the chicken MC1R gene (Kerje et al. 2003), includes all sites previously shown to be associated with melanic phenotypes in birds. We performed polymerase chain reactions (PCRs) in 25 μl volumes, with 0.1 μl AmpliTaq DNA polymerase (Applied Biosystems, Foster City, California), 2.5 μl 10X Tris buffer with MgCl2 (Applied Biosystems), 1.5 μl dNTPs (each dNTP 150 μM), 1.5 μl (600 nM) of each primer and 2.5 μl (approx. 50 ng) of template DNA using a PTC-200 thermal cycler (MJ Research, Waltham, Massachusettes). PCR thermocycling conditions were: (i) an initial denaturing step of 94 °C for 2 min; (ii) 35 cycles of the following: 30 s at 94 °C, 45 s at 62.5 °C and 90 s at 72 °C; and (iii) a 5 min extension step at 72 °C. After purifying amplicons with PEG precipitation, we performed sequencing reactions using an ABI Prism cycle sequencing kit (Applied Biosystems) with the PCR primers and the internal sequencing primers: lcorMSHR73—5′SGCRTAGAAGATGGTGATGTAGC 3′ and lcorMSHR74—5′GTBGACCGCTACATCACCAT 3′. We purified sequencing products using AutoSeq G-50 (Amersham, Uppsala, Sweden) spin columns and visualized them using an ABI 3100 Genetic Analyzer. All sequence data generated in this study were deposited in GenBank (accession numbers DQ388308–DQ388331).
(c) Genetic analyses
We aligned sequences from both strands by eye using SEQUENCHER, v. 4.1 (Genecodes, Ann Arbor, Michigan). In the chromatograms of three individuals (table 2), we noted double peaks at single sites that were approximately half the height of neighbouring peaks. We coded individuals as heterozygous if such double peaks were noted in both strands (MacDougall-Shackleton et al. 2003). We aligned Lepidothrix MC1R sequences with G. gallus (GenBank—AY220305), C. flaveola (GenBank—AF362605 and AF362598), A. caerulescens (AY521209 and AY521182) and S. parasiticus (GenBank—AY521214 and AY521217) MC1R sequences to confirm identity of the amplicons and to translate nucleotide sequences to amino acid sequences. Alignment with these other avian sequences allowed us to examine sequences for indels and stop codons indicative of pseudogenes, neither of which was observed. This comparative analysis also allowed for the examination of variation at nucleotide sites that were previously shown to be associated with melanic plumage in other birds.
Table 2.
MC1R sequence polymorphism data for 23 L. coronata individuals. Asterisks indicate agreement with the consensus sequence. All variation among the four L. coronata alleles is synonymous with respect to the amino acid sequence. Heterozygous sites are denoted with standard IUPAC one-letter codes (W=A/T). Genotype refers to allelic combinations. Sites highlighted in grey are putatively functional nucleotide sites in other avian taxa. Substitutions at site 253 are associated with plumage polymorphism in snow geese (A. caerulescens) (Mundy et al. 2004). Substitutions at site 274 are associated with plumage polymorphism in bananaquits (Coereba flaveola) (Theron et al. 2001). Substitutions at site 689 are associated with plumage polymorphism in Arctic skuas (S. parasiticus; Mundy et al. 2004). All L. coronata individuals are monomorphic at these three sites. Nucleotide sites are numbered with reference to the chicken MC1R sequence. Lepidothrix serena is included as an outgroup.
| nucleotide site | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 2 | 2 | 3 | 4 | 6 | 6 | ||||||
| 7 | 9 | 0 | 5 | 7 | 5 | 1 | 7 | 8 | ||||
| voucher number | 8 | 1 | 4 | 3 | 4 | 4 | 1 | 8 | 9 | plumage | genotype | genbank accession number |
| consensus | C | G | C | G | G | C | A | C | A | |||
| B-28461 | T | * | T | * | * | * | * | * | * | black | 1/1 | DQ388308 |
| B-16077 | T | * | T | * | * | * | * | * | * | black | 1/1 | DQ388309 |
| RWD17053 | * | * | * | * | * | * | * | * | * | black | 2/2 | DQ388310 |
| B-3105 | * | * | * | * | * | * | * | * | * | black | 2/2 | DQ388313 |
| B-4657 | * | * | * | * | * | * | * | * | * | black | 2/2 | DQ388314 |
| B-4545 | * | * | * | * | * | * | * | * | * | black | 2/2 | DQ388315 |
| B-10492 | * | * | * | * | * | * | * | * | * | intermediate | 2/2 | DQ388316 |
| B-27578 | * | * | * | * | * | * | * | * | * | intermediate | 2/2 | DQ388318 |
| B-27832 | * | * | * | * | * | * | * | * | * | intermediate | 2/2 | DQ388319 |
| B-5394 | * | * | * | * | * | * | * | * | * | green | 2/2 | DQ388311 |
| B-5487 | * | * | * | * | * | * | * | * | * | green | 2/2 | DQ388312 |
| B-40543 | * | * | * | * | * | * | * | * | * | green | 2/2 | DQ388317 |
| B-901 | * | * | * | * | * | * | * | * | * | green | 2/2 | DQ388320 |
| B-433693 | * | * | * | * | * | * | * | * | * | green | 2/2 | DQ388321 |
| B-433692 | * | * | * | * | * | * | * | * | * | green | 2/2 | DQ388322 |
| B-433689 | * | * | * | * | * | * | * | * | * | green | 2/2 | DQ388323 |
| B-433691 | * | * | * | * | * | * | W | * | * | green | 2/3 | DQ388324 |
| B-433694 | * | * | * | * | * | * | W | * | * | green | 2/3 | DQ388325 |
| B-9269 | * | * | * | * | * | * | W | * | * | green | 2/3 | DQ388326 |
| B-9306 | * | * | * | * | * | T | T | * | * | green | 4/4 | DQ388327 |
| B-31333 | * | * | * | * | * | * | T | * | * | green | 3/3 | DQ388328 |
| B-9125 | * | * | * | * | * | * | T | * | * | green | 3/3 | DQ388329 |
| B-9177 | * | * | * | * | * | * | T | * | * | green | 3/3 | DQ388330 |
| L. serena | T | A | * | * | * | * | * | T | * | n.a | n.a | DQ388331 |
We tested for evidence of selection at MC1R by calculating Tajima's D statistic (Tajima 1989) using the program DnaSP 3.53 (Rozas & Rozas 1999).
3. Results
(a) MC1R sequence variation and its association with male plumage colour
A total of 810 bp of the MC1R gene was sequenced for 23 L. coronata individuals corresponding to nucleotide positions 70–880 of the G. gallus MC1R gene (Kerje et al. 2003). Four sites were variable within L. coronata, resulting in a total of four MC1R alleles, none of which appeared to be recombinants based on the four gamete test (Hudson & Kaplan 1985; table 2). All the substitutions were synonymous and none showed any association to the two plumage colour groups (table 2). Thus, we found no evidence that sequence variation at MC1R is associated with plumage colour variation in L. coronata.
Sequence variation at MC1R was largely consistent with phylogeographic patterns inferred from mtDNA sequence variation (Cheviron et al. 2005). Two synapomorphic nucleotide sites (78 and 204) support the split between the cis-Andean (east of the Andes) and trans-Andean (west of the Andes) lineages, which is the basal split inferred from mtDNA (Cheviron et al. 2005). One site (411) varies across the plumage zone of intergradation, and the transition is largely concordant with the southern edge of the zone of intergradation. Variation at this site, however, is not associated with plumage colour as we sampled pure green individuals that were homozygous for each allele (table 2).
(b) Nature of selection at MC1R in L. coronata
Values of Tajima's D did not differ significantly from zero (−0.4168, p>0.10), thus we failed to reject a null hypothesis of neutral variation. However, we noted that since only a few variable sites were observed, our statistical power to detect selection was low.
4. Discussion
Despite its apparent role in plumage and pelage melanism in several other vertebrate taxa (reviewed in Mundy 2005), we found no evidence that sequence variation at MC1R is associated with male plumage variation in L. coronata. We did not detect any non-synonymous differences among MC1R sequences of green and black plumaged birds, and the synonymous variation at MC1R we did uncover showed no association with plumage colour (table 2). In snow geese (A. caerulescens) and bananaquits (C. flaveola), single non-synonymous substitutions Val85→Met85 and Glu92→Lys92, respectively, are associated with melanic plumage (Theron et al. 2001; Mundy et al. 2004). All of the blue-crowned manakins sampled here were monomorphic for the amino acids associated with non-melanistic plumage in these taxa (Val and Glu; table 2), demonstrating that genes other than MC1R underlie variation in plumage melanism in L. coronata. The pattern was similar at a third putatively functional MC1R site. In Arctic skuas, a non-synonymous substitution (Arg230→His230) is associated with melanic plumage (Mundy et al. 2004). Again, all of the manakins screened here were monomorphic at this site, but in this case, the amino acid associated with melanistic plumage, histidine, was fixed. This result is particularly noteworthy because Mundy et al. (2004) suggested the Arg230→His230 substitution was significant because a histidine at the homologous site in rock pocket mice (Chaetodipus intermedius) is also associated with melanistic pelage (Hoekstra & Nachman 2003; Nachman et al. 2003). These data show clearly that a histidine at this amino acid position does not confer melanic plumage in all avian genetic backgrounds.
To our knowledge, this is only the second documented case of a lack of a relationship between melanic plumage and MC1R sequence variation. These data, in conjunction with the negative results reported in Phylloscopus warblers (MacDougall-Shackleton et al. 2003), demonstrate that in some cases, genes other than MC1R underlie melanic plumage polymorphism, suggesting that variation exists in the genetic basis of melanistic plumage traits. Indeed, such variation has already been demonstrated in rock pocket mice, within which melanic pelage is correlated with MC1R variation in some populations, but not in others (Hoekstra & Nachman 2003).
Since we did not sequence the first 23 and last 20 codons of MC1R, we cannot rule out entirely the presence of functionally important variation in these regions. However, the gene portion sequenced here contained all of the sites previously shown to be functionally important in birds, all of which lie in trans-membrane or cytoplasmic domains (Theron et al. 2001; Kerje et al. 2003; Mundy et al. 2004; Mundy 2005). The unsequenced regions are not parts of these domains, and we are unaware of any studies that have reported functionally important sites in these unsequenced regions in any taxon. Thus, it is unlikely that functionally important variation would exist in these regions.
First-year male L. coronata are green, resembling females, and probably acquire their definitive adult plumage during their second year. This ontogenetic pattern suggests a regulatory gene may influence adult male plumage polymorphism. Moreover, the diversity of intermediate plumage types within the zone of intergradation suggests that male plumage colour is a polygenic trait. Together, these observations indicate the potential roles of molecular genetic and development pathways in the control of plumage trait variation in this taxon. Many avian species exhibit similar ontonogenic patterns with males developing definitive plumage later in life. Since adult plumage is the target of sexual selection in many species, further studies of taxa with delayed plumage maturation are needed.
Over 100 loci are known to influence levels of pigmentation in vertebrates (Smyth 1990; Bennett & Lamoreux 2003). Future studies exploring other candidate genes, especially those with regulatory functions are likely to provide great insight into our understanding of the evolution of avian plumage colour.
Acknowledgments
We wish to thank the following institutions for loaning tissue samples and specimens used in this study: the American Museum of Natural History, The Field Museum of Natural History, the Louisiana State University Museum of Natural Science. Constructive criticisms from Matthew D. Carling, Ellen D. Ketterson, and three anonymous reviewers greatly improved the manuscript. Funding for this project was provided by the Frank M. Champman Memorial Fund of the American Museum of Natural History (Z.A.C.) and NSF grants DEB-0332093 and DBI-0400797 (R.T.B.).
References
- Andersson L. Melanocortin variants with phenotypic effects in horse, pig, and chicken. Ann. N. Y. Acad. Sci. 2003;994:313–318. doi: 10.1111/j.1749-6632.2003.tb03195.x. [DOI] [PubMed] [Google Scholar]
- Bennett D.C, Lamoreux M.L. The color loci of mice—a genetic century. Pigment Cell Res. 2003;16:333–344. doi: 10.1034/j.1600-0749.2003.00067.x. 10.1034/j.1600-0749.2003.00067.x [DOI] [PubMed] [Google Scholar]
- Brumfield R.T, Jernigan R.W, McDonald D.B, Braun M.J. Evolutionary implications of divergent clines in an avian (Manacus; Aves) hybrid zone. Evolution. 2001;55:2070–2087. doi: 10.1111/j.0014-3820.2001.tb01322.x. [DOI] [PubMed] [Google Scholar]
- Cheviron Z.A, Hackett S.J, Capparella A.P. Complex evolutionary history of a Neotropical lowland forest bird (Lepidothrix coronata) and its implications for historical hypotheses of the origin of Neotropical avian diversity. Mol. Phylogenet. Evol. 2005;36:336–357. doi: 10.1016/j.ympev.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Doucet S.M, Shawkey M.D, Rathburn M.K, Mays H.L, Montgomerie R. Concordant evolution of plumage colour, feather microstructure and a melanocortin receptor gene between mainland and island populations of a fairy-wren. Proc. R. Soc. B. 2004;271:1663–1670. doi: 10.1098/rspb.2004.2779. 10.1098/rspb.2004.2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eizirik E, Yuhki N, Johnson W.E, Menotti-Raymond M, Hannah S.S, O'Brien S.J. Molecular genetics and evolution of melanism in the cat family. Curr. Biol. 2003;13:448–453. doi: 10.1016/s0960-9822(03)00128-3. 10.1016/S0960-9822(03)00128-3 [DOI] [PubMed] [Google Scholar]
- Haffer J. Speciation in some Amazonian forest birds. J. Ornithol. 1970;111:285–331. 10.1007/BF01653396 [Google Scholar]
- Hoekstra H.E, Nachman M.W. Different genes underlie adaptive melanism in different populations of rock pocket mice. Mol. Ecol. 2003;12:1185–1194. doi: 10.1046/j.1365-294x.2003.01788.x. 10.1046/j.1365-294X.2003.01788.x [DOI] [PubMed] [Google Scholar]
- Hoekstra H.E, Drumm K.E, Nachman M.W. Ecological genetics of adaptive color polymorphism in pocket mice: geographic variation in selected and neutral genes. Evolution. 2004;58:1329–1341. doi: 10.1111/j.0014-3820.2004.tb01711.x. [DOI] [PubMed] [Google Scholar]
- Hudson R.R, Kaplan N.L. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerje S, Lind J, Schutz K, Jensen P, Andersson L. Melanocortin 1-receptor (MC1R) mutations are associated with plumage colour in chicken. Anim. Genet. 2003;34:241–248. doi: 10.1046/j.1365-2052.2003.00991.x. 10.1046/j.1365-2052.2003.00991.x [DOI] [PubMed] [Google Scholar]
- Kijas J.M.H, Wales R, Tornsten A, Chardon P, Moller M, Andersson L. Melanocortin receptor 1 (MC1R) mutations and coat color in pigs. Genetics. 1998;150:1177–1185. doi: 10.1093/genetics/150.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall-Shackleton E.A, Blanchard L, Gibbs H.L. Unmelanized plumage patterns in old world leaf warblers do not correspond to sequence variation at the melanocortin-1 receptor locus (MC1R) Mol. Biol. Evol. 2003;20:1675–1681. doi: 10.1093/molbev/msg186. 10.1093/molbev/msg186 [DOI] [PubMed] [Google Scholar]
- Mundy N.I. A window on the genetics of evolution: MC1R and plumage colouration in birds. Proc. R. Soc. B. 2005;272:1633–1640. doi: 10.1098/rspb.2005.3107. 10.1098/rspb.2005.3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy N.I, Badcock N.S, Hart T, Scribner K, Janssen K, Nadeau N.J. Conserved genetic basis of a quantitative plumage trait involved in mate choice. Science. 2004;303:1870–1873. doi: 10.1126/science.1093834. 10.1126/science.1093834 [DOI] [PubMed] [Google Scholar]
- Nachman M.W, Hoekstra H.E, D'Agostino S.L. The genetic basis of adaptive melanism in pocket mice. Proc. Natl Acad. Sci. 2003;100:5268–5273. doi: 10.1073/pnas.0431157100. 10.1073/pnas.0431157100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omland K.E, Lanyon S.E. Reconstructing plumage evolution in orioles (Icterus): repeated convergence and reversal in patterns. Evolution. 2000;54:2119–2133. doi: 10.1111/j.0014-3820.2000.tb01254.x. [DOI] [PubMed] [Google Scholar]
- Price T.D. Sexual selection and natural selection in bird speciation. Phil. Trans. R. Soc. B. 1998;353:251–260. 10.1098/rstb.1998.0207 [Google Scholar]
- Rosenblum E.B, Hoekstra H.E, Nachman M.W. Adaptive reptile color variation and the evolution of the MC1R gene. Evolution. 2004;58:1794–1808. doi: 10.1111/j.0014-3820.2004.tb00462.x. [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSp version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Smyth J.R. Genetics of plumage, skin, and pigmentation in chickens. In: Crawford R.D, editor. Poultry breeding and genetics. Elsevier Science; New York, NY: 1990. [Google Scholar]
- Snow D. Family Pipridae (manakins) In: del Hoyo J, Elliot A, Christie D, editors. Handbook of Birds of the World: contigas to pipits and wagtails. vol. 9. Lynx Edicions; Barcelona, Spain: 2004. pp. 110–169. [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi S, Suzuki H, Yabuuchi M, Takahashi S. A possible involvement of the melanocortin-1 receptor in regulating feather pigmentation in the chicken. Biochim. Biophys. Acta. 1996;1308:164–168. doi: 10.1016/0167-4781(96)00100-5. [DOI] [PubMed] [Google Scholar]
- Theron E, Hawkins K, Bermingham E, Ricklefs R.E, Mundy N.I. The molecular basis of an avian plumage polymorphism in the wild: a melanocortin-1-receptor point mutation is perfectly associated with the melanic plumage morph of the bananaquit, Coereba flaveola. Curr. Biol. 2001;11:550–557. doi: 10.1016/s0960-9822(01)00158-0. 10.1016/S0960-9822(01)00158-0 [DOI] [PubMed] [Google Scholar]