Abstract
Metal ions are critical for the proper folding of RNA, and the GAAA tetraloop–receptor is necessary for the optimal folding and function of many RNAs. We have used NMR to investigate the role of metal ions in the structure of the tetraloop–receptor in solution. The NMR data indicate native tertiary structure is formed under a wide range of ionic conditions. The lack of conformational adaptation in response to very different ionic conditions argues against a structural role for divalent ions. Nuclear Overhauser effects to cobalt hexammine and paramagnetic relaxation enhancement induced by manganese ions were used to determine the NMR structures of the tetraloop receptor in association with metal ions, providing the first atomic-level view of these interactions in the solution state. Five manganese and two cobalt hexammine ions could be localized to the RNA surface. The locations of the associated metal ions are similar, but not identical to, those of previously determined crystal structures. The sites of association are in general agreement with nonlinear Poisson–Boltzmann calculations of the electrostatic surface, emphasizing the general importance of diffusely associated ions in RNA tertiary structure.
Keywords: NMR, RNA tertiary structure, GAAA tetraloop, 11-nucleotide receptor, metal ions
INTRODUCTION
RNA plays a central role in gene expression (Gesteland et al. 2006). The many unique properties of RNA derive from its ability to fold into complex three-dimensional structures, mediated by specific RNA tertiary interactions. However, the polyanionic nature of RNA results in long-range electrostatic forces that, in the absence of ions, would be counteractive to RNA tertiary structure formation. The importance of ion–RNA interactions for RNA tertiary folding have long been recognized (Stein and Crothers 1976a,b; Leroy and Gueron 1977; Leroy et al. 1977; Draper 2004).
Loop–receptor interactions are necessary for proper folding and catalysis of a number of RNAs (Jaeger et al. 1994; Murphy and Cech 1994; Pley et al. 1994; Costa and Michel 1995, 1997; Tanner and Cech 1995; Khvorova et al. 2003). The most well-studied and ubiquitous loop–receptor interaction is an A-minor interaction between the GAAA tetraloop and a conserved 11-nucleotide receptor found in Group I and II introns and RNase P (Jaeger et al. 1994; Murphy and Cech 1994; Costa and Michel 1995; Tanner and Cech 1995; Cate et al. 1996; Costa et al. 1997). In all tetraloop–receptor structures solved to date, the GAAA tetraloop binds by stacking upon an adenosine platform and hydrogen bonding to a complementary minor groove pocket within the receptor. We recently determined the NMR structure of the GAAA tetraloop–receptor interaction (Davis et al. 2005). The NMR structure was determined in 9 mM MgCl2, and was facilitated by a rationally designed homodimeric construct, which results in a complex that tightly associates through the coupling of two loop–receptor interactions (Fig. 1; Jaeger et al. 2001; Davis et al. 2005). Within experimental error, there is no detectable difference between the NMR structure of the homodimeric tetraloop receptor and the structures of tetraloop–receptor interactions within other larger RNA systems studied by crystallography (Cate et al. 1996; Golden et al. 1998; Juneau et al. 2001; Adams et al. 2004a,b). Therefore, the homodimeric tetraloop–receptor construct recapitulates the native interaction observed in many RNA structures and provides a tractable system for the study of RNA tertiary structure by NMR. Solution NMR provides a means to investigate the RNA structure under a wide range of ionic conditions, which may not be possible with crystallographic approaches.
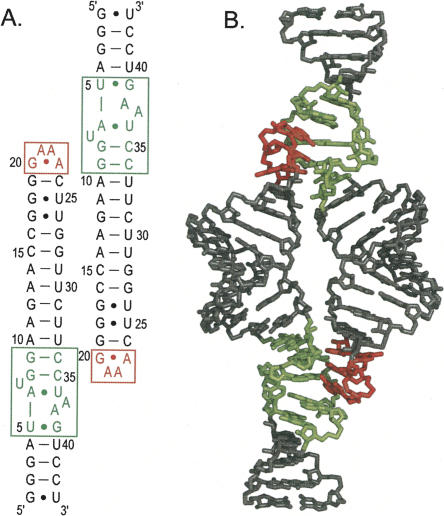
FIGURE 1.
Sequence and structure of the tetraloop–receptor RNA construct used in these studies. (A) Secondary structure. (B) Previously determined three-dimensional structure (PDB ID 2ADT). The tetraloop is highlighted in red and the receptor in green.
Metal ions are important for the tetraloop–receptor interaction, but it is not clear as to whether ions play an integral structural role or are just required for electrostatic shielding. Previous NMR data were obtained in the presence of 9 mM magnesium, and no complex formation was detected at low concentration (<100 mM) monavalent ions. X-ray crystallography of group I introns has revealed three metal ions associated with the bound tetraloop–receptor complex, including: (1) a magnesium directly coordinated to the G250 phosphate oxygen within the receptor (Cate et al. 1996; Juneau et al. 2001; Adams et al. 2004a,b); (2) a monovalent ion coordinated to the guanosine immediately below the adenosine platform nucleotides (Cate et al. 1996; Basu et al. 1998), and (3) a cobalt hexammine binding site at the tandem G–U wobble pairs in the major groove of the tetraloop stem (Cate et al. 1996; Juneau et al. 2001). EPR studies have revealed a magnesium dependence for tetraloop docking to the receptor (Qin et al. 2005). Recent fluorescence resonance energy transfer (FRET) experiments show a broad metal ion dependence for tetraloop docking, with charge of the ion being the most important factor for determining the concentration of ion required for tetraloop docking (Downey et al. 2006).
Metal ion associations with RNA are difficult to monitor in solution. Magnesium and potassium, the dominant counterions for RNA folding in vivo, are unfortunately spectroscopically invisible. NMR chemical shift perturbation mapping is a sensitive, but indirect, means of detecting ion association with RNA (Butcher et al. 2000; Huppler et al. 2002; Sigel et al. 2004; Fan et al. 2005), and can be difficult to interpret if ion binding simultaneously induces conformational change. Direct methods for detecting sites of metal ion association include Nuclear Overhauser effects (NOEs) to cobalt hexammine, and manganese-induced paramagnetic relaxation enhancement (PRE) (Bertini and Luchinat 1986; Kieft and Tinoco 1997; Gonzalez and Tinoco 1999; Butcher et al. 2000). There have only been a few instances where ions have been localized to an RNA structure using NMR (Kieft and Tinoco 1997; Gonzalez and Tinoco 1999; Butcher et al. 2000). To our knowledge, the tetraloop receptor is the only RNA helical packing motif that has been solved independently by both X-ray crystallography and NMR, and there have been no atomic-level comparisons of metal ion binding to RNA tertiary structures in the solution and crystalline states.
We wished to investigate the role of metal ions in tetraloop–receptor-mediated tertiary structure formation in solution, and sought to determine the NMR structure of the tetraloop–receptor complex with associated ions. Our results indicate that the tetraloop receptor forms a native complex under a wide variety of ionic conditions with no detectable conformational adaptation. This is the first study to investigate the ion dependence of the tetraloop–receptor interaction at the atomic level, and the results indicate that general electrostatic shielding is sufficient for native complex formation. We report the NMR structures of the tetraloop–receptor complex in the presence of (1) magnesium and cobalt hexammine and (2) magnesium and manganese. There are two distinct areas of cobalt hexammine association and five regions of manganese association. The ions localize to pockets within the highly electronegative major groove, in general agreement with nonlinear Poisson–Boltzmann calculations of the solvent-accessible electrostatic surface. Comparisons with previously solved crystal structures reveal that the two methods give highly similar, but nonidentical views of metal ion association with RNA.
RESULTS
Ionic dependence of the tetraloop–receptor interaction
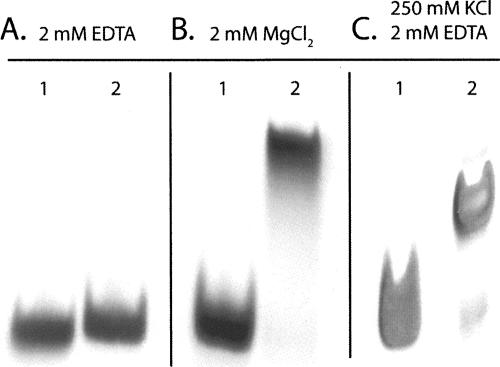
Electrophoretic mobility shift assays indicate that the homodimeric tetraloop–receptor complex forms in the presence and absence of magnesium (Fig. 2). In the presence of 250 mM KCl and 2 mM EDTA, the relative mobility of the GAAA tetraloop–receptor RNA is consistent with formation of the homodimer. These data are also in agreement with recent FRET results of a tethered tetraloop–receptor complex that docks in the presence of potassium (K 1/2 = 200 mM) without divalent cations (Downey et al. 2006). Since crystal structures have revealed several ions closely associated with the tetraloop receptor (Cate and Doudna 1996; Cate et al. 1996; Basu et al. 1998; Golden et al. 1998; Adams et al. 2004a,b), it is plausible that structural adaptation may accompany complex formation in the absence of magnesium.
FIGURE 2.
GAAA tetraloop–receptor complex formation analyzed by nondenaturing gel electrophoresis. Lane 1: A21U, A22G double mutant (monomeric) RNA. Lane 2: GAAA tetraloop–receptor RNA. Gels conditions: 90 mM Tris-borate (pH 7.6) and (A) 2 mM EDTA, (B) 2 mM MgCl2, and (C) 250 mM KCl and 2 mM EDTA. The relative mobilities of lanes 1 and 2 within each panel can be compared directly, but the relative mobilities in A, B, and C cannot be directly compared due to the different electrophoretic conditions.
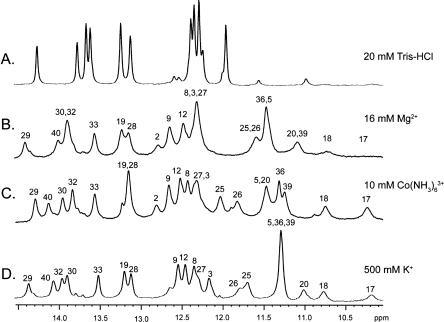
The tetraloop–receptor interaction can be efficiently monitored by NMR, using a homodimeric complex (Fig. 1; Davis et al. 2005). We investigated the ionic dependence of this interaction by monitoring the imino proton chemical shifts as a function of added potassium, magnesium, and cobalt hexammine (Fig. 3). The resonance assignments of the imino peaks were confirmed by 2D nuclear Overhauser effect spectroscopy [(NOESY); data not shown]. In low ionic strength (20 mM) Tris–HCl buffer, the RNA is monomeric with no evidence of tetraloop–receptor formation (Fig. 3A). Nearly complete complex formation was observed upon direct addition of 16 mM magnesium or 10 mM cobalt hexammine to 1 mM RNA samples (Fig. 3B,C; note that millimolar concentrations of multivalent ions are limiting when directly added to RNA that is also millimolar in concentration). NMR spectra identical to Figure 3B were also obtained by dialyzing the 1 mM RNA sample against an excess (1 L) of 2 mM magnesium (conditions corresponding to those in Fig. 2B; data not shown). Additionally, dialyzing the RNA against 500 mM KCl and 2 mM EDTA resulted in complete complex formation (Fig. 3D). Nearly complete complex formation was also observed after dialysis against 250 mM KCl and 2 mM EDTA (corresponding to electrophoretic conditions in Fig. 2C; data not shown). These data are consistent with FRET studies that indicate the tetraloop–receptor interaction can form in potassium or cobalt hexammine (Downey et al. 2006).
FIGURE 3.
GAAA tetraloop–receptor complex formation as followed by 1D NMR. 1D 1H NMR spectra of the GAAA tetraloop–receptor RNA in: (A) 20 mM Tris-HCl (pH 7.0), (B) 16 mM MgCl2 and 20 mM Tris-HCl (pH 7.0), (C) 10 mM Co(NH3)6Cl3, 2 mM EDTA, and 20 mM Tris-HCl (pH 7.0), and (D) 500 mM KCl, 2 mM EDTA, and 20 mM Tris-HCl (pH 7.0). Base identity of imino proton resonances is labeled above the peaks.
While the above data indicate the tetraloop–receptor interaction forms under a variety of ionic conditions, there are interesting differences in the 1D NMR spectra (Fig. 3). For example, the imino peaks of G17 and G18, arising from the tandem G–U wobble pairs adjacent to the tetraloop, are sharp in cobalt hexammine and potassium but are severely line broadened in magnesium (Fig. 3B,C,D). The line broadening can be attributed to an increased proton exchange rate catalyzed by the primary amine of Tris, since it is not observed in the absence of Tris buffer (Davis et al. 2005). The observation that Tris buffer does not increase the exchange rate of the G17 and G18 imino protons in the presence of cobalt hexammine and potassium suggests that the tandem G–U wobbles are either selectively stabilized, less solvent accessible, or both, when these ions are present. Indeed, tandem G–U wobble pairs have been previously observed to bind potassium and cobalt hexammine but not magnesium (Fan et al. 2005). A cobalt hexammine molecule was observed to bind at this position in crystal structures (Cate and Doudna 1996; Cate et al. 1996). Therefore, the NMR data are consistent with selective binding of cobalt hexammine and potassium, but not magnesium, to the tandem G–U wobble pairs in the tetraloop–receptor complex. Additionally, significant chemical shift changes are associated with the different ionic conditions. Since chemical shifts are highly sensitive reporters of the local chemical environment, the observed changes could be due to differences in metal ion localization, or to subtle changes in structure.
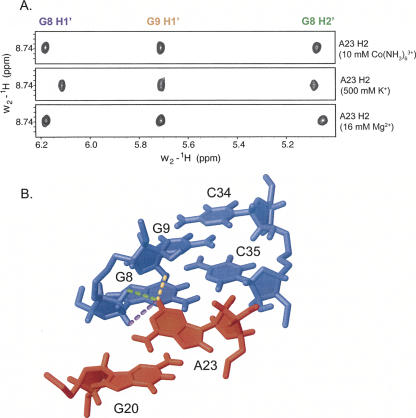
The intensity of the NOE is a very sensitive reporter of structural change, owing to its strong distance dependence (r−6). Intermolecular NOEs between the tetraloop and receptor were analyzed for the different ionic conditions. All resolved intermolecular NOEs observed previously (Davis et al. 2005) were observed in the presence of cobalt hexammine and potassium, including 17 out of 19 assigned intermolecular NOEs in the presence of cobalt hexammine, and 16 out of 19 assigned intermolecular NOEs observed in the presence of potassium (unassigned intermolecular NOEs were in regions of spectral overlap). We could find no significant difference in the intensities or integrated volumes of the intermolecular NOEs, which were essentially identical in each of the ionic conditions. For example, the A23 base from the tetraloop directly contacts the receptor, and identical intermolecular NOEs are observed when the complex is formed in 10 mM cobalt hexammine, 500 mM potassium, or 16 mM magnesium (Fig. 4). The identical NOE intensities are observed despite chemical shift changes. Therefore, we conclude that the chemical shift changes are due entirely to differences in the associated ions, rather than changes in the RNA structure. These data suggest that the RNA interface between the tetraloop and its receptor does not change in response to ionic conditions, within the detectable limit of the NOE data (∼1–2 Å).
FIGURE 4.
Representative intermolecular NOEs observed upon GAAA tetraloop–receptor complex formation. (A) Strips from 2D NOESY spectra showing intermolecular NOEs from the tetraloop to the receptor in presence of 10 mM Co(NH3)6 3+, 500 mM K+, and 16 mM Mg2+. (B) Portion of the tetraloop–receptor complex (PDB ID 2ADT) with the tetraloop shown in red and receptor in blue. Dashed lines represent intermolecular NOEs shown in (A) from A23H2 to: G8H1′ (violet), G9H1′ (orange), and G8H2′ (green).
Structure determination with associated metal ions
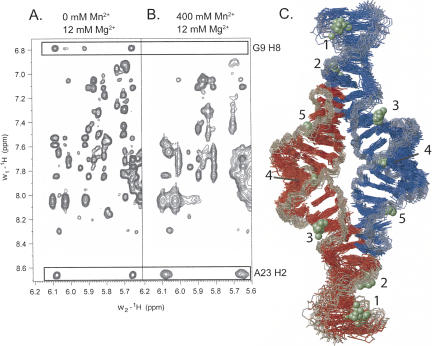
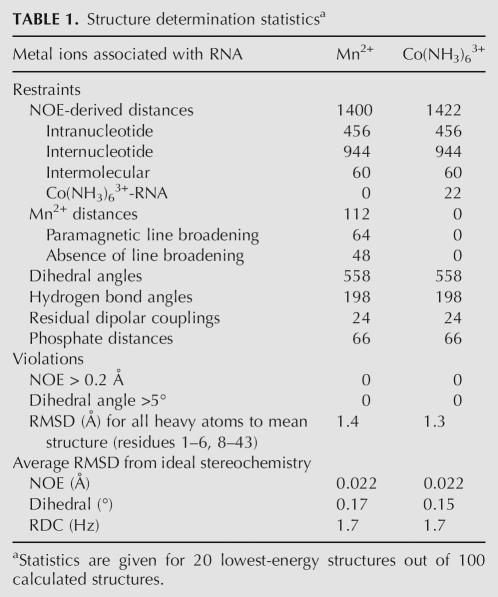
Manganese-induced paramagnetic relaxation enhancement causes line broadening of NMR resonances with a distance dependence of r−6 (Bertini and Luchinat 1986). Therefore, manganese-induced PRE can be translated into distance restraints in order to localize associated manganese ions to an RNA structure (Butcher et al. 2000). Selective line broadening of proton resonances occurs at manganese concentrations in the 100 μM range (Fig. 5A,B). This concentration is less than the RNA concentration, indicating that the manganese ions are in fast exchange on the NMR timescale. Manganese (400 μM) is a 4–10-fold higher concentration than the amount typically required to observe selective line broadening in other RNAs in the absence of divalent ions (Ott et al. 1993; Allain and Varani 1995; Hansen et al. 1999; Butcher et al. 2000), but the manganese ions must compete with 12 mM magnesium in this case. 2D NOESY data were quantified in the presence and absence of manganese, and proton resonances broadened to <75% of their original peak height were scored as line broadened. In this manner, 32 protons per subunit (64 per homodimer) were identified as positions of manganese association and incorporated as distance restraints for structure calculations as described in Materials and Methods. Due in part to the observed fast exchange rates, the ions were initially modeled as hexahydrates Mn(H2O)6 2+.
FIGURE 5.
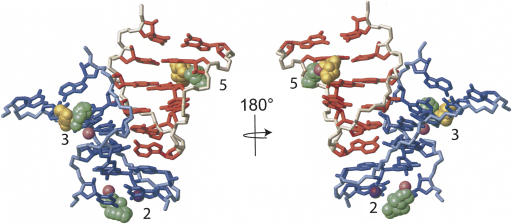
Structure determination of the GAAA tetraloop–receptor RNA with associated manganese ions. 2D NOESY data in the absence (A) and the presence (B) of Mn2+. (A) 0 μM Mn2+, 12 mM MgCl2. (B) 400 μM Mn2+, 12 mM MgCl2. Boxed regions highlight NOEs involving G9 H8 and A23 H2, shown for comparison. The G9 NOEs are scored as selectively broadened, while the A23 NOEs are not. (C) Superimposition of the 20 lowest-energy structures of the GAAA tetraloop–receptor RNA with associated manganese ions. RNA strands are colored in blue and red, with the backbone highlighted in gray. Manganese ions are shown as green spheres. Five regions of manganese ion association per monomer are numbered.
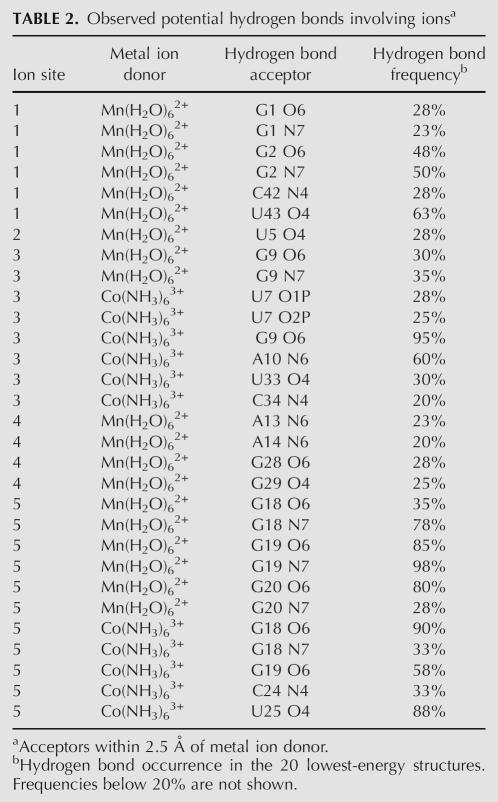
The PRE data can be fully satisfied by a minimum number of five manganese ions (Fig. 5C). All manganese interactions were defined by six to seven PRE restraints (see Table 2 below), which were sufficient to uniquely position the ions with respect to the RNA. All PRE restraints could be fully satisfied with hexahydrated manganese ions. Structure calculations using these restraints converged with an RMSD of 1.4 Å for 20 out of 100 structures calculated (Table 1). The 20 lowest-energy structures are shown (Fig. 5C); note that since there are 20 structures, there are also 20 ions superimposed at each region of manganese association (Fig. 5C). The clustering of the 20 ions on the 20 structures gives a visual indication of the precision with which NMR can be used to locate the ions.
TABLE 2.
Observed potential hydrogen bonds involving ionsa
TABLE 1.
Structure determination statisticsa
The first region of manganese localization is at the 5′-GGG end of the RNA and is a known site of manganese association (Fig. 5C, site 1; Allain and Varani 1995). The second area of manganese association is just below the AA platform in the receptor, in the same region previously shown to bind potassium (Fig. 5C, site 2; Cate et al. 1996; Basu et al. 1998). The third region is also in the receptor at the tandem G–C pairs, and was observed in the crystal structure as a directly coordinated magnesium ion (Fig. 5C, site 3; Cate et al. 1996). The fourth region of manganese association is buried in the middle of the helical region (Fig. 5C, site 4). The fifth region of manganese association is at the tandem G–U wobble pairs (G17–U26, G18–U25) (Fig. 5C, site 5), and corresponds to the same region that binds cobalt hexammine in the crystal structure (Cate et al. 1996) and in the isolated helix in solution (Kieft and Tinoco 1997).
The observed manganese association at the AA platform monovalent ion binding site was somewhat surprising. Therefore, we wished to determine if a potassium ion could competitively inhibit manganese association at this site. The RNA was dialyzed into 140 mM KCl, 2 mM MgCl2, and 20 mM Tris at pH 7.0, and manganese was titrated into the sample to a final concentration of 400 μM. However, no significant change in the degree of line broadening was observed (data not shown). Therefore, we conclude that manganese can effectively compete with potassium for interactions with the AA platform.
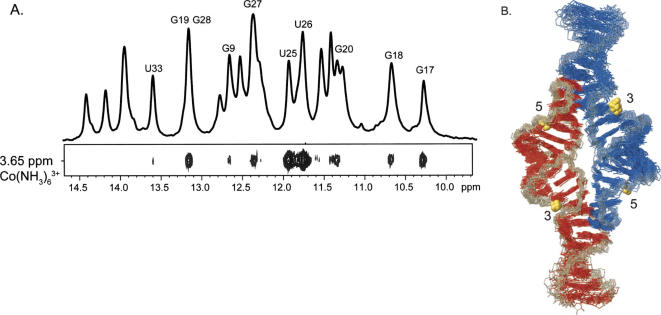
Cobalt hexammine is a trivalent mimic of hexahydrated magnesium, and is useful for probing metal ion sites by NMR (Kieft and Tinoco 1997; Butcher et al. 2000). The amine groups of cobalt hexammine do not exchange, precluding inner sphere coordination. NOEs between the RNA and cobalt hexammine were used to determine the locations of cobalt hexammine association. Upon addition of 2 mM cobalt hexammine, 22 NOEs were observed between cobalt hexammine and the RNA (Fig. 6A; Table 2). Using these NOE restraints, structures were calculated and the 20 lowest-energy structures converged to 1.3 Å (Fig. 6B; Table 1). The cobalt hexammine NOEs define two distinct regions of cobalt hexammine association. The first region is at the tandem G–C pairs in the receptor (Fig. 6B, site 3). An innersphere coordinated magnesium ion was observed to bind to this site by crystallography (Cate et al. 1996). The second region, defined by 18 NOEs, is at the tandem G–U wobbles and has been observed previously by crystallography and NMR (Fig. 6B, site 5; Cate and Doudna 1996; Cate et al. 1996; Kieft and Tinoco 1997). The cobalt hexammine ions are associated in the same regions as manganese ions at sites 3 and 5 (Fig. 5C). The observed interactions between the ions and the bases of the RNA are observed as potential hydrogen bonds between acceptor groups on the bases and the amines or waters surrounding the ions (Table 2).
FIGURE 6.
Structure determination of the GAAA tetraloop–receptor RNA with associated cobalt hexammine ions. (A) Top: 1D NMR spectrum of imino protons. Bottom: section of a 2D NOESY displaying NOEs between Co(NH3)6 3+ and RNA. NOE assignments are indicated above the corresponding peak in the 1D spectrum. (B) Superimposition of the 20 lowest-energy structures of the GAAA tetraloop–receptor RNA with associated cobalt hexammine ions. RNA strands are colored as in Figure 5. Cobalt hexammine ions are shown as gold spheres. Regions of ion binding are numbered relative to the corresponding manganese ion association sites in Figure 5.
DISCUSSION
A comparison of the NMR and crystal structures of the tetraloop–receptor interaction reveals that the three metal ions observed by NMR are in highly similar locations to those observed in the crystal structures (Fig. 7; Cate et al. 1996; Golden et al. 1998; Juneau et al. 2001; Adams et al. 2004a,b). The NMR structure shows both cobalt hexammine and manganese associate at the tandem G–U wobbles (Fig. 7, site 5) in a nearly identical location to the cobalt hexammine or magnesium ions observed in the crystal structures (Cate and Doudna 1996; Juneau et al. 2001). The manganese associated below the AA platform (Fig. 7, site 2) is in the vicinity of a potassium binding site (Basu et al. 1998), but is closer to the electron density observed by Juneau et al. (2001). The NMR data suggest a divalent ion can localize to the AA platform, although the NMR observed manganese ions are on average 6.9 Å away from the potassium binding site. This difference may be due to the very different coordination preferences and geometries of potassium versus manganese. Both manganese and cobalt hexammine associate at the tandem G–C pairs in the receptor, which were previously observed to be directly coordinated by magnesium (Fig. 7, site 3; Cate et al. 1996), indicating that a directly coordinated ion at tandem G–C pairs is not required for tetraloop–receptor formation.
FIGURE 7.
Tetraloop–receptor interaction with metal ions observed by NMR and by X-ray crystallography. Superimposition of the 20 lowest-energy NMR structures (with cobalt hexammine ions and with manganese ions) and the three crystal structures (PDB ID 1GID, 1HR2, and 1U6B) over the tetraloop–receptor interaction. For simplicity, only the RNA from the lowest-energy structure with manganese ions is shown, with the tetraloop region in red and the receptor in blue. Ions from all five structures are displayed, including the superimposition of 20 ions from 20 NMR structures. Metal ion association sites are numbered, corresponding to numbers in Figures 5 and 6. Site 2 contains manganese (green, this study), magnesium (pink, PDB ID 1HR2), and potassium (violet, PDB ID 1U6B). Site 3 contains manganese (green, this study), cobalt hexammine (gold, this study), and magnesium (pink, PDB ID 1GID). Site 5 contains manganese (green, this study), cobalt hexammine (gold, this study), and cobalt hexammine (pink, PDB ID 1GID).
Differences in cobalt hexammine and manganese association can be attributed to the methods used to identify locations of individual metal ions. Manganese-induced PRE is a more sensitive method, since only 500 μM manganese was required to identify five distinct metal ion interaction sites, whereas 2 mM cobalt hexammine identified two sites of preferred ion localization. Increased concentrations of cobalt hexammine detected many nonspecific interactions and resulted in NOEs to every nucleotide on the RNA (data not shown). The overlap between the manganese and cobalt hexammine interaction sites indicates that the type of interacting ion is far less important than general electrostatic shielding, and the electrostatic surface of the molecule plays a dominant role in determining the sites of ion association.
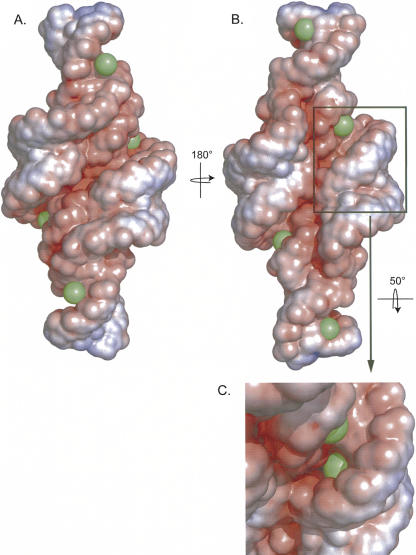
Nonlinear Poisson–Boltzmann calculations of the electrostatic surface show that the regions of the RNA populated with metal ions are the highly electronegative and accessible major grooves (Fig. 8) as expected for diffusely bound ions according to the thermodynamic framework of Misra and Draper (2001) and Misra et al. (2003). Additionally, a highly electronegative region is observed where the backbones of the two helices come in close proximity in the very center of the molecule (Fig. 8A). These backbone resonances are unassigned due to spectral overlap, so we cannot exclude that a metal ion may interact with this region of the molecule as well. A strip of intense electronegative surface is also visible down the center of the molecule, where the minor grooves come in close contact due to the tetraloop–receptor interaction (Fig. 8B). In this case, the resonances are assigned and there is no NOE or PRE evidence for cobalt hexammine or manganese associating with this region of the molecule. Most of the minor groove surface is occupied by the tetraloop–receptor interaction, such that the available electronegative surface of the minor groove is narrow and much smaller than the major groove, and no electron density for ion interactions were observed in the minor groove regions of the tetraloop–receptor crystal structures (Cate and Doudna 1996; Juneau et al. 2001; Adams et al. 2004a,b). Interestingly, a manganese ion is nearly completely buried in the very electronegative center of the major groove, in the middle of the two helices at G28/U29 (Fig. 5C, site 4; Fig. 8C). The RNA sequence in this region derives from the NMR construct and does not correspond to an intron crystal structure.
FIGURE 8.
Electrostatic surface potential of the lowest-energy NMR structure with associated Mn2+. Electrostatic potential of the RNA displayed on the solvent accessible surface with the following scale: red = −9 kT/e, white = −2 kT/e, and blue = 5 kT/e. Manganese ions are shown as green spheres with the radius of a hexahydrated ion. (B) Rotation (180°) of A. (C) Detailed view of the RNA tilted 50° on the x-axis to display the Mn2+ ion bond in the middle of the helix (not visible in other views).
In summary, we demonstrate that it is possible to localize ions to a moderately sized (30 kDa) RNA complex by NMR. It is striking that the ion binding sites observed in solution by NMR correspond quite well to those observed by crystallography. A limitation of the manganese-induced PRE method is the inability to distinguish the hydration state of the ion; however, all of the PRE distance restraint data in this case could be satisfied by fully hydrated manganese ions, and cobalt hexammine binding to sites 3 and 5 provides further support for the ability of fully hydrated ions to bind at these sites. With respect to the RNA structures, there is no difference, within the experimental uncertainty (RMSD = 1.3–1.4 Å, Table 1), between the structures calculated in the presence of manganese and cobalt hexammine. This is also illustrated by the intensities of the intermolecular NOEs between the tetraloop and receptor, which are unchanged regardless of whether the complex is formed in 10 mM cobalt hexammine, 500 mM potassium, or 16 mM magnesium (Fig. 4). These data indicate that the RNA undergoes no detectable conformational adaptation in response to very different ionic conditions. Finally, the structural comparison in Figure 7 suggests that changing the position of the ion is energetically more favorable than altering the RNA structure.
MATERIALS AND METHODS
Sample preparation
RNA was prepared by in vitro transcription from a chemically synthesized DNA oligonucleotide template (Integrated DNA Technologies) using His6-tagged T7 polymerase and was purified as previously described (Huppler et al. 2002; Reiter et al. 2003; Davis et al. 2005). Purified RNA was resuspended into the appropriate dialysis buffer, and dialyzed against 1 L of the same buffer with three buffer exchanges at 12-h intervals. “Metal ion-free” RNA was prepared by extensive dialysis against 2 mM EDTA and 20 mM Tris–HCl, pH 7.0. Final RNA concentrations were between 0.8 and 1.5 mM RNA.
Polyacrylamide gel electrophoresis
RNA samples for nondenaturing PAGE were diluted to 0.2 mM RNA in TBE (90 mM Tris–borate, 2 mM EDTA, pH 7.6), TBE+KCl (TBE plus 250 mM KCl), or TB+MgCl2 (90 mM Tris–borate, pH 7.6, 2 mM MgCl2) and mixed with 10× glycerol dye (50% glycerol 0.25% bromophenol blue 0.25% xylene cyanol). RNA samples were added to 10% (19:1) acrylamide/bisacrylamide nondenaturing electrophoresis gel in TBE, TBE+KCl or TB+MgCl2 buffer. Gels were run at 4°C at 14 watts for 4–6 h. RNA was visualized using UV absorption and stained using toluodine blue. A previously described monomeric A21U, A22G double mutant RNA, was used as a reference (Davis et al. 2005).
NMR spectroscopy
NMR spectra were collected at the National Magnetic Resonance Facility at Madison (NMRFAM) on Bruker 600 and 750 MHz instruments and Varian 600, 800, and 900 MHz instruments. Spectrometers are equipped with either cryogenic single Z-axis gradient probes or room temperature triple axis HCN probes. NMR data were analyzed using XWINNMR 2.6 software (Bruker Biospin) and NMRpipe (http://spin.niddk.nih.gov/bax/software/NMRPipe; Delaglio et al. 1995). Resonance assignments and spectral comparisons were completed using Sparky (http://www.cgl.ucsf.edu/home/sparky) and structures were analyzed using MOLMOL (Koradi et al. 1996). Nonlinear Poisson–Boltzmann calculations of the electrostatic surface potentials were calculated using QNIFFT (Chin et al. 1999) and visualized using the APBS plugin in PyMOL (http://www.delanoscientific.com; Baker et al. 2001).
Exchangeable proton resonances were assigned by analysis of 2D NOESY (150 msec mixing time), 2D 1H-15N HMQC, and 2D 1H-15N HSQC spectra in 90% H2O and 10% D2O at 10°C, and compared to previously assigned chemical shifts and NOEs (Davis et al. 2005). Nonexchangeable proton resonances were assigned by reference to 2D NOESY (25, 50, 100, 150, and 200 msec mixing time), TOCSY, 2D 1H-13C HSQC, 3D 1H-13C-1H HCCH COSY, and 3D NOESY-HMQC spectra in 99.99% D2O at 30°C as previously described (Davis et al. 2005). Water suppression for samples in 90% H2O was achieved by either a 1–1 spin echo pulse sequence (Bax et al. 1987) or a double pulse field gradient spin-echo sequence (Hwang and Shaka 1995; Davlvit 1998).
Complex formation by addition of metal ions was followed by analysis of imino proton chemical shifts using 1D 1H NMR. MgCl2, KCl, NaCl, and Co(NH3)6Cl3 were titrated into RNA samples treated with 2 mM EDTA and dialyzed extensively against 20 mM Tris–HCl (pH 7.0). 1 M MgCl2, 3 M KCl, 3M NaCl, and 125 mM Co(NH3)6Cl3 were titrated into 1 mM RNA until >95% of the RNA was in dimer form, with final metal ion concentrations of 16 mM MgCl2, 500 mM KCl, and 10 mM Co(NH3)6Cl3. For the 500 mM KCl sample, the RNA was dialyzed against 500 mM KCl, 2 mM EDTA, and 20 mM Tris–HCl (pH 7.0).
Manganese-induced paramagnetic relaxation enhancement and NOEs to cobalt hexammine were used to identify locations of metal ion association. MnCl2 was added stepwise to 1 mM RNA in 12 mM MgCl2, corresponding to final Mn2+ concentrations of: 10 μM, 25 μM, 50 μM, 100 μM, 200 μM, and 500 μM. Manganese-induced PRE was followed using 1D 1H NMR, 2D 1H NOESY and 2D 1H TOCSY. An additional Mn2+ titration was performed with the RNA complex equilibrated with 140 mM KCl, 2 mM MgCl2, and 20 mM Tris–HCl (pH 7.0). 125 mM Co(NH3)6Cl3 was added stepwise to 1 mM RNA containing 10 mM MgCl2 until the final concentration of Co(NH3)6Cl3 was 2 mM. NOEs between Co(NH3)6 3+ and the RNA were obtained by 2D 1H-1H NOESY in 90% H2O.
RDCs were measured using XWINNNMR (Bruker) software by calculating the difference between the one-bond 1H-15N couplings for isotropic and partially aligned samples (Davis et al. 2005). PALES software (http//:spin.niddk.nih.gov/bax/software/PALES; Zweckstetter and Bax 2000) was used to estimate the value for the axial (Da) and the rhombic (R) components of the alignment tensor. An extensive grid search was then used to determine optimal Da and R values (Clore et al. 1998). Da and R values of 35 Hz and 0.6, respectively, were used for both the manganese and cobalt hexammine containing structure calculations.
Structure calculations
Interproton NOE distances were estimated from the integrated peak volumes obtained from the 2D NOESY data. NOEs were analyzed from nonexchangeable 2D NOESY spectra at 25, 50, 100, and 200 msec mixing times. NOE peak volumes were measured using Sparky (http://www.cgl.ucsf.edu/home/sparky), and distances were estimated by comparing the peak volumes to the average pyrimidine H5–H6 NOE volume corresponding to a fixed distance of 2.4 Å. Distances were grouped into three classes: strong (1.8–3.0 Å), medium (1.8–4.5 Å), and weak (3.0–6.0 Å). All NOEs between Co(NH3)6 3+ and the RNA were restrained to 3.2–7.2 Å, corresponding to the distance from the RNA proton to the center of the cobalt atom. Mn(H2O)6 2+ distances based on the PRE measurements were restrained to 2.0–7.0 Å for proton resonances displaying significant manganese-induced line broadening. Line broadening due to Mn2+-induced PRE was determined to be significant if the NOE intensities were reduced to <25% of their original intensity. Preliminary sets of structures were calculated with different numbers of ions, in order to find the minimum number of ions required to satisfy the Co(NH3)6 3+ NOEs and Mn2+ PRE data. Structures with Co(NH3)6 3+ and Mn(H2O)6 2+ were calculated separately. From the preliminary calculations, it was found that four Co(NH3)6 3+ ions (two for each monomer) were minimally required to satisfy the observed NOEs and were restrained to two protons at site 3 and nine protons at site 5. Ten Mn(H2O)6 2+ ions (five for each monomer) were minimally required to fit the PRE data and were restrained to 31 protons (six to seven at each site). To reduce Mn(H2O)6 2+ localization to regions that did not exhibit line broadening, 24 protons for which no line broadening was observed were restrained to be >7.0 Å away from manganese. The backbone torsion angles (α, β, δ, ε, γ, and ζ) for residues in the helical regions (1–4, 10–19, 24–33, and 40–43) were constrained to A-form values (±20°) (Saenger 1984), consistent with 2D NOESY and HNN COSY data. Additionally A-form phosphate backbone restraints for these regions were incorporated as previously reported (Davis et al. 2005). The backbone for the tetraloop and the receptor regions were left unrestrained. Nucleotides with strong H1′–H2′ couplings and a H1′–H3′ (A6 and A37) couplings as observed in a 1H-1H TOCSY experiment with a 45-msec mixing time were constrained as C2′-endo, while nucleotides with weak H1′–H2′ couplings (U7 and G43), and no visible H1′–H3′ couplings were left unrestrained. All other nucleotides were constrained as C3′−endo (±20°). Analysis of H1′-aromatic NOEs from the 2D NOESY experiment (25-msec mixing time) indicated that most nucleotides fell into the anti-range, and were therefore constrained to have χ angles of −160° ± 15°. Nucleotides 7 and 37 had relatively intense H2′ to aromatic NOE cross-peaks; therefore, their χ angles were left unrestrained. Weak planarity restraints (10 kcal mol−1 Å−2) were used for bases involved in base pairing.
CNS 1.1 (Brunger et al. 1998) was used to calculate 100 initial structures from an extended conformation with random initial velocities using NOE, dihedral, and H-bonding restraints as previously described (Davis et al. 2005). The structures were subjected to 60 psec (15-fsec time steps) of restrained molecular dynamics in torsion angle space, followed by 90 psec of slow cooling. Finally, 30 psec (5-fsec time steps) of restrained molecular dynamics in Cartesian coordinate space were preformed. The structures were refined against RDCs as previously described (Davis et al. 2005) using XPLOR-NIH (Schwieters et al. 2003), and metal ions were added at this stage [Mn(H2O)6 2+ and Co(NH3)6 3+]. Structures were refined by cooling from 3000 K to 100 K in 58 cycles of restrained Cartesian coordinate space corresponding to a total of 28 psec. The force constant for RDCs was increased from 0.001 to 0.2 kcal mol−1 Hz−2. Five hundred steps of energy minimization using the Powell algorithm followed simulated annealing.
Atomic coordinates
Coordinates and restraints for structure calculations have been deposited in the RCSB (PDB ID codes 2I7Z and 2I7E). Chemical shifts have been deposited into BioMagResBank (accession number 6652).
ACKNOWLEDGMENTS
This work was supported by NSF Grant MCB-0445613 to S.E.B. J.H.D. received predoctoral support from the University of Wisconsin Molecular Biophysics Training Grant (T32 GM08293). This study made use of the National Magnetic Resonance Facility at Madison, which is supported by National Institutes of Health grants P41RR02301 (Biomedical Research Technology Program, National Center for Research Resources) and P41GM66326 (National Institute of General Medical Sciences). Equipment in the facility was purchased with funds from the University of Wisconsin, the National Institutes of Health (P41GM66326, P41RR02301, RR02781, and RR08438), the National Science Foundation (DMB-8415048, OIA-9977486, and BIR-9214394), and the U.S. Department of Agriculture. The authors thank Ryan Marcheschi, Nicholas Reiter, David Staple, and Dipali Sashital for many helpful discussions.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.268307.
REFERENCES
- Adams, P.L., Stahley, M.R., Gill, M.L., Kosek, A.B., Wang, J., Strobel, S.A. Crystal structure of a group I intron splicing intermediate. RNA. 2004a;10:1867–1887. doi: 10.1261/rna.7140504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, P.L., Stahley, M.R., Kosek, A.B., Wang, J., Strobel, S.A. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004b;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- Allain, F.H., Varani, G. Divalent metal ion binding to a conserved wobble pair defining the upstream site of cleavage of group I self-splicing introns. Nucleic Acids Res. 1995;23:341–350. doi: 10.1093/nar/23.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, N.A., Sept, D., Joseph, S., Holst, M.J., McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu, S., Rambo, R.P., Strauss-Soukup, J., Cate, J.H., Ferre-D'Amare, A.R., Strobel, S.A., Doudna, J.A. A specific monovalent metal ion integral to the AA platform of the RNA tetraloop receptor. Nat. Struct. Biol. 1998;5:986–992. doi: 10.1038/2960. [DOI] [PubMed] [Google Scholar]
- Bax, A., Sklenar, V., Clore, G.M., Gronenborn, A.M. Water suppression in two-dimensional spin-locked NMR experments using a novel phase-cycling procedure. J. Am. Chem. Soc. 1987;109:6511–6513. [Google Scholar]
- Bertini, I., Luchinat, C. NMR of paramagnetic molecules in biological systems. Benjamin/Cummings; Menlo Park, CA: 1986. [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Butcher, S.E., Allain, F.H., Feigon, J. Determination of metal ion binding sites within the hairpin ribozyme domains by NMR. Biochemistry. 2000;39:2174–2182. doi: 10.1021/bi9923454. [DOI] [PubMed] [Google Scholar]
- Cate, J.H., Doudna, J.A. Metal-binding sites in the major groove of a large ribozyme domain. Structure. 1996;4:1221–1229. doi: 10.1016/s0969-2126(96)00129-3. [DOI] [PubMed] [Google Scholar]
- Cate, J.H., Gooding, A.R., Podell, E., Zhou, K., Golden, B.L., Kundrot, C.E., Cech, T.R., Doudna, J.A. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- Chin, K., Sharp, K.A., Honig, B., Pyle, A.M. Calculating the electrostatic properties of RNA provides new insights into molecular interactions and function. Nat. Struct. Biol. 1999;6:1055–1061. doi: 10.1038/14940. [DOI] [PubMed] [Google Scholar]
- Clore, G.M., Gronenborn, A.M., Tjandra, N. Direct structure refinement against residual dipolar couplings in the presence of rhombicity of unknown magnitude. J. Magn. Reson. 1998;131:159–162. doi: 10.1006/jmre.1997.1345. [DOI] [PubMed] [Google Scholar]
- Costa, M., Michel, F. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J. 1995;14:1276–1285. doi: 10.1002/j.1460-2075.1995.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, M., Michel, F. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: Comparison with in vivo evolution. EMBO J. 1997;16:3289–3302. doi: 10.1093/emboj/16.11.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, M., Deme, E., Jacquier, A., Michel, F. Multiple tertiary interactions involving domain II of group II self-splicing introns. J. Mol. Biol. 1997;267:520–536. doi: 10.1006/jmbi.1996.0882. [DOI] [PubMed] [Google Scholar]
- Davis, J.H., Tonelli, M., Scott, L.G., Jaeger, L., Williamson, J.R., Butcher, S.E. RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop–receptor complex. J. Mol. Biol. 2005;351:371–382. doi: 10.1016/j.jmb.2005.05.069. [DOI] [PubMed] [Google Scholar]
- Davlvit, C. Efficient multiple-solvent suppression for the study of the interactions of organic solvents with biomolecules. J. Biomol. NMR. 1998;11:437–444. [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Downey, C.D., Fiore, J.L., Stoddard, C.D., Hodak, J.H., Nesbitt, D.J., Pardi, A. Metal ion dependence, thermodynamics, and kinetics for intramolecular docking of a GAAA tetraloop and receptor connected by a flexible linker. Biochemistry. 2006;45:3664–3673. doi: 10.1021/bi0520941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper, D.E. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, Y., Gaffney, B.L., Jones, R.A. RNA GG×UU motif binds K+ but not Mg2+ J. Am. Chem. Soc. 2005;127:17588–17589. doi: 10.1021/ja0555522. [DOI] [PubMed] [Google Scholar]
- Gesteland, R., Cech, T., Atkins, J. The RNA world. 3rd ed. Cold Spring Harbor Laboratory Press; Woodbury, NY: 2006. [Google Scholar]
- Golden, B.L., Gooding, A.R., Podell, E.R., Cech, T.R. A preorganized active site in the crystal structure of the Tetrahymena ribozyme. Science. 1998;282:259–264. doi: 10.1126/science.282.5387.259. [DOI] [PubMed] [Google Scholar]
- Gonzalez R.L., Jr., Tinoco I., Jr. Solution structure and thermodynamics of a divalent metal ion binding site in an RNA pseudoknot. J. Mol. Biol. 1999;289:1267–1282. doi: 10.1006/jmbi.1999.2841. [DOI] [PubMed] [Google Scholar]
- Hansen, M.R., Simorre, J.P., Hanson, P., Mokler, V., Bellon, L., Beigelman, L., Pardi, A. Identification and characterization of a novel high affinity metal-binding site in the hammerhead ribozyme. RNA. 1999;5:1099–1104. doi: 10.1017/s1355838299990738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppler, A., Nikstad, L.J., Allmann, A.M., Brow, D.A., Butcher, S.E. Metal binding and base ionization in the U6 RNA intramolecular stem–loop structure. Nat. Struct. Biol. 2002;9:431–435. doi: 10.1038/nsb800. [DOI] [PubMed] [Google Scholar]
- Hwang, T.L., Shaka, A.J. Water suppression that works. Excitation sculpting using arbitrary waveforms and pulse field gradients. J. Magn. Reson. A. 1995;112:275–279. [Google Scholar]
- Jaeger, L., Michel, F., Westhof, E. Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J. Mol. Biol. 1994;236:1271–1276. doi: 10.1016/0022-2836(94)90055-8. [DOI] [PubMed] [Google Scholar]
- Jaeger, L., Westhof, E., Leontis, N.B. TectoRNA: Modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 2001;29:455–463. doi: 10.1093/nar/29.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneau, K., Podell, E., Harrington, D.J., Cech, T.R. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA–solvent interactions. Structure. 2001;9:221–231. doi: 10.1016/s0969-2126(01)00579-2. [DOI] [PubMed] [Google Scholar]
- Khvorova, A., Lescoute, A., Westhof, E., Jayasena, S.D. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- Kieft, J.S., Tinoco I., Jr. Solution structure of a metal-binding site in the major groove of RNA complexed with cobalt (III) hexammine. Structure. 1997;5:713–721. doi: 10.1016/s0969-2126(97)00225-6. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., Wuthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Leroy, J.L., Gueron, M. Electrostatic effects in divalent ion binding to tRNA. Biopolymers. 1977;16:2429–2446. doi: 10.1002/bip.1977.360161108. [DOI] [PubMed] [Google Scholar]
- Leroy, J.L., Gueron, M., Thomas, G., Favre, A. Role of divalent ions in folding of tRNA. Eur. J. Biochem. 1977;74:567–574. doi: 10.1111/j.1432-1033.1977.tb11426.x. [DOI] [PubMed] [Google Scholar]
- Misra, V.K., Draper, D.E. A thermodynamic framework for Mg2+ binding to RNA. Proc. Natl. Acad. Sci. 2001;98:12456–12461. doi: 10.1073/pnas.221234598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra, V.K., Shiman, R., Draper, D.E. A thermodynamic framework for the magnesium-dependent folding of RNA. Biopolymers. 2003;69:118–136. doi: 10.1002/bip.10353. [DOI] [PubMed] [Google Scholar]
- Murphy, F.L., Cech, T.R. GAAA tetraloop and conserved bulge stabilize tertiary structure of a group I intron domain. J. Mol. Biol. 1994;236:49–63. doi: 10.1006/jmbi.1994.1117. [DOI] [PubMed] [Google Scholar]
- Ott, G., Arnold, L., Limmer, S. Proton NMR studies of manganese ion binding to tRNA-derived acceptor arm duplexes. Nucleic Acids Res. 1993;21:5859–5864. doi: 10.1093/nar/21.25.5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pley, H.W., Flaherty, K.M., McKay, D.B. Model for an RNA tertiary interaction from the structure of an intermolecular complex between a GAAA tetraloop and an RNA helix. Nature. 1994;372:111–113. doi: 10.1038/372111a0. [DOI] [PubMed] [Google Scholar]
- Qin, P.Z., Feigon, J., Hubbell, W.L. Site-directed spin labeling studies reveal solution conformational changes in a GAAA tetraloop receptor upon Mg(2+)-dependent docking of a GAAA tetraloop. J. Mol. Biol. 2005;351:1–8. doi: 10.1016/j.jmb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Reiter, N.J., Nikstad, L.J., Allmann, A.M., Johnson, R.J., Butcher, S.E. Structure of the U6 RNA intramolecular stem–loop harboring an S(P)-phosphorothioate modification. RNA. 2003;9:533–542. doi: 10.1261/rna.2199103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenger, W. Principles of nucleic acids. Springer-Verlag; New York: 1984. [Google Scholar]
- Schwieters, C.D., Kuszewski, J.J., Tjandra, N., Clore, G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Sigel, R.K., Sashital, D.G., Abramovitz, D.L., Palmer, A.G., Butcher, S.E., Pyle, A.M. Solution structure of domain 5 of a group II intron ribozyme reveals a new RNA motif. Nat. Struct. Mol. Biol. 2004;11:187–192. doi: 10.1038/nsmb717. [DOI] [PubMed] [Google Scholar]
- Stein, A., Crothers, D.M. Conformational changes of transfer RNA. The role of magnesium(II) Biochemistry. 1976a;15:160–168. doi: 10.1021/bi00646a025. [DOI] [PubMed] [Google Scholar]
- Stein, A., Crothers, D.M. Equilibrium binding of magnesium(II) by Escherichia coli tRNAfMet. Biochemistry. 1976b;15:157–160. doi: 10.1021/bi00646a024. [DOI] [PubMed] [Google Scholar]
- Tanner, M.A., Cech, T.R. An important RNA tertiary interaction of group I and group II introns is implicated in gram-positive RNase P RNAs. RNA. 1995;1:349–350. [PMC free article] [PubMed] [Google Scholar]
- Zweckstetter, M., Bax, A. Prediction of sterically induced alignment in a dilute liquid crystalline phase: Aid to protein structure determination by NMR. J. Am. Chem. Soc. 2000;122:3791–3792. [Google Scholar]