Abstract
Coordination between separate pathways may be facilitated by the requirements for common protein factors, a finding congruent with the link between proteins regulating DNA replication with other important cellular processes. We report that the smallest of Drosophila origin recognition complex subunits, Orc6, was found in embryos and cell culture localized to the cell membrane and cleavage furrow during cell division as well as in the nucleus. A two-hybrid screen revealed that Orc6 interacts with the Drosophila peanut (pnut), a member of the septin family of proteins important for cell division. This interaction, mediated by a distinct C-terminal domain of Orc6, was substantiated in Drosophila cells by coimmunoprecipitation from extracts and cytological methods. Silencing of Orc6 expression with double-stranded RNA resulted in a formation of multinucleated cells and also reduced DNA replication. Deletion of the C-terminal Orc6–peanut interaction domain and subsequent overexpression of the Orc6 mutant protein resulted in the formation of multinucleated cells that had replicated DNA. This mutant protein does not localize to the membrane or cleavage furrows. These results suggest that Orc6 has evolved a domain critical mainly for cytokinesis.
Keywords: DNA replication, cytokinesis, peanut
The origin recognition complex (ORC), a heteromeric six-subunit protein, is a central component for eukaryote DNA replication. ORC binds to DNA at replication origin sites and serves as a scaffold for assembly of other key initiation factors such as cdc6, cdt1, the MCM complex, and cdc45 (1). In Drosophila (2, 3), both biochemical and genetic data support its role as an initiator protein. Mutants, homozygous for ORC2, ORC3, or ORC5 defects all die in larval stages, as large maternal ORC stores are depleted. In the terminal stages, there is a dramatic decrease in DNA replication and cellular proliferation (4–7).
In addition to its well documented role in the initiation of DNA replication, ORC is involved in other functions. Some of these activities directly link cell-cycle progression with DNA replication (8), whereas other functions seem distinct from replication. For example, in budding yeast, ORC participates in the establishment of transcriptionally repressed domains at the silent mating type loci, HMR and HML (see ref. 9 for review). In Drosophila, ORC interacts with HP1 protein and therefore may influence the establishment or maintenance of heterochromatin (10, 11). The Latheo gene product, which is dORC3, seems to be involved in ion transport at neuromuscular junctions (6).
The Orc6 gene is the least conserved of the ORC subunits, and amino acid alignments with the budding yeast Orc6 and the metazoan smallest subunit show no statistically significant homologies. The Drosophila (12) and Homo sapiens (13) Orc6 subunits are homologues and are similar in size to the Schizo-saccharomyces pombe counterpart (14), all of which are considerably smaller than the Saccharomyces cerevisiae Orc6. Moreover, the human Orc6 homologue does not seem to be tightly associated with the other subunits (13, 15), but when expressed in the baculovirus system with the other ORC genes, the protein does join a six-subunit complex (15).
We reported that the Drosophila Orc6 is an essential component of the complex, because it is required for DNA binding, and an ORC(1-5) complex could not complement an ORC-depleted extract for DNA replication (12). We were particularly intrigued by the high levels of free Orc6 in embryonic and cultured cell extracts given that all other subunits could be detected biochemically only in association with the others. A considerable fraction of this pool was cytoplasmic as judged by cytological methods, and the protein was detected proximal to the cytoplasmic membranes. In this work, we show that Orc6 interacts with the Drosophila Pnut protein, a member of the septin family of proteins important for cell division. Silencing of Orc6 expression by RNA interference (RNAi) caused an apparent block to cytokinesis as binucleated cells appeared rapidly, with loss of DNA replication occurring later. We also show that the Orc6–Pnut interaction is mediated by a distinct C-terminal domain of Orc6. Deletion of this domain and subsequent overexpression of the Orc6 mutant protein resulted in the formation of binucleated cells that had replicated DNA. These results suggest that Orc6 contains a domain important for cytokinesis.
Methods
Yeast Two-Hybrid Screen. The Interactor trap/yeast two-hybrid system has been described (16). The plasmid pEG202 was used to generate a full-length Orc6-lexA DNA binding domain fusion gene (pLexAdOrc6). The tester strain was EGY48, which has a chromosomal LEU2 gene modified such that it has a UAS with LexA sites that allow for selection for viability when cells are plated on medium lacking Leu, and a resident plasmid pSH18–34 containing a lacZ fusion gene also with LexA sites that allows discrimination based on color when the yeast is grown on medium containing 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-gal). Such cells transformed with plexAdOrc6 were transformed with a cDNA Drosophila embryonic library (Clontech MATCHMAKER library) carried by the vector pJG4–5; this vector carries an inducible yeast GAL1 promoter to express Drosophila fusion proteins with an acidic activation domain. We also used a cDNA library representing ovary expressed genes (Roger Brent, Molecular Biosciences Institute, Berkeley, CA). We screened =500,000 colonies from the embryo library and 250,000 colonies from the ovary library; 49 positive cDNAs representing 16 different cDNAs were found in total, and the peanut cDNA was represented in each screen. Transformants with both Orc6 and Drosophila cDNA library proteins were first analyzed for their ability to grow on medium lacking Leu in a presence of galactose (Gal) to assure the expression of library proteins. Transformants were further tested on a medium with X-gal in the presence of Gal and leucine.
GFP–ORC Constructs and Immunofluorescence. GFP–ORC fusion genes were prepared as described (12). C-terminal mutant Orc6 constructs were designed by using the PCR technique. Transfection of the Drosophila L2 Schneider cells was done according to the manufacturer's (Invitrogen) recommendations. Cells were fixed by using 1–3% paraformaldehyde and subsequently subjected to immunofluorescent microscopy (Zeiss Axioplan, ×100 magnification). Confocal microscopy was performed by using a Zeiss LSM 510 microscope. Immunostaining of the Drosophila embryos was performed as described (10) by using affinity-purified antibody raised against Drosophila Orc6 and monoclonal antibody against Pnut protein (17).
Protein Expression and Immunoprecipitation. His-tagged Drosophila WT Orc6 protein and Orc6 mutant proteins were expressed in Escherichia coli (XA-90) by using pQE-30 vector and purified according to the manufacturer's (Qiagen, Valencia, CA) recommendations. A Qiagen expression kit was used for subcloning and expression. For precipitation of His-tagged proteins, cobalt and nickel beads were used. Five nanograms of each recombinant protein per 1 μl of extract was used. After incubation with 20 μlof Drosophila L2 extract for 1 h, proteins were precipitated. Precipitated material was analyzed for presence of Pnut protein during immunoblot experiment with α-Pnut antibody, and about one-half of the precipitated material was analyzed for Pnut.
For immunoprecipitation experiments, both Drosophila embryonic extract and L2 cell extract were used. Antibodies raised against Drosophila Orc6 subunit were affinity-purified. Immunoprecipitated material was separated by using SDS/PAGE gel and transferred to a poly(vinylidene difluoride) membrane. The membrane was then incubated with mouse monoclonal antibody raised against Drosophila Pnut protein (17). Protein bands were visualized by subsequent enhanced chemiluminescence (ECL) assay (Amersham Biosciences).
RNAi Assay. Double-stranded RNAs (dsRNAs) were obtained by using the Megascript kit from Ambion. Orc6 primers (5′-CGGCCAGTGAATTGTTTAATACGACTCACTATAGGGACTACCTTAATAGAACAGTTAA-3′ and 5′-CGGCCAGTGAATTGTTTAATACGACTCACTATAGGGCTAAGCCTCGAGAAGCTGGCT-3′) flanked with T7 promoter were used. Drosophila L2 cells were transfected with Orc6 dsRNA or control luciferase dsRNA (20 μg of RNA per well of a six-well dish) and harvested after 24, 48, or 72 h. RNAi efficiency was tested by immunoblotting with Orc6 antibody.
Orc6 C-Terminal Peptides. Orc6 C-terminal peptide corresponding to the last 71 aa (186-257) was synthesized. Peptide (Orc6-71-C) was labeled with biotin and incubated with 25 μl of Drosophila embryonic (EE) or L2 whole cell extract (L2E) for 1 h at 4°C. The biotin-labeled peptide was incubated with Drosophila embryonic whole-cell extract and then reisolated by using paramagnetic beads coupled with streptavidin (Promega). The bound material was subjected to SDS/PAGE and analyzed by Western blotting for the presence of Drosophila Pnut protein.
Results and Discussion
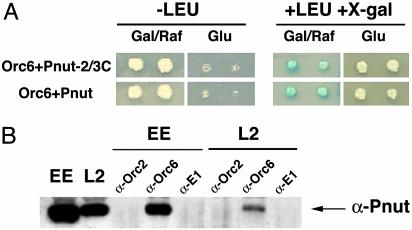
To obtain more information on Orc6 function, apart from its functions in the complex, we performed a screen of a Drosophila cDNA library, searching for new interacting proteins by employing the interactor/trap yeast two-hybrid technique (16). Full-length Drosophila Orc6 was subcloned into a plasmid carrying a LexA DNA-binding domain cassette. The resulting lexA–Orc6 fusion was used as a bait to probe for interacting proteins present in a Drosophila cDNA expression library. Several independent positives were selected; among the positives, two clones contained C-terminal fragments of the Drosophila peanut (pnut) gene (amino acids 145–539). We also cloned the full-length pnut gene into pJG4–5 vector to test whether it would interact with Orc6 in yeast. Both full-length and truncated genes behaved similarly in the yeast two-hybrid system, and representative results of such assays are presented as Fig. 1A.
Fig. 1.
Orc6 interact with Pnut protein. (A) Orc6 and Pnut interact in a yeast two-hybrid system. Transformants carrying both Orc6 and Drosophila Pnut protein were first analyzed for their ability to grow on medium lacking Leu in the presence of Gal. Transformants were further tested on a medium with X-gal, in the presence of Gal and leucine. Both full-length Drosophila Pnut protein and the truncated Pnut (two-thirds from C terminus, amino acids 145–539) were found to interact with Orc6. (B) Pnut protein can be immunoprecipitated from Drosophila embryonic extract (EE) or L2 cell extract (L2) by using antibodies raised against Drosophila Orc6 subunit. Antibodies raised against Drosophila Orc2 (2) and BPV E1 protein (34) were used as controls. Two microliters of extracts was loaded in lanes EE and L2, and one-third of the immunoprecipitation from 20 μl of extract was loaded in the other lanes. We estimate that 15% of Pnut coprecipitates with Orc6.
Septins are polymerizing proteins with a common GTPase activity and were first discovered in S. cerevisiae but now seem to be ubiquitous in fungi and animals (18–20). Pnut is one of the five Drosophila septins identified to the date (17, 20). Genetic data showed that the proteins play diverse roles in organization of the cell cortex and in cytokinesis. At the molecular level, the role of GTP binding and hydrolysis by septins is unclear and is probably not required for filament formation (21). Thus, formation of filaments at a cleavage furrow or in the cytoplasm during interphase may be an activity of septins independent of other roles for the proteins, perhaps in intracellular signaling, because they seem to be more homologous to the ras superfamily members than to other GTPases (20). In many cell types examined to date, the septins form rings at the site of the cleavage furrow and septin mutants in S. cerevisiae (22) and Drosophila (17) are defective in cytokinesis. In Drosophila, Pnut is an essential protein. In pnut mutants, cells of the imaginal disk tissues fail to proliferate and instead develop clusters of large multinucleated cells, consistent with an important role in cytokinesis. In S. cerevisiae, septins are required for proper localization of bud site-selection markers Bud3p and Bud4p and of the subunits of the chitin synthase III complex (23–25). These and other data led to the hypothesis that the septins function as a scaffold on which other proteins assemble along the cytoplasmic side of the cleavage furrow (26, 27). How septins themselves localize is unknown; adaptor proteins likely recruit them to specific targets (21).
Whole-cell extracts from Drosophila embryos (0–12 h of development) and L2 tissue culture cells were subjected to immunoprecipitation with polyclonal antibody raised against the Drosophila Orc6 subunit. Coprecipitated material was analyzed by Western blotting for the presence of Pnut protein. As shown in Fig. 1B, polyclonal anti-Orc6 serum coimmunoprecipitated Pnut together with Orc6 protein from both Drosophila embryonic and L2 tissue culture extracts. No Pnut signal was detected in control reactions with polyclonal antibody against either the Drosophila Orc2 subunit or BPV E1 protein. The negative results with the Orc2 sera imply that it is the pool of Orc6 unassociated with the other ORC proteins that interacts with Pnut.
Although we could easily detect coimmunoprecipitation of Pnut with anti-Orc6 antibodies, the reciprocal experiment showed that only a very minor fraction of Orc6 was precipitated by monoclonal antibodies raised against Pnut protein (data not shown). This result was perhaps due to epitope masking in the Orc6–Pnut complex, coupled with the fact that this particular Pnut monoclonal antibody (17) worked best in immunostaining and rather poorly in immunoprecipitations (data not shown).
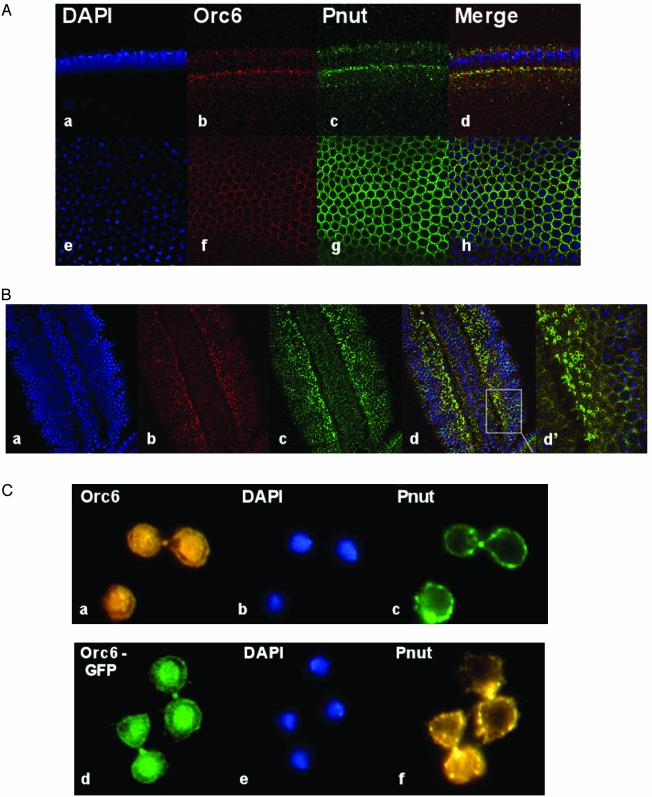
We used both Drosophila embryos at different stages of development and L2 cells to determine whether Orc6 and Pnut proteins colocalize. At the stage of cellularization that occurs after the 13th nuclear division, the Pnut signal became apparent at the advancing membrane front and especially at the cytoplasmic connections between cells and yolk. These yolk plugs maintain actin, myosin, anillin, and the septins in a surrounding ring (28). Orc6 was indeed found with Pnut at these locations (Fig. 2A). Later, as cell membranes grew and eventually reached a full depth, Orc6 was also localized at the membrane locations together with Pnut protein. Orc6 also colocalized with Pnut in 3.5- to 4-h-old embryos (stages 8 and 9; Fig. 2B). At these stages, shortly after gastrulation, cells enter cell cycle 15 when neuroblasts delaminate from the ectoderm. Both Drosophila Orc6 and Pnut proteins were found at the cleavage furrows between dividing cells in the presumptive neurogenic ectoderm (Fig. 2B; see in particular the merge signals showing rings in Fig. 2Bd′).
Fig. 2.
Orc6 and Pnut colocalize in Drosophila cells. (A) Immunofluorescence studies of Orc6 and Pnut in Drosophila embryos at the beginning of the cellularization (=2 h of development). Drosophila embryos were fixed with 1% paraformaldehyde and stained as described in Methods. Antibody against Drosophila Orc6 (3) and Pnut protein (17) were used for staining. 4′,6-diamidino-2-phenylindole (DAPI) staining and merged images are also presented. a–d represent a longitudinal view; e–h represent the corresponding tangential view. (B) Immunofluorescence studies of Orc6 and Pnut in Drosophila embryos at =3.5–4 h of development. DAPI (a), α-Orc6 (b), and α-Pnut (c) antibodies were used for staining. A merged image and a magnified section (d and d′) are also presented. (C) Pnut and Orc6 colocalize in Drosophila L2 cells. Drosophila L2 cells were fixed with 2% paraformaldehyde and stained as described (10). Drosophila Orc6 and Pnut antibodies were used, and images of endogenous proteins are shown (a and c). Localization of ectopically expressed GFP–Orc6 fusion protein in L2 cells is also presented (d), along with the endogenous Pnut detection (f). Overexpression of GFP–Orc6 fusions and staining has been described (12).
Double staining of Drosophila L2 tissue culture cells with Orc6 and Pnut antibodies (Fig. 2C) also showed a colocalization in dividing cells. In addition to its anticipated nuclear localization, endogenous Orc6 was localized to the cell membranes together with Pnut protein. Moreover, in mitotic cells Orc6 and Pnut colocalized at the cleavage furrow of the dividing cells (Fig. 2C). Transient ectopic expression of the GFP–Orc6 fusion protein again showed distinct nuclear and membrane localization of the protein (Fig. 2Cd). Similar GFP fusions with ORC1 and two genes elicited only nuclear signals (ref. 12 and data not shown), and cytology with ORC2 antibodies detected only a nuclear stain in embryos, as reported (10).
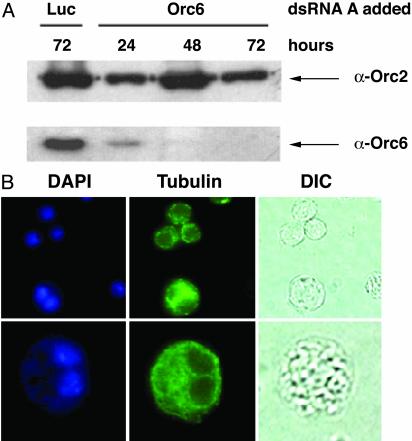
Because pnut mutations in Drosophila result in a cytokinesis defect, we explored the role of Orc6 in this process by depletion with RNAi. After transfection of the appropriate double-stranded RNA into an asynchronous population of Drosophila cells, immunoblot analysis of treated L2 cells revealed that the level of Orc6 protein was greatly reduced by 24 h and almost completely lost by 72 h (Fig. 3A). The level of Orc2 protein was not significantly decreased in cells treated with Orc6 dsRNA, and the cells transfected with luciferase dsRNA as a control showed normal levels of Orc2 and Orc6 proteins during the time course of this experiment (Fig. 3A, lane 1, and data not shown). Immunostaining of transfected cells with anti-Orc6 antibody showed a concomitant disappearance of the Orc6 signal (data not shown). DNA replication in cells treated with Orc6 dsRNA also decreased over time. BrdUrd incorporation was detected in 70–80% of these cells after the first 24 h of incubation, similar to what was observed with untreated cells. However, the fraction of cells incorporating the precursor nucleotide dropped continuously (40–50% at 48 h and 10–15% at 72 h). The most striking phenotype observed after the transfection of L2 cells with Orc6 dsRNA was the rapid appearance of binucleated cells (Fig. 3B). The number of multinucleated cells observed increased from a background of <0.2% in the population to 5% after the first 24 h and reached =30% after 72 h of transfection. Prolonged periods of the Orc6 depletion by RNAi resulted in a decrease of cell proliferation and increased cell death. This multinucleated phenotype was not observed in vivo for lethal mutations of ORC subunits 2, 3, or 5 (4–7). Moreover, dsRNA for ORC2, although effective for decreasing BrdUrd incorporation, showed no efficacy for elevating the number of binucleated cells above the background. However, such defects were observed in RNAi-based studies of Drosophila passenger proteins INCENP and Aurora B (29). From our data, we speculate that Orc6 participates in some aspects of the cell division cycle that influences cytokinesis. Furthermore, the kinetics of cytokinesis defects suggest that the cytokinetic function is more sensitive to small changes in the Orc6 pool than is DNA replication.
Fig. 3.
Silencing of Orc6 in Drosophila L2 cells by dsRNA causes the multinucleation. (A) Immunoblotting of whole-cell extract from Drosophila L2 cells transfected with Orc6 dsRNA or control luciferase dsRNA (Luc) (20 μg of RNA per well of a six-well dish) and harvested after 24, 48, or 72 h. RNAi efficiency was tested with the Orc6 antibody. An immunoblot with Orc2 was used as a loading control. The levels of Orc2 and Orc6 proteins after transfection with luciferase did not change during 3 days of study. (B) Drosophila L2 cells, transfected with Orc6 dsRNA as described above, were stained with DAPI and α-tubulin antibody after 72 h. Up to 30% of cells were multinucleated at that time. Differential interference contrast microscopy images (DIC) are also shown.
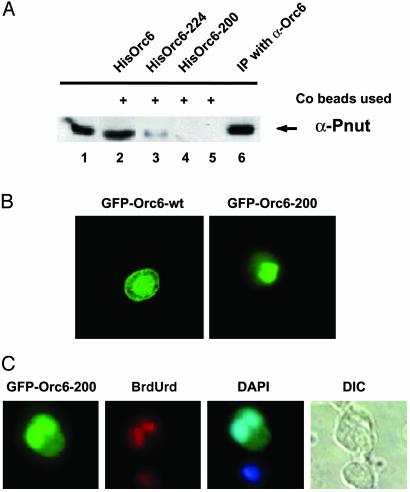
As noted before (12), the C-terminal 25 aa of Drosophila Orc6 contains a leucine-rich region that may mediate protein–protein interactions through an amphipathic helix. A similar motif, thought to be important for protein–protein interaction, is also found in the Pnut protein and is present in most of the septins described to date (20). We tested the notion that this region of Orc6 is important for Pnut interaction with a series of Orc6 C-terminal deletion mutants. Purified WT and mutant proteins expressed in E. coli as His-tagged fusions were tested for their ability to precipitate Pnut protein from the Drosophila cell culture extracts. Proteins were precipitated by using cobalt beads (Qiagen) that can bind selectively His-tagged proteins. Precipitated material was analyzed by employing a Western immunoblotting assay (Fig. 4A) and the α-Pnut antibody for detection. In contrast to the WT Orc6 His-tagged protein, Orc6 mutant protein lacking the terminal 57 aa (Orc6–200) failed to precipitate Pnut from the Drosophila extracts. Orc6 protein truncated at the terminal 33 aa (Orc6–224) was able to precipitate Pnut protein but less efficiently than the intact protein (Fig. 4A, lane 3). Thus, the terminal leucine-rich section of Orc6 is important for Pnut interaction but not likely the sole mediator. Equivalent results were obtained with Drosophila embryonic extracts (data not shown).
Fig. 4.
The C-terminal domain of Orc6 is essential for interaction with Pnut protein and cell division. (A) His-tagged purified Drosophila WT Orc6 protein (HisOrc6, lane 2) and Orc6 mutant proteins (HisOrc6–224, lane 3, and HisOrc6–200, lane 4) were incubated with Drosophila L2 extracts. Precipitated with cobalt beads, material was analyzed for presence of Pnut protein during immunoblot experiment with α-Pnut antibody. No recombinant protein was added in lane 5; 1 μl of L2 extract was loaded on lane 1. Lane 6 shows Pnut protein precipitated from L2 extract by antibody raised against Drosophila Orc6 protein. (B) Orc6–200 deletion mutant protein does not localize to cell membrane in L2 cells after overexpression. L2 cells expressing recombinant GFP fusions of Orc6-WT and Orc6–200 deletion mutant are presented. (C) Overexpression of Orc6–200 causes the formation of multinucleated cells, which are still able to incorporate BrdUrd. Cells were labeled with BrdUrd for 20 h after transfection with the GFP-fusion Orc6–200 mutant protein and stained with anti-BrdUrd antibody. Presented images show the GFP signal, BrdUrd incorporation, DAPI staining, and DIC images from left to right. These data and those with other mutants are presented quantitatively in Table 1.
We asked whether these deletion mutants might have dominant-negative effects if expressed in cultured cells. The results are summarized in the Table 1. Overexpression of WT Orc6 protein did not produce any noticeable effect on either cell morphology or the ability of the cells to replicate DNA in side-by-side comparison to nontransfected cells. However, the overexpression of C-terminal Orc6 mutants resulted in an elevated number of cells with multiple nuclei (5–7% of transfected cells for Orc6–224 and up to 30% for Orc6–200), as shown in Table 1 and Fig. 4C. Overexpression of the Orc6–200 allele also resulted in a loss of membrane localization (Fig. 4B). Cells carrying GFP–Orc6–224 or GFP–Orc6–200 were able to incorporate BrdUrd during a 20-h labeling period at the level of nontransfected cells or cells transfected with GFP–Orc6 WT, as judged by intensity of staining and the fraction of cells with signal (Table 1 and Fig. 4C). Further, by using our recombinant ORC system (13), the ORC6–200 allele, when expressed with the other subunits, incorporated effectively into the complex and thus could compete with the WT gene for complex formation when it is overexpressed (data not shown). This result suggests that the core replication domain of Orc6 was not affected by these mutations, although the cytokinetic function as measured by the presence of binucleated cells was lost. The antimorphic nature of alleles such as GFP–ORC6–200 indicates that the replication domain may also contain critical functions for cytokinesis, but that in the absence of the C terminus the defective protein interferes with the process. For example, the ORC6–200 may bind and sequester a protein important for releasing ORC6 from chromatin to transport the protein to the membrane locations. Overexpression of Orc6–163 mutant protein, on the other hand, resulted in large cells that did not incorporate BrdUrd. Moreover, transfection of L2 cells with Orc6–163 apparently had a toxic effect on cells and resulted in decreased proliferation and increased cell death.
Table 1. Ectopic expression of Drosophila Orc6 in L2 cells.
| Drosophila Orc6 protein | BrdUrd incorporation, % nuclei | Multinucleated cells, % cells |
|---|---|---|
| Orc6-WT | 70-80 | 0-0.2 |
| Orc6-224 | 70-80 | 5-7 |
| Orc6-200 | 70-80 | 25-30 |
| Orc6-163 | 2-5 | ND* |
| Control† | 70-80 | 0-0.2 |
Not determined; abnormal cell morphology obscured nuclei.
No transfection control; cells have endogenous ORC.
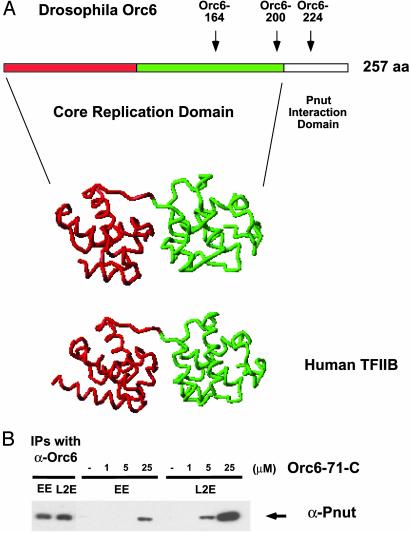
A reasonable extrapolation from these data is that a C-terminal domain of Orc6, perhaps beginning around amino acid 200 and progressing from there toward the C terminus, defines a Pnut interaction domain and a membrane-proximal localization function critical for cytokinesis. To probe the domain organization of Orc6 in silico, we used a web-based method for protein fold prediction that employs 1D and 3D sequence profiles coupled with secondary structure and solvation potential information (ref. 30; www.sbg.bio.ic.ac.uk/~3dpssm). With this program, we compared Drosophila and human Orc6 sequences with known protein structures. Unexpectedly, we found that the predicted Orc6 structure over much of its length was homologous to the structure of the human TFIIB transcription factor bound to the DNA in a complex with TBP (31). The E value, an inverse measure of the program's reliability for this alignment, was commensurate with a certainty of >99.9% for the human Orc6 and 99.2% for the Drosophila Orc6 homologue. Two points aside from the hypothetical nature of this modeling are emphasized here. (i) The break in the predicted TFIIB homology domain is at amino acid 203, and the C-terminal amino acids of Orc6 do not fit into this fold, in rather good agreement with our biochemical and cell-based genetic assays. Deletion alleles map an approximate break in functional activities to this region. (ii) A recent report (32) also presented data consistent with a cleavage furrow localization of human Orc6 and function in cytokinesis. Ablation of human ORC6 expression in cultured cells via the RNAi method also leads to a rapid appearance of binucleated cells and a decrease in DNA replication. The domain structure homology predictions are noted as even higher for the human protein. These points and a view of the predicted homology with TFIIB are illustrated in Fig. 5A.
Fig. 5.
Drosophila Orc6 protein may have structural homology to TFIIB, and the C terminus separate from this region is sufficient to bind Pnut. (A) The predicted core replication domain (amino acids 1–203) and Pnut interacting domains of Orc6 are shown in the line. The endpoints of the mutants used in this study are indicated by arrows. Human TFIIB has a two-domain structure indicated by the red and green, and a similar structure is predicted for ORC6. (B) An Orc6 C-terminal peptide corresponding to the last 71 amino acids (186–257) interacts with Pnut protein. Peptide (Orc6–71-C) was labeled with biotin and incubated with 25 μlof Drosophila embryonic (EE) or L2 whole-cell extract (L2E) for 1 h at 4°C. The micromolar concentration of the peptide is indicated above the lines. The peptide was precipitated by streptavidin paramagnetic beads (Promega), and one-third of the material was analyzed for Pnut. The amount of Pnut precipitated by endogenous Orc6 from an equivalence of 8 μl is shown alongside (EE and L2E).
We asked whether the C-terminal region of Orc6 was both necessary and sufficient for Pnut interaction as anticipated from the hypothesis that this region defines a discrete domain of the protein. A peptide corresponding to the last 71 aa of Drosophila Orc6 protein was synthesized, linked by a spacer to biotin, and used to investigate this point. The biotin-labeled peptide was incubated with Drosophila embryonic whole-cell extract and then reisolated by using paramagnetic beads coupled with streptavidin (Promega). The bound material was subjected to SDS/PAGE and analyzed by Western blotting for the presence of Drosophila Pnut protein. The Orc6 C-terminal peptide was able to precipitate Pnut from the extract with a concentration as low as 5–10 μmol (Fig. 5B). Streptavidin beads alone or a control peptide with a similar amino acid composition and length linked to avidin served as negative controls. We conclude that this C-terminal region is a distinct domain of the protein. The putative structural homology between ORC6 and TFIIB can be tested, for example, by physical methods, and if borne out it would bring forth the notion that certain proteins involved in the initiation of replication coevolved with proteins important for transcription. In this context, it is intriguing that archael organisms have a single gene encoding an Orc1 family member and a TFB (TFIIB homologue) (33); perhaps the respective encoded proteins interact.
The primary question raised by these findings might be posed as follows: Does the role of Orc6 in cytokinesis actually link the regulation of DNA replication to this late step in cell division? A priori, we might envision that the first steps toward building a prereplication complex in early G1 or late telophase might be tied to the successful completion of cytokinesis. Orc6 molecules at the cleavage furrow might participate in some event during cytokinesis and then after execution shuttle to chromosomes perhaps with other proteins. This shuttling might make dependent the completion of a cytokinetic function to the start of a new round of replication. This model posits a late step in cytokinesis for ORC6 that might couple the cytokinetic and DNA replication pathways. Alternatively, Orc6 may participate in some early role in cytokinesis assisting in a targeting function for septins in metazoans, thus potentially linking assembly of such septin rings at the cleavage furrow to the completion of DNA replication.
Acknowledgments
We thank Dr. Tom Alber (University of California, Berkeley) for guiding us to the structure database for Orc6 homologies, Dr. Steven Brenner and Emma Hill (University of California, Berkeley) for detailed discussions relevant to this search, and Dr. Barbara Meyer for her comments on the manuscript. Dr. David King (Howard Hughes Medical Institute) synthesized the peptides used in Fig. 5. This work was supported by American Cancer Society Grant IRG-60-001-44, National Cancer Institute Grant CA-13148-31 (both to I.N.C.), and National Institutes of Health Grant CA30490 (to M.B.).
Abbreviations: ORC, origin recognition complex; RNAi, RNA interference; dsRNA, double-stranded RNA; X-gal, 5-bromo-4-chloro-3-indolyl β-d-galactoside; DAPI, 4′,6-diamidino-2-phenylindole.
References
- 1.Bell, S. P. & Dutta, A. (2002) Annu. Rev. Biochem. 71, 333–374. [DOI] [PubMed] [Google Scholar]
- 2.Gossen, M., Pak, D. T., Hansen, S. K., Acharya, J. K. & Botchan, M. R. (1995) Science 270, 1674–1677. [DOI] [PubMed] [Google Scholar]
- 3.Chesnokov, I., Gossen, M., Remus, D. & Botchan, M. (1999) Genes Dev. 13, 1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landis, G., Kelley, R., Spradling, A. C. & Tower, J. (1997) Proc. Natl. Acad. Sci. USA 94, 3888–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loupart, M. L., Krause, S. A. & Heck, M. S. (2000) Curr. Biol. 10, 1547–1556. [DOI] [PubMed] [Google Scholar]
- 6.Pinto, S., Quintana, D. G., Smith, P., Mihalek, R. M., Hou, Z. H., Boynton, S., Jones, C. J., Hendricks, M., Velinzon, K., Wohlschlegel, J. A., et al. (1999) Neuron 23, 45–54. [DOI] [PubMed] [Google Scholar]
- 7.Pflumm, M. F. & Botchan, M. R. (2001) Development (Cambridge, U.K.) 128, 1697–1707. [DOI] [PubMed] [Google Scholar]
- 8.Wuarin, J., Buck, V., Nurse, P. & Millar, J. B. (2002) Cell 111, 419–431. [DOI] [PubMed] [Google Scholar]
- 9.Shore, D. (2001) Curr. Biol. 11, R816–R819. [DOI] [PubMed] [Google Scholar]
- 10.Pak, D. T., Pflumm, M., Chesnokov, I., Huang, D. W., Kellum, R., Marr, J., Romanowski, P. & Botchan, M. R. (1997) Cell 91, 311–323. [DOI] [PubMed] [Google Scholar]
- 11.Huang, D. W., Fanti, L., Pak, D. T., Botchan, M. R., Pimpinelli, S. & Kellum, R. (1998) J. Cell Biol. 142, 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chesnokov, I., Remus, D. & Botchan, M. (2001) Proc. Natl. Acad. Sci. USA 98, 11997–12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhar, S. K. & Dutta, A. (2000) J. Biol. Chem. 275, 34983–34988. [DOI] [PubMed] [Google Scholar]
- 14.Moon, K. Y., Kong, D., Lee, J. K., Raychaudhuri, S. & Hurwitz, J. (1999) Proc. Natl. Acad. Sci. USA 96, 12367–12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vashee, S., Simancek, P., Challberg, M. D. & Kelly, T. J. (2001) J. Biol. Chem. 276, 26666–26673. [DOI] [PubMed] [Google Scholar]
- 16.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (2001) Current Protocols in Molecular Biology (Wiley, New York).
- 17.Neufeld, T. P. & Rubin, G. M. (1994) Cell 77, 371–379. [DOI] [PubMed] [Google Scholar]
- 18.Longtine, M. S., DeMarini, D. J., Valencik, M. L., Al-Awar, O. S., Fares, H., De Virgilio, C. & Pringle, J. R. (1996) Curr. Opin. Cell Biol. 8, 106–119. [DOI] [PubMed] [Google Scholar]
- 19.Trimble, W. S. (1999) J. Membr. Biol. 169, 75–81. [DOI] [PubMed] [Google Scholar]
- 20.Field, C. M. & Kellogg, D. (1999) Trends Cell Biol. 9, 387–394. [DOI] [PubMed] [Google Scholar]
- 21.Kinoshita, M., Field, C. M., Coughlin, M. L., Straight, A. F. & Mitchison, T. J. (2002) Dev. Cell 3, 791–802. [DOI] [PubMed] [Google Scholar]
- 22.Hartwell, L. H. (1971) Exp. Cell Res. 69, 265–276. [DOI] [PubMed] [Google Scholar]
- 23.Chant, J. & Pringle, J. R. (1995) J. Cell Biol. 129, 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanders, S. L. & Herskowitz, I. (1996) J. Cell Biol. 134, 413–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeMarini, D. J., Adams, A. E., Fares, H., De Virgilio, C., Valle, G., Chuang, J. S. & Pringle, J. R. (1997) J. Cell Biol. 139, 75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longtine, M. S., Fares, H. & Pringle, J. R. (1998) J. Cell Biol. 143, 719–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Longtine, M. S., Theesfeld, C. L., McMillan, J. N., Weaver, E., Pringle, J. R. & Lew, D. J. (2000) Mol. Cell. Biol. 20, 4049–4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glotzer, M. (2001) Annu. Rev. Cell Dev. Biol. 17, 351–386. [DOI] [PubMed] [Google Scholar]
- 29.Adams, R. R., Maiato, H., Earnshaw, W. C. & Carmena, M. (2001) J. Cell Biol. 153, 865–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelley, L. A., MacCallum, R. M. & Sternberg, M. J. (2000) J. Mol. Biol. 299, 499–520. [DOI] [PubMed] [Google Scholar]
- 31.Tsai, F. T. & Sigler, P. B. (2000) EMBO J. 19, 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prasanth, S. G., Prasanth, K. V. & Stillman, B. (2002) Science 297, 1026–1031. [DOI] [PubMed] [Google Scholar]
- 33.Bult, C. J., White, O., Olsen, G. J., Zhou, L., Fleischmann, R. D., Sutton, G. G., Blake, J. A., FitzGerald, L. M., Clayton, R. A., Gocayne, J. D., et al. (1996) Science 273, 1058–1073. [DOI] [PubMed] [Google Scholar]
- 34.Sun, S., Thorner, L., Lentz, M., MacPheerson, P. & Botchan, M. (1990) J. Virol. 64, 5093–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]