Abstract
We have identified and cloned a human liver cDNA encoding an unusual mosaic polyprotein, called polyserase-I (polyserine protease-I). This protein exhibits a complex domain organization including a type II transmembrane motif, a low-density lipoprotein receptor A module, and three tandem serine protease domains. This unusual modular architecture is also present in the sequences predicted for mouse and rat polyserase-I. Human polyserase-I gene maps to 19p13, and its last exon overlaps with that corresponding to the 3′ UTR of the gene encoding translocase of mitochondrial inner membrane 13. Northern blot analysis showed the presence of a major polyserase-I transcript of 5.4 kb in human fetal and adult tissues and in tumor cell lines. Analysis of processing mechanisms of polyserase-I revealed that it is synthesized as a membrane-associated polyprotein that is further processed to generate three independent serine protease units. Two of these domains are proteolytically active against synthetic peptides commonly used for assaying serine proteases. These proteolytic activities of the polyserase-I units are blocked by serine protease inhibitors. We show an example of generation of separate serine protease domains from a single translation product in human tissues and illustrate an additional mechanism for expanding the complexity of the human degradome, the entire protease complement of human cells and tissues.
Proteases comprise a growing group of structurally and functionally diverse proteins that share a common property: the ability to hydrolyze peptide bonds (1). To date, >500 different human proteases and protease homologs have been identified (2, 3). Functional studies involving this large group of enzymes were originally aimed at analyzing their role in nonspecific reactions associated with protein catabolism. However, over recent years, it has been widely recognized that proteolysis represents a fundamental mechanism for regulating many events on which cell life and death depend. Thus, proteases through their ability to perform highly selective and limited cleavage of specific substrates may control the appropriate intracellular or extracellular localization of multiple proteins, the ectodomain shedding from membrane-bound proteins, or the activation and inactivation of cytokines, growth factors, and peptide hormones (2, 3). These proteolytic processing events influence essential biological processes including cell-cycle progression, morphogenesis, tissue remodeling, cell proliferation, cell migration, angiogenesis, and apoptosis (4–8). Furthermore, alterations in the structure, function, and regulation of proteases underlie several human diseases such as cancer, arthritis, cardiovascular disorders, and neurodegenerative diseases (9–13).
The increasing complexity of human proteolytic systems has made necessary the introduction of global concepts and approaches to try to clarify the multiple questions that have arisen in the protease field. Thus, we have coined the term degradome to define the complete set of proteases that are produced at a specific moment or circumstance by a cell, tissue, or organism (2). Recently, and as part of our studies focused on trying to get a complete view of the human degradome, we observed the presence of three putative serine protease genes closely linked at a region of the short arm of chromosome 19. In this work, we report the finding that these three predicted independent genes are in fact part of a unique gene encoding a human polyprotease that we have tentatively called polyserase-I. We also show that polyserase-I undergoes a series of proteolytic processing events to yield three independent serine protease domains. This is an example of generation of independent serine protease domains in human tissues from a single translation product and illustrates an additional strategy for increasing the complexity of the human degradome.
Materials and Methods
Molecular Cloning. A computer search of human genome databases allowed the identification of three regions in the human DNA contig NT_011245 (chromosome 19p13), which after conceptual translation, showed the presence of structural hallmarks for serine proteases. Then, a PCR-based approach to clone cDNAs for these proteases was designed. The strategy first involved the use of specific oligonucleotides derived from the central putative protease gene present in the cluster. The sequences of the designed primers Polys2f, Polys2f-nd, Polys 2r, and Polys2r-nd are indicated in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. The PCR was performed in a GeneAmp 2400 PCR system (Perkin–Elmer Life Sciences) for 40 cycles of denaturation (94°C, 20 s), annealing (64°C, 15 s), and extension (68°C, 60 s). The amplified PCR product (439 bp) from a human liver cDNA library was cloned and sequenced. The 5′ and 3′ ends of the cloned cDNA were extended by successive rounds of rapid amplification of cDNA ends (RACE) by using RNA from human liver and the Marathon cDNA amplification kit (CLONTECH). Sequence analysis of the RACE-extended cDNA clones led us to conclude that the two additional protease genes present at the 5′ and 3′ ends of the central protease gene of the NT_011245 DNA contig were in fact part of a single gene encoding a protein with three protease-like domains. Once the start and stop codons of this large gene were located, the full-length cDNA was obtained by PCR using the primers polysATG and polysEND (Table 1). PCR conditions were as above, but with 180 s of extension.
Northern Blot Analysis. Nylon filters containing poly(A)+ RNA of diverse human tissues were prehybridized at 42°C for 3 h in 50% formamide, 5× standard saline phosphate/EDTA (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA), 10× Denhardt's solution, 2% SDS, and 100 μg/ml denatured herring sperm DNA. Hybridization was performed with a 0.5-kb radiolabeled probe amplified from the cDNA clone with oligonucleotides northF and northR (Table 1). After hybridization for 20 h under the same conditions used for prehybridization, filters were washed with 0.1× SSC, 0.1% SDS for 2 h at 50°C, and autoradiographed.
Production and Purification of Recombinant Proteins. Two cDNA contructs encoding the first or the second serine protease domains of the cloned cDNA were generated by PCR amplification using the oligonucleotides mod1f, mod1r, mod2f, and mod2r (Table 1). The PCR amplifications were performed for 25 cycles of denaturation (95°C, 15 s), annealing (59°C, 10 s), and extension (68°C, 50 s). The PCR products were cloned in the SmaI site of the pGEX-3X expression vector (Amersham Pharmacia Biosciences). The resulting vectors, pGEX-3X-serase-1 and pGEX-3X-serase-2, were transformed into BL21(DE3)pLysE Escherichia coli cells, and expression was induced by isopropyl β-d-thiogalactoside. The recombinant proteins were purified on a glutathione-Sepharose 4B column and used for enzymatic assays.
Protease Assays. The putative enzymatic activity of the recombinant GST-serase-1 and GST-serase-2 proteins was analyzed by using the synthetic fluorescent substrates N-t-Boc-Gln-Ala-Arg-7-amino-4-methylcoumarin (AMC), N-t-Boc-Gln-Gly-Arg-AMC, N-t-Boc-Ala-Pro-Ala-AMC, and N-t-Boc-Ala-Phe-Lys-AMC. Routine assays were performed at 37°C at substrate concentration of 100 μM in an assay buffer containing 50 mM Tris·HCl (pH 8.0), 20 mM NaCl, and 2.5% Me2SO. The fluorometric measurements were made in a MPF-44A Perkin–Elmer spectrofluorometer (λex = 360 nm, λem = 460 nm). Kinetic studies were performed by using different concentrations of the fluorogenic peptide (5–50 μM) in 100 μl of assay buffer containing 25 nM of recombinant proteases, and peptide hydrolysis was measured as the increase in fluorescence at 37°C for different times. Initial velocities and kcat/Km ratio were calculated as described (14). For inhibition assays, the recombinant proteins were preincubated for 120 min at 37°C with 2 mM synthetic or 100 nM endogenous proteinase inhibitors (Calbiochem), and the hydrolyzing activity of polyserase domains was determined by fluorometric measurements as above.
Membrane Immunolocalization. Full-length polyserase-I cDNA tagged with an hemagglutinin (HA) epitope was subcloned into pcDNA3 expression vector and transfected into COS-7 cells. Fluorescent detection was performed by incubating the slides with mAb 12CA5 (Roche Molecular Biochemicals) against HA or with anti-serase-2 polyclonal antibody, followed by another incubation with goat anti-mouse fluoresceinated antibody or a rhodamine-conjugated anti-rabbit antibody. Slides were analyzed with a confocal laser microscope, Leica TCS-SP2-AOBS.
Western Blot Analysis. The polyserase-I cDNA tagged with a FLAG epitope at the 5′ end and an HA epitope at the 3′ end was transfected into HeLa and COS-7 cells. Polyclonal antibodies against the first and second protease modules produced in E. coli were raised in rabbits following standard procedures. We also prepared rabbit polyclonal antibodies against a synthetic peptide derived from the third protease module (FDVYGDP-KQWAAF; positions 870–882) and conjugated to maleimide-activated keyhole limpet hemocyanin. For Western blot analysis, proteins were separated on SDS/PAGE, blotted onto nitrocellulose membranes, and incubated with the different antibodies. To obtain membrane-enriched fractions, HeLa and COS-7 cells were scraped from the plates, and membrane fractions were prepared as described (15).
Results
Isolation and Characterization of a Human Liver cDNA Encoding Polyserase-I, a Serine Polyprotease. A search of the human genome databases looking for novel protease genes led us to the identification of a DNA contig containing three regions putatively encoding new serine proteases. However, detailed sequence analysis of these three regions revealed that they were very close to each other, suggesting that they could be part of a single gene containing different protease modules. To explore this possibility, we performed PCR amplifications with specific oligonucleotides derived from the central protease module, followed by successive rounds of 5′ and 3′ rapid amplifications of cDNA ends. This strategy led us to generate a cDNA of 3,180 bp, which after cloning and sequencing, revealed the presence of a single RNA transcript containing the coding information for the three predicted serine protease domains (Fig. 1). Computer analysis of this cDNA sequence demonstrated that it codes for a protein of 1,059 aa, with a calculated molecular weight of 114,020. The proposed AUG initiation codon is located in a context (GCCAUGG) that matches perfectly the Kozak consensus sequence for translation initiation (16) and is immediately preceded by an in-frame UGA stop codon, strongly suggesting that this AUG corresponds to the true initiation site for this protein. Domain analysis using the smart tool and the tmhmm (transmembrane helices Markov model) program, indicated that the protein possesses a type II transmembrane segment and a low-density lipoprotein receptor (LDLR) domain. Following this domain, the three protease domains are easily recognized: serase-1 (for serine protease-1) from positions 190 to 433, serase-2 from 491 to 733, and serase-3 from 816 to 1059 (Fig. 6, which is published as supporting information on the PNAS web site). Sequence alignment also shows that serases-1 and -2 contain the putative catalytic Ser residue within the consensus motif Gly-Asp-Ser-Gly-Gly, whereas serase-3 contains an Ala residue (Gly-Asp-Ala-Gly-Gly) instead of the Ser residue in the active site, indicating that this third module should be catalytically inactive (Fig. 6). The presence of Asp residues six amino acids before the catalytic serines in serases-1 and -2 is indicative that both have trypsin-like specificity with preference for substrates containing basic residues in the P1 position. Additional structural hallmarks of serine proteases are also conserved in the identified serases (Fig. 6). Thus, the sequence Ser-Trp-Gly proposed to interact with the side chains of the substrates for a proper orientation of the scissile bond is present in the three serases. The His and Asp residues necessary for catalytic activity are also conserved (His-243 and Asp-292, His-544 and Asp-592, and His-868 and Asp-915, respectively), as well as the six Cys residues involved in the formation of three disulfide bonds in the catalytic region. As in other serine proteases, a fourth disulfide bond is predicted to be formed between Cys-191 located at the prodomain of serase-1 and Cys-312 of the catalytic domain of this domain. Similar bonds would occur between Cys-492 and Cys-612 of serase-2 and between Cys-817 and Cys-935 of serase-3. The formation of the first of these predicted disulfide bonds would imply that the catalytic domains of the polyprotease should still remain bound at the cell surface even after cleavage at the activation sites.
Fig. 1.
Structure of polyserase-I. TM, transmembrane domain; LDLR, LDLR A module; serase, serine protease domain.
All of these structural features present in the identified human protein are also conserved in its putative mouse and rat orthologs, whose sequence was deduced by using the human sequence as query in mouse and rat genome databases (Fig. 6). The gene encoding the rat ortholog is located in chromosome 7 and that for the mouse polyprotein in chromosome 10, in a region syntenic to chromosome 19p13 where the human gene is located. Both rodent orthologs are ≈80% identical to the human enzyme. Further analysis of the predicted sequence for the identified human polyprotease indicated a maximum percentage of identities with matriptase/MT-SP1 (46% with serase-1, 43% with serase-2, and 48% with serase-3), and matriptase 2 (45% with serase-1, 40% with serase-2, and 48% with serase-3), both being members of the TTSP (type II transmembrane serine proteases) family of proteolytic enzymes. To perform a more detailed analysis of the relationship between the identified protein and other serine proteases, we first generated a phylogenetic tree of members of the TTSP family (Fig. 7, which is published as supporting information on the PNAS web site). The dendrogram allows the classification of these proteases into six different subfamilies. Interestingly, the three protease-like domains of the polyprotease fit perfectly within the matriptase subgroup, previously composed of matriptase and matriptase-2. Alignments of these protease domains with all of the remaining members of the human TTSP family as well as with different serine proteases (Fig. 8, which is published as supporting information on the PNAS web site) also confirmed the relationship between the polyprotease and members of the TTSP family of serine proteases. In accordance with this structural analysis, we conclude that the cloned human liver cDNA encodes a novel membrane-bound polyprotein with three putative serine protease modules, which we propose to call polyserase-I (from polyserine protease-I) (GenBank accession no. AJ488946). It is also remarkable that the last exon of the human polyserase-I gene, which begins at position 3050 of the cDNA, overlaps in antisense orientation with part of the 3′ UTR of the translocase of mitochondrial inner membrane 13 (TIMM13) gene, which has been found implicated in human deafness dystonia syndrome (17).
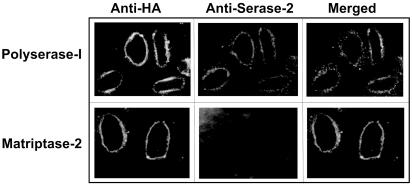
Membrane Localization of Polyserase-I. To provide experimental support to the proposal that polyserase-I is anchored to the plasma membrane, we transfected COS-7 cells with a cDNA construct for this protein containing the HA epitope at the C-terminal end. Transfected cells were then analyzed by immunofluorescence with a mouse mAb (12CA5) specific for this epitope. As can be seen in Fig. 2, a fluorescent pattern surrounding the cell surface was visualized in transfected cells producing polyserase-I. A similar pattern was observed when COS-7 cells were immunostained with an antiserum raised against the serase-2 domain of polyserase-I (Fig. 2). Cells transfected with a cDNA for HA-tagged matriptase-2, a well established membrane-bound serine protease (15, 18), were used as a control of this experiment (Fig. 2). Based on these membrane-localization experiments, we can conclude that polyserase-I is a bona fide member of the growing family of TTSP membrane-bound serine proteinases (19–21) and can be classified as a novel member of the subfamily of matriptases (15, 18, 22).
Fig. 2.
Membrane localization of polyserase-I. Immunofluorescent detection of polyserase-I in COS-7 cells with monoclonal anti-HA and polyclonal anti-serase-2 antibodies. Cells transiently transfected with matriptase-2-HA were used as positive control. Detection of anti-HA and anti-serase-2 antibodies was carried out with a fluoresceinated anti-mouse antibody and a rhodamine-conjugated anti-rabbit antibody, respectively.
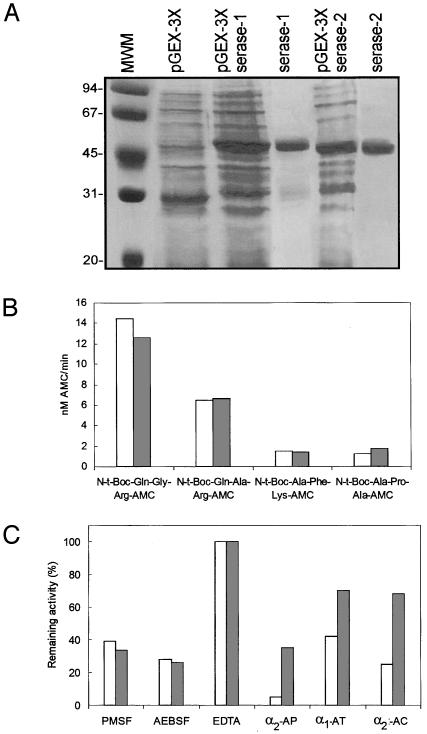
Production of the Recombinant Serase-1 and Serase-2 Domains of Polyserase-I in E. coli and Analysis of Their Enzymatic Properties. To analyze the enzymatic properties of serase-1 and serase-2, we expressed the two predicted catalytically active domains of this membrane-bound polyprotease in bacterial cells. After induction of the transformed bacteria, protein bands of the expected size (52 and 51 kDa, respectively) were detected by SDS/PAGE analysis of the bacterial protein extracts (Fig. 3A). These recombinant fusion proteins were purified by using glutathione-Sepharose chromatography, and the soluble GST-serase-1 and GST-serase-2 proteins eluted from the affinity column were directly used for enzymatic analysis. As described for other GST-fusion proteins (15), we observed that the fusion proteins were apparently autoactivated after incubation at 37°C, showing the presence of an additional 26-kDa band that likely corresponds to the respective catalytic domains after proteolytic release of the GST-moiety (data not shown). Proteolytic activity of the recombinant enzymes was analyzed by using a panel of synthetic quenched fluorescent peptides commonly used for assaying serine proteinases. Both enzymes hydrolyzed the peptides N-t-Boc-Gln-Ala-Arg-AMC and N-t-Boc-Gln-Gly-Arg-AMC, whereas N-t-Boc-Ala-Phe-Lys-AMC and N-t-Boc-Ala-Pro-Ala-AMC were not significantly hydrolyzed by the recombinant proteins (Fig. 3B). Catalytic activities of the two recombinant serases were substantially abolished with the serine protease inhibitors PMSF and 4-(2-aminoethyl)benzenesulfonyl fluoride, but not with EDTA (Fig. 3C). We next evaluated the possibility that endogenous proteinase inhibitors could block the activity of protease domains of polyserase-I. As shown in Fig. 3C, α2-antiplasmin extensively inhibited the hydrolyzing activity of serase-1 against N-t-Boc-Gln-Gly-Arg-AMC and also had a significant inhibitory activity against the serase-2 domain. Other proteinase inhibitors such as α1-antitrypsin and α1-antichymotrypsin also showed some inhibitory activity against serase-1 but had only a minor effect on serase-2 (Fig. 3C). To further characterize the catalytic activity of the protease domains of polyserase-I, we also performed a kinetic study using N-t-Boc-Gln-Gly-Arg-AMC as substrate. To this purpose, the recombinant serase-1 and serase-2 domains were incubated with different concentrations of the fluorogenic substrate, and the respective kcat/Km were deduced as described (14). The observed kcat/Km of serase-1 and serase-2 for N-t-Boc-Gln-Gly-Arg-AMC at pH 8.0 were 6.1 × 104 M–1·s–1 and 5.1 × 103 M–1·s–1, respectively. These values are similar to those determined for mouse matriptase when using N-t-Boc-Gln-Ala-Arg-AMC as substrate (kcat/Km = 6.85 × 103 M–1·s–1) (23), but lower than those for human matriptase when using the substrate Spectrozyme tPA (kcat/Km = 6.9 × 106 M–1·s–1) (20).
Fig. 3.
Production of recombinant serase-1 and serase-2 and analysis of their enzymatic activities. (A) Five microliters of bacterial extracts transformed with pGEX-3X (lane 2) or pGEX-3X-serase-1 (lane 3), pGEX-3X-serase-2 (lane 5), and purified serases (lanes 4 and 6) was analyzed by SDS/PAGE. The sizes of the molecular weight markers (MWM) (lane 1) are indicated on the left. (B) The indicated fluorescent peptides were incubated with purified serases, and fluorometric measurements were made at λex = 360 nm and λem = 460 nm (serase-1, white bars; serase-2, gray bars). (C) Inhibition assays with the purified serases and the substrate N-t-Boc-Gln-Gly-Arg-AMC in the presence or absence of the indicated inhibitors. AP, α2-antiplasmin; AT, α1-antitrypsin; AQ, α1-antichymotrypsin; AEBSF, 4-(2-aminoethyl)benzenesulfonyl fluoride.
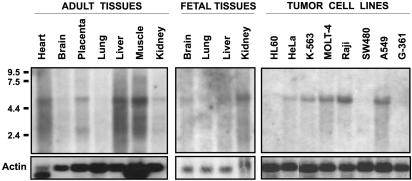
Analysis of Polyserase-I Expression in Human Tissues. To study the pattern of polyserase-I expression in human tissues, we hybridized Northern blots containing poly(A)+ RNA prepared from different tissues and tumor cell lines, with a probe corresponding to the 5′ end of the isolated cDNA. A band of ≈5.4 kb was detected in a panel of fetal human tissues, including kidney, liver, lung, and brain (Fig. 4), and in a variety of tumor cell lines. Minor transcripts of 3.8 and 2.4 kb were also detected in adult tissues, mainly in skeletal muscle, liver, placenta, and heart (Fig. 4). The major 5.4-kb band could correspond to a transcript containing the full-length cDNA described for polyserase-I, whereas minor transcripts could derive from alternative splicing events found during the course of this work (GenBank accession no. AJ488947).
Fig. 4.
Analysis of polyserase-I expression in human tissues and tumor cell lines. Northern blot analysis of polyserase-I expression in fetal and adult human tissues and a panel of tumor cell lines. Approximately 2 μg of polyadenylated RNA from the indicated tissues or tumor cell lines was hybridized with a 5′ probe of the polyserase-I full-length cDNA. The positions of the RNA markers are shown. The filters were subsequently hybridized with an actin probe to ascertain the differences in RNA loading among the different tissues.
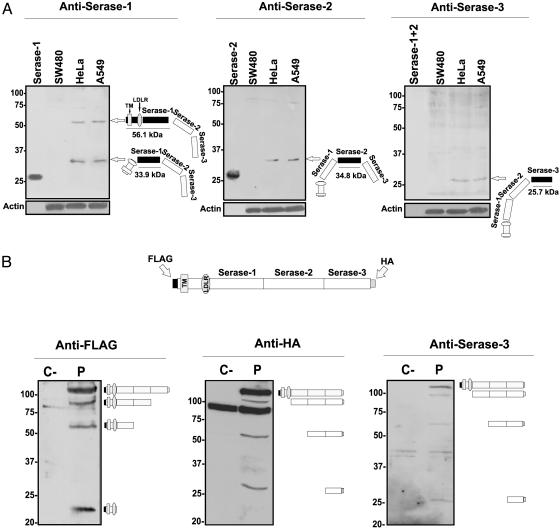
Analysis of the Posttranslational Processing of Polyserase-I. Polyclonal antibodies raised against the three protease modules were used to detect the presence of polyserase-I in HeLa and A549 cells that endogenously express this gene (Fig. 4). As can be seen in Fig. 5A, two bands of ≈55 and 32 kDa were detected by anti-serase-1 antibodies in HeLa and A549 cell extracts, but not in SW480 cells that were used as negative control in these experiments. The 55-kDa serase-1 immunoreactive band could correspond to a protein species comprising the first protease module together with the LDLR and transmembrane domains (56.1 kDa predicted), whereas the 32-kDa protein could correspond to the active form of serase-1 after cleavage by a trypsin-like protease at Arg-202 and Arg-503 (33.9 kDa predicted). Anti-serase-2 antibodies specifically detected a 35-kDa band in HeLa and A549 cells that could correspond to a processed form of serase-2 after cleavage at Arg-503 and Arg-826 activation sites (34.8 kDa predicted). Finally, anti-serase-3 antibodies recognized a band of ≈25 kDa, which closely matches the expected size of this domain after processing at Arg-826 (25.7 kDa predicted). The anti-serase-3 antibodies also detected a band of higher molecular mass that was also present in SW480 cells, and therefore it must correspond to a nonspecific immunoreactivity of these antipeptide antibodies (Fig. 5A).
Fig. 5.
Analysis of polyserase-I posttranslational processing. (A) Western blots probed with the indicated anti-serase antibodies by using extracts of SW480, HeLa, and A549 cells. Recombinant serase-1 and serase-2 were used as controls. Blots were subsequently probed with an anti-β-actin mAb to ascertain differences in loading. (B) Western blots probed with anti-FLAG, anti-HA, and anti-serase-3 antibodies by using membrane-enriched fractions of transfected COS-7 cells. Protein extracts were treated with 2-mercaptoethanol before electrophoresis. The position of each immunoreactive band is shown on the right, and molecular weight markers are indicated on the left. P, cells transfected with the full-length cDNA for polyserase-I; C–, negative control.
To further investigate the putative polyserase-I processing mechanisms, we transfected COS-7 cells with an eukaryotic expression vector for this enzyme containing a FLAG epitope at the N terminus and a HA epitope at the C terminus of the protein. Western blot analysis of membrane-enriched fractions of COS-7-transfected cells with an anti-FLAG antibody indicated that most of the protein remained intact (115 kDa), likely as a consequence of the overexpression generated by the pcDNA3 vector, which saturates the processing pathway of the polyprotein (Fig. 5B). However, additional major bands of 90 kDa (FLAG + transmembrane + LDLR + serase-1 + serase-2), 55 kDa (FLAG + transmembrane + LDLR + serase-1), and 23 kDa were also detected (Fig. 5B). This small protein species could correspond to the FLAG epitope linked to transmembrane and LDLR domains, once the serase-1 domain has been cleaved by a trypsin-like protease after Arg-202 (23.1 kDa predicted). On the other hand, the use of anti-HA antibodies detected a band of 115 kDa likely corresponding to the entire protein, and additional bands of ≈100, 55, and 25 kDa that could correspond to serase-1 + serase-2 + serase-3-HA (94.8 kDa predicted), serase-2 + serase-3-HA (61.3 kDa predicted), and serase-3-HA (26.6 kDa predicted) domains, respectively (Fig. 5B). Furthermore, when filters were hybridized with an anti-serase-3 antibody, the same pattern of specific bands as that obtained with anti-HA antibodies was observed (Fig. 5B). In all cases, similar results were obtained when HeLa cells were used instead of COS-7 cells (data not shown).
Taken together, these data indicate that polyserase-I is a membrane-bound protein that undergoes a series of proteolytic processing events that may result in the release of its three distinct protease units. These proteins would likely remain associated with the cell membrane because of the putative disulfide bonds formed between cysteine residues located in the prodomain and the catalytic domain of each serase domain. Nevertheless, we also have preliminary evidence of the presence of low amounts of the three serine protease domains in conditioned medium from cells transfected with the full-length polyserase-I cDNA (data not shown). These observations suggest that polyserase-I may undergo shedding processes similar to those reported to occur in other members of the TTSP family of membrane-associated serine proteases (23, 24), leading to the secretion of the processed products into the extracellular space.
Discussion
We have cloned a cDNA encoding a mosaic protein that shows a complex organization, including a type II transmembrane region, an LDL receptor module, and three serine protease domains. According to structural and functional features, this complex protein is a member of the TTSP family of membrane-bound serine proteases and has been called polyserase-I. The presence of several catalytic units embedded in a single polypeptide chain is very unusual, and to our knowledge, there are just two additional human proteases, angiotensin-converting enzyme (ACE) and carboxypeptidase D, that exhibit some similarities with polyserase-I in their domain architecture, but also marked differences in the posttranslational processing. ACE is a type-I membrane metalloprotease widely studied because of its key function in the renin-angiotensin system and its relevance as a therapeutic target in cardiovascular diseases (25). This enzyme contains two metalloproteinase domains generated by duplication from an ancestral gene. The two ACE domains are enzymatically active, but show different catalytic constants and interact differently with several competitive inhibitors (26–28). There are several forms of ACE with different structural organization. The endothelial or somatic form contains both metalloproteinase domains within the same polypeptide chain, whereas the testicular or germinal form contains only the C-terminal domain and derives from the use of an alternative transcription start site in intron 13 of the ACE gene (29). There is also an ileal form that contains only the N-terminal metalloprotease domain, although its origin is still unclear (30). Carboxypeptidase D is also a type-I membrane-bound enzyme that contains three metalloproteinase domains, two of them being catalytically active and showing complementary activities. The first domain exhibits an optimal activity at pH 6.3–7.5, and the second one at pH 5.0–6.5, thereby allowing this enzyme to show a broad activity against a wide range of substrates (31). The three protease units present in carboxypeptidase D are an integral part of the complete multidomain structure and are not cleaved from the unique translation product. By contrast, polyserase-I undergoes a series of proteolytic processing events that lead to the generation of three independent protease units. Two of these serine protease domains are active and show hydrolyzing activity against synthetic substrates used for assaying serine proteases. These activities are extensively inhibited by synthetic and endogenous serine protease inhibitors, providing additional evidence on the activity of these two polyserase-I domains as serine proteases. The third serine protease domain should belong to the category of protease-homologs, owing to the presence of changes in specific amino acid residues located within critical active-site regions. These inactive protease-homologs generate growing interest because of their putative roles as regulatory or inhibitory molecules, acting as dominant negatives by binding substrates through the inactive catalytic domains. Accordingly, the third serine-protease module of polyserase-I could somewhat regulate the activity of the two catalytic active domains of this polyprotease.
The polyserase-I processing described herein demonstrates a human polyprotease with the ability to generate independent protease domains from a single translation product. Furthermore, the finding that antibodies against the individual protease domains do not recognize significant amounts of the entire translation product in cells that endogenously produce polyserase-I strongly suggests that the processed form is the naturally occurring one in these cells. This situation, albeit extremely unusual, is not unprecedented in other organisms and resembles that recently described for ovochymase from Xenopus laevis, another complex mosaic protein with three serine protease domains that are released after translation of a single polyprotein product (32). Likewise, oviductin from Xenopus laevis or Bufo japonicus (33, 34) contains two serine protease domains, one of them catalytically inactive, that also undergo posttranslational proteolytic processing events to generate independent units. Both ovochymase and oviductin are polyproteases involved in fertilization processes, although they do not contain transmembrane domains and therefore do not belong to the TTSP family, as is the case of polyserase-I. Recent analysis of the genome sequence of model organisms such as Drosophila melanogaster and Caenorhabditis elegans has also revealed some predicted genes with several serine protease domains, although it is unclear whether they are integral part of a single protein or are posttranslationally cleaved into separate functional modules (35, 36).
The complex mosaic architecture of polyserase-I is absolutely conserved in the predicted sequence for its mouse and rat orthologs, although the functional advantages derived from this multidomain design are still unclear. In this work, and as a previous step to try to elucidate the physiological role of polyserase-I, we have also examined its expression pattern in human tissues. Polyserase-I expression was detected in a variety of human tissues and tumor cell lines, albeit at low levels in most cases. There was also evidence of additional transcripts generated by alternative splicing events, thereby providing an additional mechanism of generating diversity in polyserase-I. Our finding that polyserase-I belongs to the matriptase subfamily of TTSPs could suggest that polyserase-I plays functional roles similar to those previously suggested for matriptase/MT-SP1 and matriptase-2, its closest relatives in the TTSP family of membrane-bound proteases (15, 18–22). Nevertheless, further studies will be required to establish putative functional connections of polyserase-I to matriptases or other members of the growing TTSP protease family. In this regard, it is remarkable that the physiological roles of most TTSPs are still unclear, although there are some cases in which their participation in specific functions has been reported (reviewed in refs. 19 and 21). This is the case for enteropeptidase, which is involved in digestive processes through its role in the proteolytic activation of trypsinogen to trypsin (37, 38); hepsin has been proposed to be involved in liver homeostasis (39, 40); matriptase/MT-SP1 plays a role in epidermal development (41); TMPRSS3 participates in inner ear development and mutations in the gene encoding this protease cause a form of deafness (42), and finally, corin acts as a pro-atrial natriuretic peptide convertase, a cardiac hormone essential for the regulation of blood pressure (43, 44). The identification of the murine ortholog of human polyserase-I raises the possibility of generating mutant mice deficient in this polyprotease. This ongoing project may provide interesting information regarding the physiological and pathological processes, including cancer progression, in which polyserase-I may be involved.
Supplementary Material
Acknowledgments
We are grateful to all members of our laboratory for helpful comments. This work was supported by grants from Comisión Interministerial de Ciencia y Tecnología, Daiichi Fine Chemical, Fundación “La Caixa,” Gobierno del Principado de Asturias, and the European Union. The Instituto Universitario de Oncología is supported by Obra Social Cajastur-Asturias, Spain.
Abbreviations: HA, hemagglutinin; LDLR, low-density lipoprotein receptor; TTSP, type II transmembrane serine protease; ACE, angiotensin-converting enzyme; AMC, 7-amino-4-methylcoumarin.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AJ488946 and AJ488947).
References
- 1.Barrett, A. J., Rawlings, N. D. & Woessner, J. F. (1998) Handbook of Proteolytic Enzymes (Academic, San Diego).
- 2.Lopez-Otin, C. & Overall, C. M. (2002) Nat. Rev. Mol. Cell Biol. 3, 509–519. [DOI] [PubMed] [Google Scholar]
- 3.Puente, X. S., Sánchez, L. M., Overall, C. M. & Lopez-Otin, C. (2003) Nat. Rev. Genet. 4, 544–558. [DOI] [PubMed] [Google Scholar]
- 4.McCawley, L. J. & Matrisian, L. M. (2001) Curr. Opin. Cell. Biol. 13, 534–540. [DOI] [PubMed] [Google Scholar]
- 5.Sternlicht, M. D. & Werb, Z. (2001) Annu. Rev. Cell. Dev. Biol. 17, 463–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters, J. M. (2002) Mol. Cell 9, 931–943. [DOI] [PubMed] [Google Scholar]
- 7.Blasi, F. & Carmeliet, P. (2002) Nat. Rev. Mol. Cell Biol. 3, 932–943. [DOI] [PubMed] [Google Scholar]
- 8.Stennicke, H. R. & Salvesen, G. S. (2000) Biochim. Biophys. Acta 1477, 299–306. [DOI] [PubMed] [Google Scholar]
- 9.Overall, C. M. & López-Otín, C. (2002) Nat. Rev. Cancer 2, 657–672. [DOI] [PubMed] [Google Scholar]
- 10.Egeblad, M. & Werb, Z. (2002) Nat. Rev. Cancer 2, 161–174. [DOI] [PubMed] [Google Scholar]
- 11.Parks, W. C. (2002) J. Clin. Invest. 110, 613–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy, J. & Selkoe, D. J. (2002) Science 297, 353–356. [DOI] [PubMed] [Google Scholar]
- 13.Krane, S. M. (2001) J. Clin. Invest. 107, 31–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Northrop, D. B. (1999) Adv. Enzymol. Relat. Areas Mol. Biol. 73, 25–55, ix. [DOI] [PubMed] [Google Scholar]
- 15.Velasco, G., Cal, S., Quesada, V., Sanchez, L. M. & Lopez-Otin, C. (2002) J. Biol. Chem. 277, 37637–37646. [DOI] [PubMed] [Google Scholar]
- 16.Kozak, M. (1986) Cell 44, 283–292. [DOI] [PubMed] [Google Scholar]
- 17.Roesch, K., Curran, S. P., Tranebjaerg, L. & Koehler, C. M. (2002) Hum. Mol. Genet. 11, 477–486. [DOI] [PubMed] [Google Scholar]
- 18.Hooper, J. D., Campagnolo, L., Goodarzi, G., Truong, T. N., Stuhlmann, H. & Quigley, J. P. (2003) Biochem. J., in press. [DOI] [PMC free article] [PubMed]
- 19.Hooper, J. D., Clements, J. A., Quigley, J. P. & Antalis, T. M. (2001) J. Biol. Chem. 276, 857–860. [DOI] [PubMed] [Google Scholar]
- 20.Takeuchi, T., Shuman, M. A. & Craik, C. S. (1999) Proc. Natl. Acad. Sci. USA 96, 11054–11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu, Q. (2003) Curr. Top. Dev. Biol. 54, 167–206. [DOI] [PubMed] [Google Scholar]
- 22.Lin, C. Y., Anders, J., Johnson, M., Sang, Q. A. & Dickson, R. B. (1999) J. Biol. Chem. 274, 18231–18236. [DOI] [PubMed] [Google Scholar]
- 23.Cho, E. G., Kim, M. G., Kim, C., Kim, S. R., Seong, I. S., Chung, C., Schwartz, R. H. & Park, D. (2001) J. Biol. Chem. 276, 44581–44589. [DOI] [PubMed] [Google Scholar]
- 24.Afar, D. E., Vivanco, I., Hubert, R. S., Kuo, J., Chen, E., Saffran, D. C., Raitano, A. B. & Jakobovits, A. (2001) Cancer Res. 61, 1686–1692. [PubMed] [Google Scholar]
- 25.Natesh, R., Schwager, S. L., Sturrock, E. D. & Acharya, K. R. (2003) Nature 421, 551–554. [DOI] [PubMed] [Google Scholar]
- 26.Soubrier, F., Alhenc-Gelas, F., Hubert, C., Allegrini, J., John, M., Tregear, G. & Corvol, P. (1988) Proc. Natl. Acad. Sci. USA 85, 9386–9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehlers, M. R., Fox, E. A., Strydom, D. J. & Riordan, J. F. (1989) Proc. Natl. Acad. Sci. USA 86, 7741–7745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei, L., Clauser, E., Alhenc-Gelas, F. & Corvol, P. (1992) J. Biol. Chem. 267, 13398–13405. [PubMed] [Google Scholar]
- 29.Lattion, A. L., Soubrier, F., Allegrini, J., Hubert, C., Corvol, P. & Alhenc-Gelas, F. (1989) FEBS Lett. 252, 99–104. [DOI] [PubMed] [Google Scholar]
- 30.Deddish, P. A., Wang, J., Michel, B., Morris, P. W., Davidson, N. O., Skidgel, R. A. & Erdos, E. G. (1994) Proc. Natl. Acad. Sci. USA 91, 7807–7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novikova, E. G., Eng, F. J., Yan, L., Qian, Y. & Fricker, L. D. (1999) J. Biol. Chem. 274, 28887–28892. [DOI] [PubMed] [Google Scholar]
- 32.Lindsay, L. L., Yang, J. C. & Hedrick, J. L. (1999) Proc. Natl. Acad. Sci. USA 96, 11253–11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindsay, L. L., Wieduwilt, M. J. & Hedrick, J. L. (1999) Biol. Reprod. 60, 989–995. [DOI] [PubMed] [Google Scholar]
- 34.Hiyoshi, M., Takamune, K., Mita, K., Kubo, H., Sugimoto, Y. & Katagiri, C. (2002) Dev. Biol. 243, 176–184. [DOI] [PubMed] [Google Scholar]
- 35.Krem, M. M. & Di Cera, E. (2001) EMBO J. 20, 3036–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ross, J., Jiang, H., Kanost, M. R. & Wang, Y. (2003) Gene 304, 117–131. [DOI] [PubMed] [Google Scholar]
- 37.Rutgeerts, L. & Eggermont, E. (1976) Tijdschr. Gastroenterol. 19, 231–246. [PubMed] [Google Scholar]
- 38.Lu, D., Yuan, X., Zheng, X. & Sadler, J. E. (1997) J. Biol. Chem. 272, 31293–31300. [DOI] [PubMed] [Google Scholar]
- 39.Wu, Q., Yu, D., Post, J., Halks-Miller, M., Sadler, J. E. & Morser, J. (1998) J. Clin. Invest. 101, 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu, I. S., Chen, H. J., Lee, Y. S., Huang, P. H., Lin, S. R., Tsai, T. W. & Lin, S. W. (2000) Thromb. Haemostasis 84, 865–870. [PubMed] [Google Scholar]
- 41.List, K., Haudenschild, C. C., Szabo, R., Chen, W., Wahl, S. M., Swaim, W., Engelholm, L. H., Behrendt, N. & Bugge, T. H. (2002) Oncogene 21, 3765–3779. [DOI] [PubMed] [Google Scholar]
- 42.Scott, H. S., Kudoh, J., Wattenhofer, M., Shibuya, K., Berry, A., Chrast, R., Guipponi, M., Wang, J., Kawasaki, K., Asakawa, S., et al. (2001) Nat. Genet. 27, 59–63. [DOI] [PubMed] [Google Scholar]
- 43.Yan, W., Wu, F., Morser, J. & Wu, Q. (2000) Proc. Natl. Acad. Sci. USA 97, 8525–8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan, W., Sheng, N., Seto, M., Morser, J. & Wu, Q. (1999) J. Biol. Chem. 274, 14926–14935. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.