Abstract
With the goal of gaining insight into the tertiary structure of the hepatitis delta virus ribozyme, cross-linking experiments using 4-thiouridine residues introduced in either the 5′-end portion of the substrate, or at seven strategic positions within the ribozyme, were performed. Mapping of the newly formed covalent bonds in cross-linked species obtained under various conditions, as well as using several mutated ribozymes, permitted monitoring of the formation of the ribozyme–substrate complex as the ribozyme proceeded along the folding pathway. In order to aid visualization of the tertiary structure transformation, an in silico animation of the “on” folding pathway was developed. In combination with those of the cleavage assays of structured substrates, these data shed light on the key contribution of the L3 loop in the formation of an active tertiary complex.
Keywords: ribozyme, hepatitis delta virus, cross-linking, RNA structure–function, folding pathway
INTRODUCTION
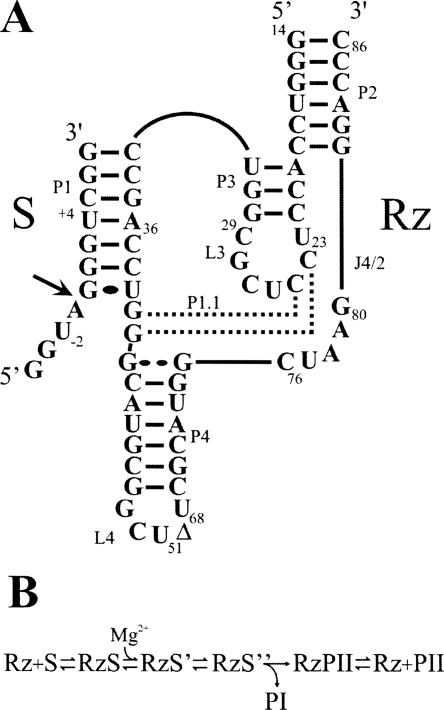
Hepatitis delta virus (HDV) possesses a single-stranded circular RNA genome of 1.7 kb that replicates through a double rolling-circle mechanism. Both genomic and antigenomic HDV RNAs possess self-cleaving RNA motifs that generate 2′-3′-cyclic phosphate and 5′-hydroxyl termini (i.e., HDV ribozymes) (for reviews, see Shih and Been 2002; Bergeron et al. 2003). Trans-acting HDV ribozymes have been developed by removing one junction, thereby producing a complex composed of two species, one possessing the substrate (S) sequence and the other the catalytic HDV ribozyme (Rz) (see Fig. 1A). According to the double pseudoknot model the secondary structure of the HDV ribozyme consists of one stem (P1), two pseudoknots (P2 and P1.1), two stem–loops (P3–L3 and P4–L4), and three single-stranded junctions (J1/2, J1/4, and J 4/2). The P3 stem length is critical, suggesting that it positions the L3 loop in a precise conformation in the catalytic center. In the antigenomic-derived HDV ribozyme the L3 loop is composed of seven nucleotides (U23CCUCGC29; see Fig. 1A). Both the J1/4 junction and the L3 loop are single-stranded in the initial stages of folding, but are subsequently involved in the formation of the P1.1 pseudoknot that consists of two Watson–Crick base pairs involving pairing between the cytosines in positions 24 and 25 with the guanosines in positions 40 and 41.
FIGURE 1.
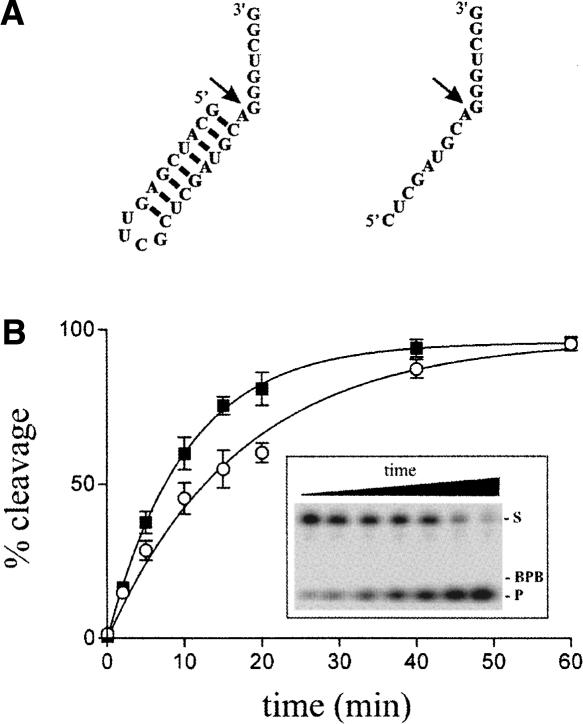
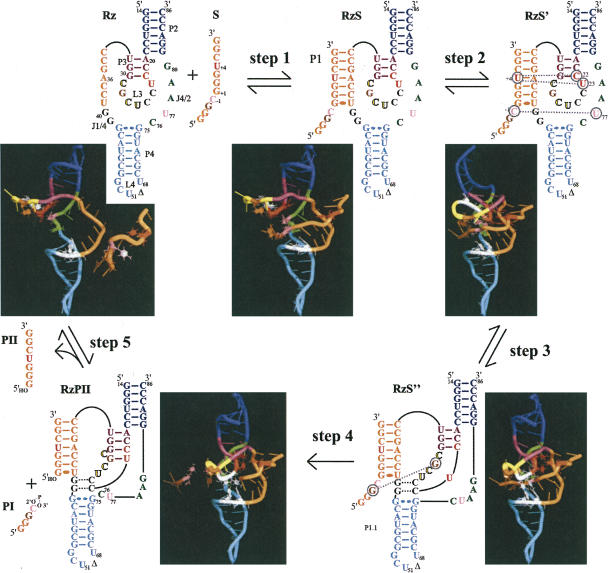
Structure and proposed kinetic pathway of the HDV ribozyme. (A) Secondary structure and nucleotide sequence of the HDV ribozyme derived from the antigenomic hepatitis delta virus genome. The numbering system is that of Shih and Been (2002). The Δ in the L4 loop indicates the P4 deletion compared to the natural variants. The wobble base pair at the bottom of the P1 stem is indicated by a single large dot, while the homopurine base pair at the top of the P4 stem is represented by two large dots. The base pairs of the P1.1 pseudoknot are indicated by the dotted lines. The arrow indicates the cleavage site. (B) Proposed kinetic pathway of the HDV ribozyme. S, Rz, PI, and PII represent the substrate, the ribozyme, and the 5′ and 3′ products, respectively.
Both X-ray diffraction and nuclear magnetic resonance experiments have provided high-resolution information on the tertiary structure of the HDV ribozyme (Ferré D'Amaré et al. 1998; Tanaka et al. 2002; Ke et al. 2004). Briefly, the catalytic core is formed by two coaxial helices resulting from the stacking of the P1–P1.1–P4 stems and of the P2–P3 stems. Moreover, these studies revealed the complex network of interactions that has to take place in the catalytic center of the HDV ribozyme in order to generate an active ribozyme. Several other biochemical approaches have also been used to identify the important chemical groups within the catalytic center. For example, chemically synthesized ribozymes with site-specific functional modifications in the sugar residues permitted identification of 10 2′-hydroxyl groups of the ribose moieties that contribute to the catalytic activity of an antigenomic-derived HDV ribozyme (Fiola and Perreault 2002). However, little is known regarding the chronology of this network of interactions.
Crystal structures and computer-assisted molecular dynamic simulations have focused on the conformational transition between the pre- and post-cleavage structures (Ke et al. 2004; Krasovska et al. 2005, 2006). Prior to formation of the active tertiary complex, three steps of the folding pathway have been identified (Fig. 1B). Initially, the ribozyme (Rz) binds its substrate (S) via formation of the P1 stem, yielding an RzS complex. This step is magnesium independent, and requires at least the presence of monovalent ions (Ananvoranich and Perreault 2000). Second, upon the addition of magnesium ions, the P1 stem is involved in an essential structural transition that produces the RzS′ complex (Ouellet and Perreault 2004). One of the consequences of this conformational transition is a P3–L3 conformational change within the catalytic center (i.e., docking of the substrate). Third, the P1.1 pseudoknot is formed, through two Watson–Crick base pairs, between the J1/4 junction and the L3 loop generating the RzS″ complex (Ferré D'Amaré et al. 1998; Wadkins et al. 1999; Nishikawa and Nishikawa 2000; Deschênes et al. 2003). It is only after these structural transitions have taken place that the chemical step, which includes an acid–base catalysis involving the highly conserved cytosine at position 76, takes place (Perrotta et al. 1999; Bevilacqua et al. 2004; Das and Piccirilli 2005). The catalytic and structural metal ion sites important to the ribozyme have been classified within a multichannel reaction mechanism (Nakano et al. 2003), the latter being supported by recent molecular dynamic simulations suggesting structural and potential chemical roles for ions (Krasovska et al. 2006).
The structural evolution of the HDV ribozyme's catalytic core along the folding pathway remains largely unknown. In fact, little information exists about the order of interactions required in order to produce an active tertiary complex. Most of this information was obtained from cross-linking experiments using 4-thiouridine (s4U), a photoactivable nucleotide that, upon UV irradiation (>300 nm), can form a covalent bond to a stacked, neighboring nucleotide (∼3 Å) (Favre et al. 1998). Substrates that include an s4U either in position –1 (adjacent to the scissile phosphate), +4 (in the middle of the P1 stem), or +8 (at the 3′ end of the P1 stem) were used to determine the position of the binding domain of the substrate within the catalytic center of an antigenomic, trans-acting, HDV ribozyme (Ouellet and Perreault 2004). In this report, we employ the same strategy in order to gain further insight into the interactions that lead to the formation of active RzS complexes. Results from the cross-linking experiments performed with s4U residues introduced in either position –2 of the substrate are presented. Cross-linking experiments using mixed RNA–DNA substrates including an s4U deoxyribonucleotide in position –2 have been reported previously, but using an HDV ribozyme of different sequence that can favor the formation of an alternative conformation and which exhibited significantly less catalytic activity (Bravo et al. 1996). Additionally, we use s4U ribonucleotides under several different conditions in order to gain insights into the HDV ribozyme's folding pathway. Last, we introduce the thiouridine residue into several positions of the ribozyme as well as examining various conditions with a number of different mutants, in order to unravel the role of the L3 loop in HDV ribozyme folding.
RESULTS AND DISCUSSION
This study was performed using an antigenomic derived trans-acting HDV ribozyme for which the kinetic and thermodynamic behavior has been extensively characterized under both single- and multiple-turnover conditions (Mercure et al. 1998; Ananvoranich et al. 1999). This version of the ribozyme is composed of 57 nt, and cleaves a model substrate of 11 nt into products of 4 nt and 7 nt (see Fig. 1).
Cross-linking of the 5′-end portion of the substrate
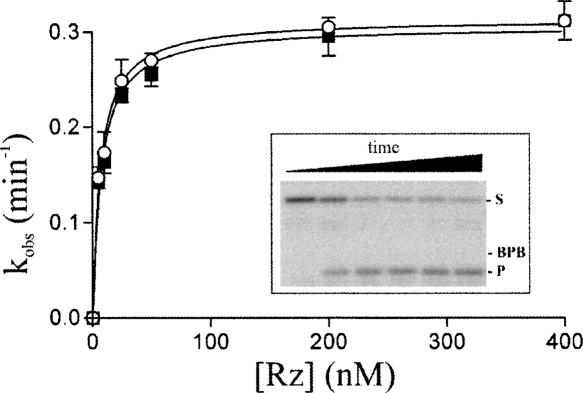
In order to gain more information from the position of the 5′-end portion of the substrate during the folding pathway, a 4-thiouridine residue was introduced in position –2. Since a substrate that includes two consecutive pyrimidines in positions –1 and –2 is not efficiently cleaved by HDV ribozyme (Deschênes et al. 2000), the cytosine in –1 was replaced by an adenosine. The integrity of the resulting substrate (i.e., Ss4U-2) was assessed for cleavage activity in comparison to that of the wild-type substrate (i.e., Swt) under single turnover conditions (Fig. 2). Plotting of the rate constants (k obs) as a function of the ribozyme concentrations demonstrated that the introduction of the s4U in position –2 did not modify the ability of the ribozyme to efficiently cleave the substrate.
FIGURE 2.
Graphical representation of the k obs for the cleavage reaction of the Swt (open circles) and the Ss4U-2 (closed squares) substrates as a function of the ribozyme concentration. The inset shows an autoradiogram of a denaturing PAGE gel of the cleavage reactions performed with the Ss4U-2 substrate and a ribozyme concentration of 200 nM. The positions of the bromophenol blue (BPB), the substrate (S), and the product (P) are indicated. From left to right, the aliquots were recovered after 2, 5, 10, 15, 20, and 40 min of reaction.
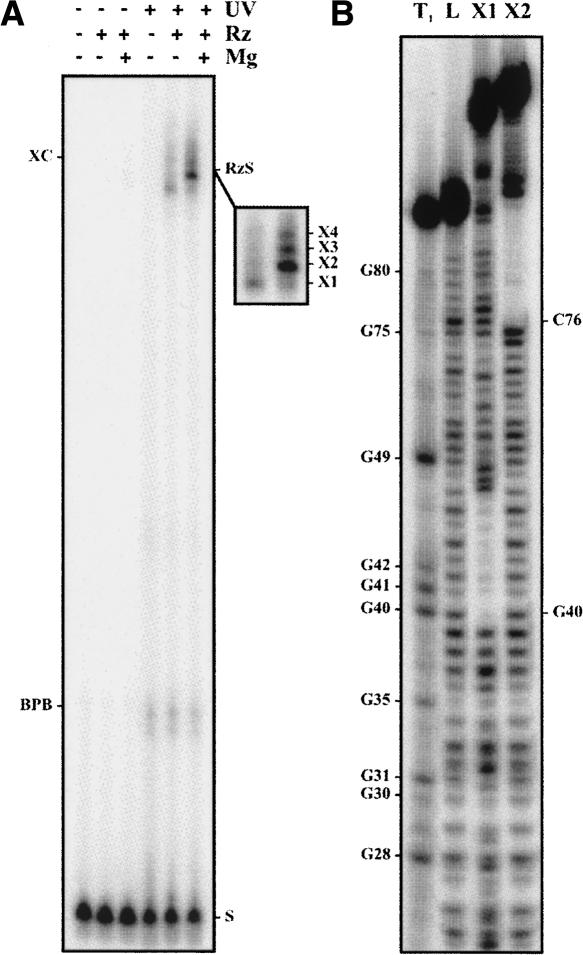
Initially, the cross-linking experiments were performed using an analog that included an s4U in position –2 (SdA-1s4U-2). The substitution of the ribonucleotide for a deoxyribonucleotide in position –1, adjacent to the scissile phosphate, prevents the cleavage from occurring (Mercure et al. 1998). As a result, the cross-linking product is not released by a cleavage event and the cross-linking yield increases. After preincubation of a trace amount of 5′ end-labeled analog and the ribozyme (500 nM) either in the presence or the absence of 10 mM MgCl2, the samples were rapidly irradiated on ice in a UV cross-linker at 366 nm and then analyzed on an 8% PAGE. RNA species with slower electrophoretic migration, which correspond to cross-linked ribozyme–analog complexes, were observed regardless of the presence or absence of magnesium. A longer migration permitted a better resolution of the four different species (see inset Fig. 3A, X1 to X4). The X1 species was the most abundant cross-linked species produced in the absence of magnesium; however, it became a minor RzS complex upon the addition of magnesium. The X4 species was obtained in trace amounts under both conditions. In the absence of magnesium, both the X2 and X3 species were found in trace amounts. Upon the addition of magnesium, the yields of both species increased dramatically, resulting in the X2 species being the most abundant.
FIGURE 3.
Results from typical cross-linking experiments. (A) Autoradiogram of an 8% PAGE gel for experiments performed using a 5′ end-labeled, noncleavable substrate (SdA-1s4U-2) and the ribozyme. The various conditions tested are indicated at the top of the gel. The positions of the xylene cyanol (XC), the bromophenol blue (BPB), and substrate (S) are indicated. The inset illustrates a longer exposition of the region including the cross-linked species. (B) Autoradiogram of a typical 10% sequencing gel performed for the NaOH mapping of the cross-linked species. Lane 1 is a partial RNase T1 hydrolysis in order to identify the guanosines, and lane 2 is the partial NaOH ladder of the ribozyme alone. Lanes 3 and 4 are partial NaOH hydrolyzes of the X1 and X2 species, respectively. The positions of the guanosines are indicated on the left, while those of the cross-links are indicated on the right.
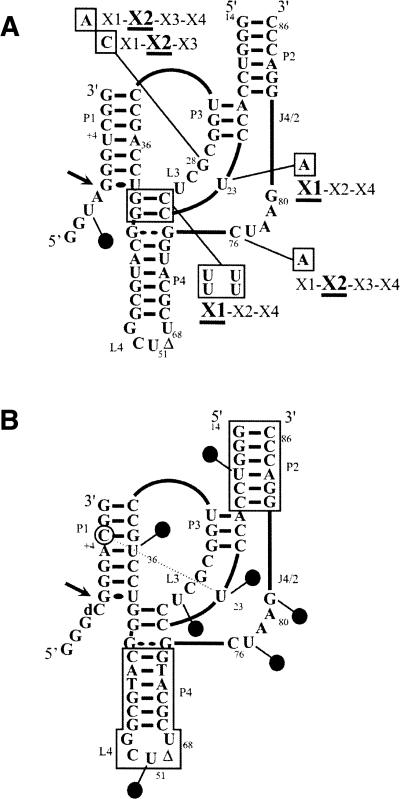
Similar results were obtained when the experiment was repeated using a trace amount of 5′ end-labeled 32P-ribozyme and an excess of nonradioactive SdA-1s4U-2 analog (data not shown). The gel bands of each lane were cut out, and the cross-linked species extracted and subsequently subjected to a partial alkaline hydrolysis and analysis on 8% sequencing gels. A typical autoradiogram for the analysis of both the X1 and X2 species obtained in the absence of magnesium is illustrated in Figure 3B. At the cross-linking site, a tri-branched structure is formed; consequently, alkaline hydrolysis of the phosphodiester bonds releases a branched RNA species that possesses a slower electrophoretic mobility in denaturing PAGE (i.e., causes a shift of all bands corresponding to the hydrolysis products generated over the cross-link sites). For example, this indicated that the X1 and X2 species resulted from cross-linking that occurred at positions G40 and C76, respectively. Similar analyses were performed with all species, and the data are reported on the HDV ribozyme's secondary structure (Fig. 4). In the absence of magnesium, a condition that permits formation of the P1 stem between the substrate and the ribozyme, but no further progress along the folding pathway, the s4U in position –2 cross-linked with G40 (X1, predominant), C76 (X2; i.e., the catalytic cytosine), and either G49 or C50 (X4) (Fig. 4A). The last cross-link with the L4 loop was not consistently observed, and was always found in a relatively small amount. The determination was not possible for X3 in the absence of magnesium, most likely because the species was not sufficiently abundant. The positions of the cross-links of the species isolated in the presence of the magnesium were also determined (Fig. 4B). The X1 and X4 species that were minor RzS complexes corresponded to cross-linking with G40 and G49/C50, respectively. The most abundant X2 species corresponded to cross-linking with C76, while the X3 species corresponded to cross-linking with G28.
FIGURE 4.
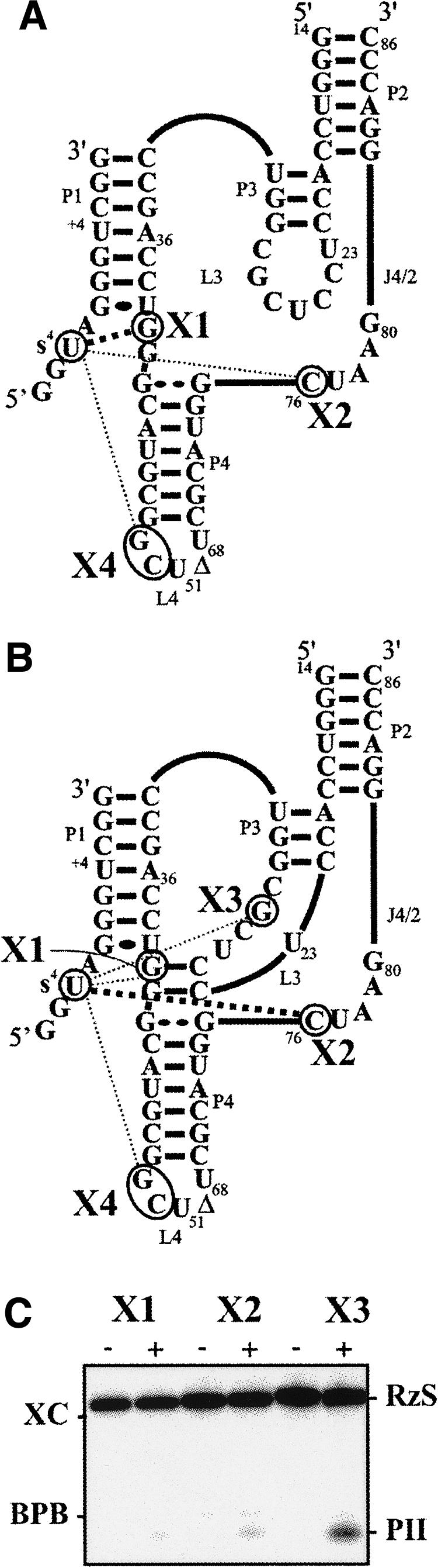
Characterization of the cross-links obtained with the SdA-1s4U-2 substrate in the absence (A) and presence of the pseudoknot (B). Schematic representation of the cross-linked sites on the secondary structure of the ribozyme obtained in both the absence and the presence of magnesium, respectively. The intensities of the dotted lines are a function of the relative abundances of the cross-linked species. (C) Autoradiogram of denaturing PAGE gels of the self-cleavage of the cross-linked RzS complexes. Cleavages of the X1, X2, and X3 cross-linked complexes were performed in the presence of urea either with (+) or without (−) magnesium. The positions of the xylene cyanol (XC), bromophenol blue (BPB), the cross-linked complexes (RzS) and the 3′-product (PII) are indicated.
A cross-linked species could correspond to the formation of either a productive ribozyme–substrate complex along the folding pathway (i.e., “on” folding pathway), or of a misfolded ribozyme–substrate complex. In order to verify whether or not the cross-linked complexes correspond to structures that include an RzS conformation compatible with being an active intermediate in the reaction pathway, each one was isolated and incubated under self-cleavage conditions. This type of approach was developed for the characterization of the cross-linked RNA species derived from the hairpin ribozyme (Pinard et al. 2001). The cross-linking experiments were repeated using a thiolated 3′ end 32pCp-labeled substrate. The radioactive 3′ end-labeled product was released in the cleavage reaction, while the 5′-end portion remained linked to the ribozyme. The patterns of cross-linked species obtained were virtually identical to those obtained above, except that all species were detected in smaller amounts because cleavage had occurred. The species X1, X2, and X3 (X4 was not abundant enough) were extracted from the gels and, initially, the self-cleavage reactions were performed in the absence of magnesium (i.e., Tris-HCl, pH 8.0, for 60 min at 37°C). Under these conditions, no self-cleavage product was detected, indicating that the cross-linked bonds were stable. The incubations were then repeated under optimal reaction conditions (i.e., plus 10 mM MgCl2). Only the X3 species exhibited self-cleavage, and even then only at a low level (data not shown). When the experiment was repeated with the prior addition of 2M urea, the X1 species did not exhibit any self-cleavage, while both the X2 and X3 species self-cleaved at levels of 2%–3% and >10%, respectively (Fig. 4C). Most likely, the X1 species did not self-cleave because the cross-link with G40 appears very early during the folding pathway, thereby preventing any further movement of the substrate within the complex. The X2 species exhibited self-cleavage in only a small proportion of the cross-linked species as the presence of a cross-link between U-2 and C76 must limit the movement required for the correct positioning of the scissile phosphate located in position –1. As a putative kinetic trap, it is likely that this intermediate is the reason why it accumulated in larger quantities. Conversely, the X3 species self-cleaved more efficiently, suggesting the presence of an RzS complex from the “on” folding pathway.
Elucidating the folding pathway
In order to gain more information on the precise time that the cross-links occurred during the folding pathway, experiments were performed using mutated ribozymes. Initially, cross-linking was performed with a mutant that did not permit the docking of the P1 stem into the catalytic center. This step is magnesium-dependent, and occurs after the formation of the P1 stem but before the formation of the P1.1 pseudoknot (Ananvoranich and Perreault 2000). In this mutant, the uridine in position 23 was mutated to an adenosine (U23A; Fig. 5A). In the presence of magnesium, the SdA-1s4U-2 was shown to cross-link with both the G41 and C76 residues (i.e., forming the X1 and X2 species). A simple U to A substitution in the L3 loop did alter the early SU-2-G40 interaction, with G40 being now replaced by G41, demonstrating that the ribozyme's folding pathway is very sensitive to L3 identity. No cross-linking was observed with G28, as was the case with the wild-type ribozyme in the absence of magnesium. Similar results were obtained when the experiment was repeated with a mutant that prevents the formation of the P1.1 pseudoknot. In this case, all four of the residues that form the pseudoknot (two GC base pairs) were substituted by four uridines (Fig. 5A). The formation of the pseudoknot has been shown to take place after P1 stem docking, and is essential for cleavage (Wadkins et al. 1999; Nishikawa and Nishikawa 2000; Deschênes et al. 2003). Clearly, these results suggest that the cross-linking between the s4U in position –2 and G28 occurs very late in the folding process. Conversely, the cross-linking with both G41 and C76 occurred at an early stage, when only the P1 stem is formed, but before the conformational transition requiring the presence of magnesium. Additionally, the experiments were performed with a C76A mutant, a mutation which completely abolishes the catalytic activity, but supports both the docking of the substrate and the formation of the P1.1 pseudoknot. In this case, the most abundant cross-linked species was X2 (i.e., the A76 cross-link), as was the case for the RzS″ complex. This result supports the idea that the cross-linking of catalytic cytosine occurred late in the folding of the active structure. Finally, the experiments were performed with G28A and G28C mutants that are completely deprived of catalytic activity. In both cases, the most abundant cross-linked species was the X2, indicating that the implication of the G28 occurred lately along the folding pathway. The intensity of the different bands was identical for the G28A and C76A, while it was significantly reduced for the C28G mutant. This difference suggests that the presence of a pyrimidine in position 28 alters the structure more compared to a purine.
FIGURE 5.
Schematic representation of the cross-linking experiments performed using various mutated and modified ribozymes. (A) Cross-link experiments were performed using the SdA-1s4U-2 substrate and either the U23A, disrupted P1.1 (i.e., possesses four U instead of two GC bp), G28A, G28C, or the C76A mutant ribozymes. The cross-linked species (X1 to X4) are identified adjacent to the mutations that are boxed. The most abundant species are underlined. (B) Identification of the seven positions where s4U residues were introduced in the RNA–DNA mixed ribozymes. The positions are identified by the circles. The deoxyribonucleotide regions are boxed. The cross-link between the positions U23 of the ribozyme and C + 5 (circled) of the substrate is identified by the dotted line.
Subsequently, seven ribozymes each including a 4-thiouridine at a different strategic position were synthesized (Fig. 5B). In order to allow efficient chemical synthesis, we used an RNA–DNA mixed ribozyme composed of 26 deoxyribonucleotides (specifically the nucleotides forming the P2 stem and the P4–L4 stem–loop) and 31 ribonucleotides (those forming the catalytic center) as a common basis. The latter mixed ribozyme catalyzed the cleavage of a small substrate with kinetic parameters virtually identical to those of the all-RNA ribozyme (Fiola and Perreault 2002). All s4U-modified ribozymes exhibited significant cleavage activity, although at different levels. The cross-linking experiments were performed using either 5′ end-labeled mixed ribozymes, or labeled SdC-1 analog (a substrate that includes a deoxyribocytosine at the cleavage site). In the absence of magnesium, neither the ribozymes alone, nor the ribozymes and substrates incubated together, resulted in the detection of species possessing a slower electrophoretic mobility upon PAGE analysis. The experiments were then repeated in the presence of magnesium. When performed in the presence of the substrate alone no cross-linked species was detected, whereas one was detected when the experiment was performed in the presence of the ribozymes. This cross-link was formed between the mixed ribozyme possessing the s4U at position 23 of the L3 loop and the cytosine located in position +5 of the substrate (Fig. 5B). The latter result is in agreement with the previously reported cross-link between the s4U located in position +4 of the substrate and U23 of the ribozyme (Ouellet and Perreault 2004). Importantly, these data confirmed that the presence of the magnesium is absolutely required for the formation of a compact tertiary structure. Moreover, the fact that most of the s4U introduced within the ribozymes were not productive is an indication that cross-linking has several requirements (e.g., distance and base stacking).
Position of the substrate's 5′-end domain
At least two different conformations have been proposed for the 5′-end portion of the substrate. According to the X-ray data, the 5′ end of the substrate makes a sharp (∼180°) turn of the backbone (at position –2) and the upstream domain extends at an angle of ∼30° to the P1 stem (Ke et al. 2004). Alternatively, MC-SYM modeling indicates that the 5′-end portion of the substrate extends outside of the catalytic site without performing a sharp turn. In this study, the formation of U-2-G40/41 cross-link supports the idea that the 5′-end portion of the substrate does not possess the backbone reversal. According to the latter structure, it should be possible to increase the space occupied by the 5′ end of the substrate without affecting the cleavage activity, while in the case of the former this should be more limiting for the formation of a backbone reversal. In other words, in the case of the crystal structure model increasing the diameter of the substrate at the 5′ end should lead to steric hindrance that would affect the cleavage activity; whereas in the other model the same modification should have no effect. In order to verify this hypothesis, 3′ end-labeled substrates, both with and without a hairpin at the 5′ end (a structure which occupies more space than its single-stranded homolog), were synthesized and their abilities to be cleaved by the ribozyme assessed (Fig. 6). The substrate possessing the hairpin was cleaved slightly less than the single-stranded homolog (0.056 min−1 vs. 0.088 min−1 in the presence 10 nM Rz). The absence of significant differences between these substrates supports the idea that the 5′ end does not form a sharp turn. The difference observed between the substrates may be due to either the conformational transition required to fold into an active tertiary RzS complex, which is the limiting step (Ananvoranich and Perreault 2000), that is slightly slower in the presence of the hairpin compared to the single-stranded substrate, or simply, to the cleavage reaction of this substrate whose rate is limited by some other step. Virtually identical results were obtained when the experiment was repeated with substrates that included the insertion of either one or two more adenosines between positions –1 and –2 (i.e., serving as a spacer pushing the hairpin, which might potentially create steric hindrance, further away). Conversely, when the hairpin started with a base pair involving the adenosine located in position –1, the level of cleavage was significantly lower (data not shown). Most likely this occurs because this situation restricts the flexibility required for the correct positioning of the adenosine with respect to the catalytic C76 for the in-line nucleophile attack.
FIGURE 6.
Cleavage assays of differently structured substrates. (A) Secondary structures and nucleotide sequences of the substrate including a 5′-end hairpin, as well as that of its single-stranded counterpart. The arrows indicate the cleavage sites. (B) Graphical representation of the cleavage percentages as a function of time for both substrates, the single-stranded substrate (closed squares) and the substrate with the 5′-end hairpin (open circles). The inset shows a typical autoradiogram of a denaturing 20% PAGE gel for the cleavage of the substrate with the 5′-end hairpin. The positions of the xylene cyanol (XC), the substrate (S), and the product (P) are indicated.
Steered, targeted dynamic molecular simulation of the folding pathway
In order to improve the visualization of the tertiary structure's transformation, we subsequently undertook to develop an in silico animation of the “on” folding pathway of the HDV ribozyme. As a “pure” dynamic molecular simulation of the complete folding pathway would necessitate a huge time scale, and given that we are primarily interested in one-way conformational changes between two intermediates, a steered, targeted dynamic molecular simulation was performed using the NAMD and VMD software packages. Due to a lack of high-resolution structures of the many conformational intermediates occurring along the folding pathway of the ribozyme, each of the five transitions was manually modeled. An initial RzS complex was modeled using MC-SYM. This initial structure was then modified by steered molecular dynamics using NAMD and VMD in order to obtain structures for the four other conformational intermediates (Fig. 7). The presence of monovalent ions is sufficient for this RzS complex to occur. Then, in the presence of magnesium, a short-lived intermediate, RzS′, has been identified in cross-linking experiments (Ouellet and Perreault 2004). The next intermediate, RzS″, includes the P1.1 pseudoknot, either followed by or concomitant with the cleavage reaction, resulting in the complex RzPII and a fast release of the 5′-end free product PI. Then, if sufficient amounts of substrate are present, the PII product can dissociate from the ribozyme and the free ribozyme (Rz) can bind a new substrate. There is no explicit chemistry reaction in this model, thereby avoiding the complexity of the acid-base catalysis as well as the dynamic locations of the magnesium ions closed to the cleavage site. Using VMD and NAMD together, interactive and targeted molecular dynamic simulations were performed in order to generate conformations missing between two discrete structures (Humphrey et al. 1996; Kalé et al. 1999). In other words, the RzS′ conformation was driven toward the RzS″ conformation artificially. Selective external forces can be applied to the molecules under a controlled environment, and the simulation can be visualized via the VMD software. For example, using the information that U+4 and C+5 stacked together with C22 and U23, a structure was obtained for the RzS′ complex. Subsequently, starting from the RzS′ complex and considering the formation of the P1.1. pseudoknot yielded a structure for the RzS″ complex. The NAMD package was then used to record the coordinates of each frame while applying harmonic restraints from one structure to another (e.g., RzS′ to RzS″). Representations of each complex are shown in Figure 7, while the animations, viewed from three different angles, are available online at http://132.210.163.235/jo/Delta_Movie.html.
FIGURE 7.
Folding pathway of an HDV ribozyme. Step 1: formation of the RzS complex (i.e., the P1 stem). Step 2: structural rearrangement involving the middle of the P1 stem that orientates the substrate in the catalytic center of the RzS′ complex. Step 3: formation of the P1.1 pseudoknot yielding a productive RzS″ complex. Step 4: cleavage reaction coupled to the fast release of PI. Step 5: dissociation of the RzPII complex into PII and free Rz. The P2 and P4 stems are blue and cyan, while the P1 stem and the substrate are orange. The internal P3 stem and the L3 loop are purple and yellow, respectively. The bases are represented by cylinders except for the stacked nucleotides (red: U + 4, C22, and U23; pink: SC-1 [scissile phosphate] and U77; brown: SG-2 and G28; and, C76 [catalytic cytosine] and the nucleotides involved in the P1.1 stem are white [in black in the 2D scheme]). All water atoms were removed for clarity.
These intermediates, although not at an atomic resolution, can be seen as being more than just a plausible set of conformations along the folding pathway as they represent key interactions and structural features reported by various laboratories (e.g., see Nishikawa and Nishikawa 2000; Pereira et al. 2002; Tanaka et al. 2002; Ke et al. 2004; Ouellet and Perreault 2004). Moreover, this animation provides the opportunity to locate the most probable places of these structural features along the folding pathway. As the use of a force field cannot accurately represent the strong (and polarizing) interaction of metal ions with RNA, and given that a recent 200-ns explicit solvent dynamic molecular simulation of the binding of both monovalent and divalent ions to both precursor and product forms of genomic HDV ribozyme (Krasovska et al. 2006), they were not included in this short-sampling steered targeted molecular dynamic simulation. The aim of this animation is clearly not to reproduce the precise energy landscape of the folding transitions, but rather to demonstrate stereochemically feasible pathways that can generate hypotheses which can then be tested experimentally. Thus, several assumptions can be taken, including: (1) starting the simulation with a ribozyme prefolded into its proper secondary structure, thereby avoiding the topological dilemma that the molecule may encounter during the folding that accompanies its transition from its primary to the secondary structure; (2) substrate approach and its nucleation to the ribozyme; (3) a forced linear pathway preventing the bouncing back and forth between intermediates; and (4) the timescales between any of the transitions are not proportional to the real timescale of folding. Previously, a “pure” two-step movie of the hammerhead ribozyme-catalyzed cleavage reaction, based on the morphing of the pre- and post-crystal structure conformations, had been reported (Dunham et al. 2003). More recently, a huge amount of computer power has been used to animate the accommodation of tRNA into the 70S ribosome in explicit solvent using NAMD software (Sanbonmatsu et al. 2005). In the present case, as little information on the high-resolution structure of the different intermediates was available, such a precise and highly computational effort was not necessary, based on our predefined assumptions (i.e., resulting in a nonatomic resolution).
The animation shows that the ribozyme goes through important conformational changes, supporting the idea that RNA is highly dynamic (Pereira et al. 2002). As a result, the ribozyme initially has a relatively relaxed structure, but becomes more compact as it proceeds along the folding pathway due mainly to the formation of additional interactions within the catalytic core (Shih and Been 2002). This animation supports the appearance of the various cross-links as the ribozyme proceeds along the folding pathway. For example, the residue in position –2 of the substrate comes in close proximity to G28 only within the RzS″ complex. Within this RzS″ complex, the C−1 of the substrate is in close proximity to the catalytic C76, but not in the case of the RzS′ complex. The cross-linking observed between position −2 of the substrate and the L4 loop (i.e., the X4 species, G49/C50), which was detected only in small amounts, is incompatible with the model structure, suggesting that it came from a misfolded RzS complex. In all of the complexes, the 5′ end of the substrate is located outside of the catalytic center in the absence of folding into a sharp turn. Moreover, the current position of the 5′-end substrate in the movie allows for a P1.1 base-pairing competition (Deschênes et al. 2003), while the sharp turn seen in the crystal structure does not allow for the possibility of base pairing with J1/4. Finally, the animation provides an interesting tool with which to establish new hypotheses. For example, the animation shows that the formation of the P1.1 stem trapped the substrate within the catalytic core, suggesting an additional key role for this pseudoknot. Therefore, this animation should inspire the design of new experiments aiming to depict in detail the folding pathway of the HDV ribozyme.
CONCLUSION AND PERSPECTIVES
The cross-linking experiments, in combination with the cleavage assays of structured substrates, provided evidence of the putative contributions of the L3 loop to the folding of the ribozyme into an active tertiary complex. The s4U located in position 23 of this loop was found to cross-link with the middle of the P1 stem (C+5), which correlates with the docking within the catalytic center. Residues C24 and C25 are involved in the formation of the P1.1 pseudoknot, a step that occurs late along the folding pathway, most likely just before, or concomitant with, the chemical attack. Based on both the structural modeling and RNase H accessibility experiments, we suggest that the formation of the P1.1 pseudoknot directly contributes to the molecular mechanism of cleavage by HDV ribozyme by trapping the region of the substrate to be cleaved within the catalytic center. Finally, the experiment performed with the s4U located in position –2 of the substrate shows that G28 was cross-linked only in the active RzS″ complex (i.e., after P1.1 formation), a late event that occurs exclusively in the “on” folding pathway. Together, these experiments implicate four of the seven nucleotides of the L3 loop. It is important to note that U23 and G28, along with the catalytic C76, are the most conserved nucleotides in the HDV ribozyme (Nehdi and Perreault 2006). Clearly, they must play an important role. Although it is tempting to propose that, together, these results shed light on the contribution of the L3 loop, it is important to remember that cross-linking data provides only an indication of the proximity of pairs of nucleotides and does not indicate a formal interaction between the residues in question.
In general, cross-linking experiments are performed to map catalytic centers. For example, the recent cross-link mapping of the native hammerhead ribozyme that permitted the loop–loop interaction suggests a different structure from the crystal one (Lambert et al. 2006). Using mutants representative of various steps along the kinetic pathway of the HDV ribozyme, information on the folding process that leads to the formation of an active RzS″ complex was obtained. The modeling and animation provide a molecular “view” of the different conformational transitions that might occur in solution. Although it does not provide any information on the time scale of each transition, nor does it present any multichannel and “off” folding pathways (i.e., including temporarily misfolded RNA), RNA dynamic design can be applied to many other RNA species, and, consequently, contributes to the emergence of guidelines and parameters for RNA folding.
MATERIALS AND METHODS
Ribozyme and substrate synthesis
Most of the ribozymes and substrates were synthesized by in vitro transcription as described previously (Mercure et al. 1998). Briefly, RNA molecules were prepared by annealing two pair of complementary oligonucleotides that included the T7 RNA promoter followed by the full-length sequence of the desired ribozyme or substrate. Using the resulting partial duplex as a template (500 pmol), in vitro transcription reactions were carried out in a final volume of 100 μL containing 15 units of RNA Guard (Amersham Biosciences), 80 mM HEPES-KOH pH 7.5, 24 mM MgCl2, 2 mM spermidine, 40 mM DTT, 5 mM of each NTP, 0.01 units of yeast pyrophosphatase (Roche Diagnostic), and 10 μg of purified T7 RNA polymerase. The samples were DNase treated, and the transcripts were purified on either 8% or 20% denaturing PAGE gels (19:1 ratio of acrylamide to bisacrylamide) using 50 mM Tris-borate pH 7.5, 7 M urea, and 1 mM EDTA solution as running buffer. The reaction products were visualized by UV shadowing, and the bands corresponding to the correct sizes were cut out. The transcripts were eluted from the gel slices, passed through G-25 Sephadex spun columns (Amersham Biosciences), the RNA was ethanol precipitated, and the quantity of nucleic acid determined by spectroscopy at 260 nm.
Substrates, analogs, and RNA–DNA ribozymes including s4U residues were purchased from Dharmacon, deprotected, and purified on PAGE gels as described above.
Radioactive labeling of RNA species
Purified substrates or ribozymes (40 pmol) were dephosphorylated using 5 units of Antarctic phosphatase as described by the manufacturer (New England BioLabs). After ethanol precipitation, the dephosphorylated RNA (5 pmol) was 5′ end-labeled by incubation with 5 units T4 polynucleotide kinase (Amersham Biosciences) in a final volume of 10 μL containing 1.6 pmol [γ-32P]-ATP (6000 Ci/mmol, Amersham Biosciences), 50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 50 mM KCl, and 5 units of T4 polynucleotide kinase (Amersham Biosciences) at 37°C for 2 h. The reactions were stopped by adding formamide dye (95% formamide, 10 mM EDTA, 0.025% bromophenol blue, and 0.025% xylene cyanol), and the RNA purified by denaturing PAGE. The bands of the correct sizes containing the 5′ end-labeled RNAs were excised, and the RNA recovered as described above.
For 3′ end-labeling, the substrates (15 pmol) were incubated for 2 h at 37°C in a final volume of 10 μL containing 50 mM Tris-HCl pH 7.8, 10 mM MgCl2, 10 mM DTT, 1 mM ATP, 10% dimethylsulfoxide, 10 pmol [32P]Cp (3000 Ci/mmol, Amersham Biosciences) and 10 units of T4 RNA ligase (New England Biolabs). The transcripts were then purified on denaturing 20% PAGE gels and recovered as described above.
Cleavage reactions
Cleavage reactions were carried out in 20 μL reaction mixtures containing 50 mM Tris-HCl (pH 7.5) and 10 mM MgCl2 at 37°C under single turnover conditions ([Rz] >> [S]). Prior to the reaction, trace amounts of either 5′ or 3′ end-labeled substrates (<1 nM) and nonradioactive ribozymes were mixed together, heated at 70°C for 1 min, snap-cooled on ice for 3 min, and then incubated at 37°C for 5 min. The cleavage reactions were initiated by the addition of MgCl2. Aliquots (2–3 μL) were removed at various times up to 1 h, and were quenched by the addition of ice-cold formamide dye buffer (10 μL). The mixtures were fractionated on denaturing 20% PAGE gels and exposed to PhosphorImager screens (Molecular Dynamics). The extent of cleavage was determined from measurements of the radioactivity present both in the substrate and in either the 5′ or 3′ product bands at each time point using the ImageQuant software. The fractions of cleaved substrate were determined, and the rate of cleavage (k obs) obtained by fitting the data to the equation At = A 0(1−e −kt), where At is the percentage of cleavage at time t, A 0 is the maximum percent cleavage (end point), and k is the rate constant (k obs). Each rate constant was calculated from at least two independent measurements.
Cross-linking experiments
Cross-linking experiments were performed in a final volume of 20 μL containing 50 mM Tris-HCl pH 7.5, 0.2 pmol ribozyme, and a trace amount of 5′ end-labeled substrate (single turnover conditions). Once the ribozyme and substrate were mixed together, but prior to the addition of magnesium (10 mM MgCl2, see below), they were heat denatured, snap-cooled, and then preincubated at 37°C. The reactions were then chilled on ice for 15 min in a 96-well plate cover. Following the addition of MgCl2 to a final concentration of 10 mM, the samples were rapidly irradiated in a UV cross-linker (UV Stratalink 2400, Stratagene) equipped with 15-W (366 nm) bulbs for 20 min on ice. Formamide dye buffer (5 μL) was then added to each sample prior to fractionation by denaturing 8% PAGE. The resulting bands were visualized using a PhosphorImager screen. With the purpose of performing self-cleavage experiments, the 5′ end-labeled S-2s4U substrates (∼5 pmol) were cross-linked to the ribozyme (15 pmol), and the resulting complexes then cut out of the gel and eluted as described above. The purified complexes were snap-cooled, subjected to self-cleavage in the presence of 50 mM Tris-HCl (pH 7.5) and 10 mM MgCl2 for 60 min at 37°C, and fractionated on 20% denaturing PAGE gels. The experiments with mixed RNA–DNA ribozymes including s4U residues were performed either in the absence (using 5 pmol of 5′-end labeled ribozymes) or the presence (using 15 pmol of nonradioactive ribozymes and 1 pmol of 5′ end SdC-1 analog) of substrate.
Mapping of the cross-links
In order to map the sites of the cross-links, the experiments were performed under multiple turnover conditions ([S] > [Rz]) using nonradioactive analogs (15 pmol SdA-1s4U-2) and 5′ end-labeled ribozymes (5 pmol). After a long electrophoretic migration, the bands corresponding to the RzS complexes were cut out and the RNA species recovered. The resulting RNA species were subjected to alkaline hydrolysis (NaOH) for 1 min, the reactions quenched on ice by the addition of 10 μL of 1 M Tris-HCl pH 7.5, the nucleic acids precipitated and then analyzed on sequencing 10% PAGE gels. The gels were prerun for 1 h at 60 W with 1× TBE buffer in both chambers, then the buffer in the lower chamber was changed to 1 M NaOAc and the gel prerun for a further 30–60 min before loading the samples. In order to permit the localization of the cross-linking sites, partial alkaline and ribonuclease T1 hydrolyzes of noncross-linked, labeled ribozymes were performed and analyzed on the same gels.
Molecular modeling
The model of the RzS complex was generated using the constraint-satisfaction program MC-Sym 3.3.2 (Major et al. 1991). A selected structure was subsequently modified by Streered Molecular Dynamics (SMD) with VMD 1.8.4 and NAMD 2.6 (Kalé et al. 1999; Humphrey et al. 1996) in order to obtain models of the Rz, RzS′, RzS″, and RzPII complexes. The simulations were performed in a common solvated environment (water sphere 52 Å in diameter) with basic dynamics and space partitioning default NAMD parameters and using the CHARMM27 nucleic acid parameters (MacKerell et al. 2000). Figures were prepared using PyMOL 0.99 software (http://pymol.sourceforge.net).
ACKNOWLEDGMENTS
We are grateful to Dominique Lévesque, Atef Nehdi, and Jonathan Perreault for interesting suggestions and reading our manuscript. This work was supported by grants from the Canadian Institute of Health Research (CIHR; Grant Number MOP-44002) to J.P.P. The RNA group is supported by grants from Université de Sherbrooke. J.P.P. holds the Canada Research Chair in Genomics and Catalytic RNA.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.263407.
REFERENCES
- Ananvoranich, S., Perreault, J.P. The kinetic and magnesium requirements for the folding of antigenomic δ ribozymes. Biochem. Biophys. Res. Commun. 2000;270:600–607. doi: 10.1006/bbrc.2000.2465. [DOI] [PubMed] [Google Scholar]
- Ananvoranich, S., Lafontaine, D.A., Perreault, J.P. Mutational analysis of the antigenomic trans-acting delta ribozyme: The alterations of the middle nucleotides located on the P1 stem. Nucleic Acids Res. 1999;27:1473–1479. doi: 10.1093/nar/27.6.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron L., Jr., Ouellet, J., Perreault, J.P. Ribozyme-based gene-inactivation systems require a fine comprehension of their substrate specificities; the case of delta ribozyme. Curr. Med. Chem. 2003;10:2589–2597. doi: 10.2174/0929867033456486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua, P.C., Brown, T.S., Nakano, S., Yajima, R. Catalytic roles for proton transfer and protonation in ribozymes. Biopolymers. 2004;73:90–109. doi: 10.1002/bip.10519. [DOI] [PubMed] [Google Scholar]
- Bravo, C., Lescure, F., Laugaa, P., Fourrey, J.L., Favre, A. Folding of the HDV antigenomic ribozyme pseudoknot structure deduced from long-range photocross-links. Nucleic Acids Res. 1996;24:1351–1359. doi: 10.1093/nar/24.7.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, S.R., Piccirilli, J.A. General acid catalysis by the hepatitis delta virus ribozyme. Nat. Chem. Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- Deschênes, P., Lafontaine, D.A., Charland, S., Perreault, J.P. Nucleotides −1 to −4 of hepatitis delta ribozyme substrate increase the specificity of ribozyme cleavage. Antisense Nucleic Acid Drug Dev. 2000;10:53–61. doi: 10.1089/oli.1.2000.10.53. [DOI] [PubMed] [Google Scholar]
- Deschênes, P., Ouellet, J., Perreault, J., Perreault, J.P. Formation of the P1.1 pseudoknot is critical for both the cleavage activity and substrate specificity of an antigenomic trans-acting hepatitis delta ribozyme. Nucleic Acids Res. 2003;31:2087–2096. doi: 10.1093/nar/gkg307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham, C.M., Murray, J.B., Scott, W.G. A helical twist-induced conformational switch activates cleavage in the hammerhead ribozyme. J. Mol. Biol. 2003;332:327–336. doi: 10.1016/s0022-2836(03)00843-x. [DOI] [PubMed] [Google Scholar]
- Favre, A., Saintome, C., Fourrey, J.L., Clivio, P., Laugaa, P. Thionucleobases as intrinsic photoaffinity probes of nucleic acid structure and nucleic acid protein interactions. J. Photochem. Photobiol. 1998;42:109–124. doi: 10.1016/s1011-1344(97)00116-4. [DOI] [PubMed] [Google Scholar]
- Ferré D’Amaré, A.R., Zhou, K., Doudna, J.A. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- Fiola, K., Perreault, J.P. Kinetic and binding analysis of the catalytic involvement of ribose moieties of a trans-acting delta ribozyme. J. Biol. Chem. 2002;277:26508–26516. doi: 10.1074/jbc.M203468200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Kalé, L., Skeel, R., Bhandarkar, M., Brunner, R., Gursoy, A., Krawetz, N., Phillips, J., Shinozaki, A., Varadarajan, K., Schulten, K. NAMD2: Greater scalability for parallel molecular dynamics. J. Comput. Phys. 1999;151:283–312. [Google Scholar]
- Ke, A., Zhou, K., Ding, F., Cate, J., Doudna, J.A. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- Krasovska, M.V., Sefcikova, J., Spackova, N., Sponer, J., Walter, N.G. Structural dynamics of precursor and product of the RNA enzyme from the hepatitis delta virus as revealed by molecular dynamics simulations. J. Mol. Biol. 2005;351:731–748. doi: 10.1016/j.jmb.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Krasovska, M.V., Sefcikova, J., Réblova, K., Schneider, B., Walter, N.G., Sponer, J. Cations and hydration in catalytic RNA: Molecular dynamics of the hepatitis delta virus ribozyme. Biophys.J. 2006;91:626–638. doi: 10.1529/biophysj.105.079368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, D., Heckman, J.E., Burke, J.M. Three conserved guanosines approach the reaction site in native and minimal hammerhead ribozymes. Biochemistry. 2006;45:7140–7147. doi: 10.1021/bi052457x. [DOI] [PubMed] [Google Scholar]
- MacKerell A.D., Jr., Banavali, N., Foloppe, N. Development and current status of the CHARM force field for nucleic acids. Biopolymers. 2000;56:257–265. doi: 10.1002/1097-0282(2000)56:4<257::AID-BIP10029>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Major, F., Turcotte, M., Gautheret, D., Lapalme, G., Fillion, E., Cedergren, R. The combination of symbolic and numerical computation for three-dimensional modeling of RNA. Science. 1991;253:1255–1260. doi: 10.1126/science.1716375. [DOI] [PubMed] [Google Scholar]
- Mercure, S., Lafontaine, D., Ananvoranich, S., Perreault, J.P. Kinetic analysis of delta ribozyme cleavage. Biochemistry. 1998;37:16975–16982. doi: 10.1021/bi9809775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, S.I., Cerrone, A.L., Bevilacqua, P.C. Mechanistic characterization of the HDV genomic ribozyme: Classifying the catalytic and structural metal ion sites within a multichannel reaction mechanism. Biochemistry. 2003;42:2982–2994. doi: 10.1021/bi026815x. [DOI] [PubMed] [Google Scholar]
- Nehdi, A., Perreault, J.P. Unbiased in vitro selection reveals the unique character of the self-cleaving antigenomic HDV RNA sequence. Nucleic Acids Res. 2006;34:584–592. doi: 10.1093/nar/gkj463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa, F., Nishikawa, S. Requirement of the canonical base pairing in the short pseudoknot structure of genomic hepatitis delta virus ribozyme. Nucleic Acids Res. 2000;28:925–931. doi: 10.1093/nar/28.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet, J., Perreault, J.P. Cross-linking experiments reveal the presence of novel structural features between a hepatitis delta virus ribozyme and its substrate. RNA. 2004;10:1059–1072. doi: 10.1261/rna.7230604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, M.J.B., Harris, D.A., Rueda, D., Walters, N.G. Reaction pathway of the trans-acting hepatitis delta virus ribozyme: A conformational change accompanies catalysis. Biochemistry. 2002;41:730–740. doi: 10.1021/bi011963t. [DOI] [PubMed] [Google Scholar]
- Perrotta, A.T., Shih, I., Been, M.D. Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science. 1999;286:123–126. doi: 10.1126/science.286.5437.123. [DOI] [PubMed] [Google Scholar]
- Pinard, R., Lambert, D., Heckman, J.E., Esteban, J.A., Gundlach C.W., IV, Hampel, K.J., Glick, G.D., Walter, N.G., Major, F., Burke, J.M. The hairpin ribozyme substrate binding-domain: A highly constrained D-shaped conformation. J. Mol. Biol. 2001;307:51–65. doi: 10.1006/jmbi.2000.4472. [DOI] [PubMed] [Google Scholar]
- Sanbonmatsu, K.Y., Joseph, S., Tung, C.S. Simulating movement of tRNA into the ribosome during decoding. Proc. Natl. Acad. Sci. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih, I.H., Been, M.D. Catalytic strategies of the hepatitis delta virus ribozymes. Annu. Rev. Biochem. 2002;71:887–917. doi: 10.1146/annurev.biochem.71.110601.135349. [DOI] [PubMed] [Google Scholar]
- Tanaka, Y., Hori, T., Tagaya, M., Sakamoto, T., Kurihara, M., Uesugi, S. Imino proton NMR analysis of HDV ribozymes: Nested double pseudoknot structure and Mg2+ ion-binding site close to the catalytic core in solution. Nucleic Acids Res. 2002;30:766–774. doi: 10.1093/nar/30.3.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadkins, T.S., Perrotta, A.T., Ferré D'Amaré, A.R., Doudna, J.A., Been, M.D. A nested double pseudoknot is required for self-cleavage activity of both the genomic and antigenomic hepatitis delta virus ribozymes. RNA. 1999;6:720–727. doi: 10.1017/s1355838299990209. [DOI] [PMC free article] [PubMed] [Google Scholar]