Abstract
Bacterial viral capsids in aqueous solution can be opened in vitro by addition of their specific receptor proteins, with consequent full ejection of their genomes. We demonstrate that it is possible to control the extent of this ejection by varying the external osmotic pressure. In the particular case of bacteriophage λ, the ejection is 50% inhibited by osmotic pressures (of polyethylene glycol) comparable to those operative in the cytoplasm of host bacteria; it is completely suppressed by a pressure of 20 atmospheres. Furthermore, our experiments monitor directly a dramatic decrease of the stress inside the unopened phage capsid upon addition of polyvalent cations to the host solution, in agreement with many recent theories of DNA interactions.
Viral capsids are often highly pressurized. In the case of double-stranded DNA bacteriophages, for example, there are large forces acting to push the genome out through the tail of the viral particle. Recent experiments (1) and theory (2, 3) have established that the forces inside these capsids reach very high values (of order 50 pN) when the genomes are fully packaged. These forces are estimated to correspond to pressures of the order of 50 atm (1 atm = 101.3 kPa; refs. 1–4). Such high values of stress result from the DNA being strongly bent and confined. Specifically, the inner diameter of the capsid is comparable to the DNA persistence length (50 nm), and the typical interaxial spacings are small enough (2.5 nm; ref. 5) that neighboring chains experience strong repulsions. It is precisely these forces that drive the ejection of the genome into the host cell after the tail of the capsid has been bound to its receptor protein in the outer cell membrane; equivalently, it is these forces that must be exerted by the motor protein responsible for packaging of the viral genome (1).
Previous in vitro experiments have demonstrated that DNA ejection is fast (occurring on a time scale of seconds) and complete when phage capsids are opened by their receptors in aqueous solution (6). The capsids are permeable to water and salt ions, so there is no difference in hydrostatic pressure between the inside and outside of the capsid, and osmotic equilibrium is also maintained (7). The pressure difference between the capsid and the solution is therefore associated only with the confinement of the DNA. When the capsid is opened, ejection proceeds until this pressure difference falls to zero. Clearly, at this point the force driving the ejection has also dropped to zero. For the in vivo situation, however, ejection does not occur into a simple salt solution. The DNA is ejected into a highly concentrated colloidal suspension, the cell cytoplasm, which is characterized by high concentrations of proteins and other macromolecular structures. In general, then, one expects this colloidal solution to give rise to a force that resists the entry of DNA (8). More explicitly, because the ejection force associated with stress inside the capsid drops monotonically as the ejection proceeds, there will be a point where it is balanced by the resisting force exerted by the cytoplasm.
To measure the ejection force as a function of the length of DNA remaining inside the capsid, we have performed experiments in which phage capsids are opened by their receptor molecules in solutions of osmotic stress polymers that cannot permeate the capsid wall. We find that the fraction of DNA ejected drops from 1 to 0 on increase of the external osmotic pressure from 0 to 20 atm. More explicitly, a buffered solution of purified phage (λ) is incubated in a solution containing its receptor protein (LamB), an endonuclease (DNase I), and an osmotic stress agent (8,000 MW polyethylene glycol, PEG8000). For any given concentration of PEG, there is a corresponding fraction of DNA that remains inside the capsids after they are opened by the receptor. All of the DNA that gets ejected is digested by the nuclease; because the capsids are impermeable to the nuclease, none of the DNA remaining inside is digested. On spinning down the system in a centrifuge, only free nucleotides remain in the supernatant. Accordingly, measurement of the 260-nm absorption by the supernatant determines directly the extent of ejection as a function of external osmotic pressure (PEG concentration).
Materials and Methods
We used bacteriophage λ EMBL3 with a 13-kb insert (9) isolated from an infected culture of Escherichia coli NM539 (10). The bacteria were grown in LB medium supplemented with 10 mM MgSO4 and 0.5% maltose. After cell lysis and treatment with DNase I and RNase A, unlysed cells and cell debris were removed by centrifugation. Phages were then precipitated in PEG8000 (10% wt/wt) and 1 M NaCl and gently resuspended in SM buffer [10 mM NaCl/10 mM MgSO4/50 mM Tris·HCl, pH 7.4/2% (vol/vol) gelatin]. The PEG and remaining cell debris were extracted from the suspension by adding chloroform, and the phage was purified by equilibrium centrifugation in cesium chloride (11). The phage band was removed and dialyzed twice against a 1,000-fold volume of the buffer; the final titer was ≈1012 virions per ml, determined by plaque assay (12). The receptor was the LamB protein purified from pop 154, a strain of E. coli K12 in which the lamB gene has been transduced from Shigella sonnei 3070 (13, 14). This protein has been shown to cause complete in vitro ejection of DNA from λ in the absence of the added solvents required with the WT E. coli receptor (15, 16). Purified LamB, solubilized from the outer membrane by using the detergent octyl-polyoxyethylene, was obtained from Alexandra Graff (Department of Physical Chemistry, University of Basel, Basel) (13).
Aliquots of λ samples containing ≈1011 virions per ml were gently dispersed in PEG8000 solutions in TM buffer (10 mM MgSO4/50 mM Tris·HCl, pH 7.4), with PEG concentrations varying between 0% and 30% by weight. DNase I was added to a final concentration of 20 μg/ml, which guaranteed complete digestion into free nucleotides of the DNA ejected from the phage. Finally, LamB protein was added in excess of the 5:1 LamB/λ ratio shown to be sufficient to open all of the phage. This ratio was established by adding LamB until we saw no further effect on the efficiency of DNA ejection (similar values for this ratio were determined elsewhere (13, 17); an octyl-polyoxyethylene concentration of 1% by volume was sufficient for all of the LamB to be solubilized (18). The samples were first incubated at 25°C for 15 min to allow phage binding to LamB, followed by DNA ejection (which should be complete within 1 min; ref. 6), and then for 1 h at 37°C to allow full digestion of the ejected DNA by DNase I. Note that the nuclease is present throughout the ejection process to avoid precipitation of the ejected DNA. Total sample volume was 200 μl. The phage capsids and their unejected DNA were removed by centrifugation (with a TLA-100 rotor, at 20°C, at 90,000 rpm for 1 h), and the amount of ejected DNA was determined by measuring the 260-nm absorbance by the nonsedimenting nucleotides left in the supernatant. A UV/VIS Hewlett-Packard single beam diode array spectrophotometer was used at 25°C with a 50-μl and 1-cm-path-length quartz cell. The blank solution used to zero the spectrophotometer consisted of PEG (at a weight fraction corresponding to each sample), TM buffer, and 1% by volume octyl-polyoxyethylene.
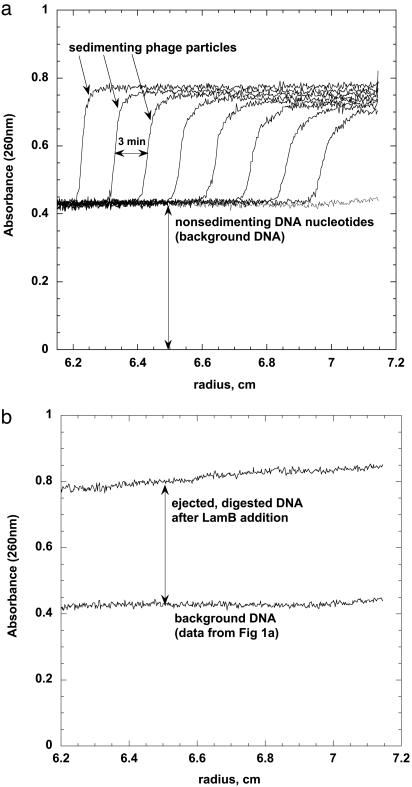
To ensure that our protocol separated phage capsids containing DNA from the ejected DNA digested by DNase I into nonsedimenting nucleotides, we used an analytical ultracentrifuge that allows the UV absorbance at 260 nm to be measured along the (radial) length of the sample as it is being spun down (Beckman Coulter XL-A analytical ultracentrifuge; data were collected at rotor speeds of 3,000–50,000 rpm in continuous mode at 20°C, using a sample volume of 300 μl). In a sample containing only phage and DNase I, i.e., no LamB or PEG, we observed a moving boundary (Fig. 1a) corresponding to the sedimenting phage capsids with their DNA fully inside. A spatially uniform background remains at long times, caused by the nonsedimenting nucleotides associated with the external DNA contaminating the phage stocks. Similar levels of external DNA in the phage stocks have also been reported (13, 15). This contamination can arise from DNA released from bacteria and phage ruptured in the course of purification and partial phage rupture over time because of the instability of the phage particles.
Fig. 1.
Analytical ultracentrifugation analysis of receptor-treated phage. Samples were centrifuged at 15,000 rpm (corresponding to 40,000 × g), and the concentration of UV-absorbing material (here DNA) was followed by scanning the cells at 260 nm. (a) Sample contains only phage and DNase I (no PEG or LamB). The moving boundary was followed with 3-min intervals and corresponds to the sedimenting phage capsids with their DNA fully inside. The remaining absorbance (uniform background line) corresponds to nonsedimenting nucleotides from the digested external contaminant DNA. (b) Sample contains phage incubated with LamB receptor and DNase I. The nonsedimenting DNA nucleotide absorbance is now almost twice as large, and no moving boundary is observed because the sedimenting empty capsids do not absorb significantly at 260 nm. All of the phage particles have been opened with LamB, and the DNA is fully ejected and digested by DNase I. For comparison, the contaminant DNA background absorbance measured and shown in a is also shown in b.
In contrast to the samples containing neither LamB nor PEG, in those with LamB (but still no PEG) the 260-nm absorbance showed only a spatially uniform value (Fig. 1b) approximately twice as large as that associated with the contaminant DNA. The enhanced signal arose from all of the phage having been opened by the LamB, with consequent complete ejection and digestion of their DNA. (Note that the empty capsids were still sedimenting in these samples, but they did not absorb significantly at 260 nm because they did not contain DNA.) The difference between the absorbance with and without LamB makes clear that DNase I does not permeate the phage. To confirm that DNA ejection is indeed complete in the absence of PEG but in the presence of 5:1 LamB, we prepared LamB-free samples in which all of the capsids were thermally ruptured (95°C for 10 min; ref. 19). The samples were then cooled, and DNase I was added. Analytical centrifugation showed that the DNA had been converted into nonsedimenting nucleotides and that the absorbance signal corresponded to that observed in samples treated with LamB instead of heat.
Results
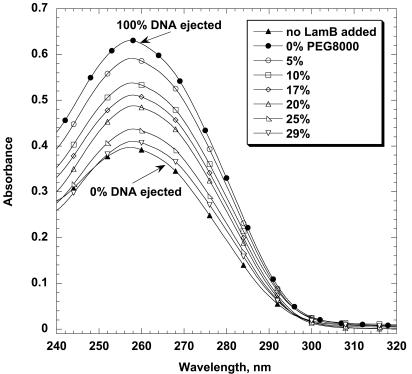
Fig. 2 shows the UV absorbance as a function of wavelength for samples with different PEG concentrations. The lowest curve corresponds to the sample in which no LamB is present, i.e., in which all of the capsids remain closed and the entire free nucleotide absorbance in the supernatant is caused by contaminant DNA. This background signal sets the zero for DNA ejection and, correspondingly, is subtracted from all of the absorbances measured for samples with LamB. The top curve, for example, is the absorbance measured for a sample in which LamB is present in excess, but no PEG has been added. Because all of the DNA is ejected under these conditions, the difference between the top and bottom curves in Fig. 2 gives directly the amount of DNA in the opened phage capsids; i.e., it defines the 100% ejection fraction. The intermediate curves correspond to increasing concentrations of PEG; the monotonic decrease in absorbance is caused by the decrease in the fraction of DNA ejected (and thereby made available for DNase digestion). We see clearly that when the concentration of PEG has reached 29%, the osmotic pressure in the external solution is sufficient to suppress completely the ejection of DNA from the phage capsids, even though they have been opened by LamB.
Fig. 2.
UV absorbance as a function of wavelength for samples with different PEG8000 concentrations. In all samples, the phage capsids with unejected DNA inside have been precipitated by centrifugation after DNase I addition, and only the absorbance of nonsedimenting DNA nucleotides is measured. The lowest curve corresponds to the sample where no LamB is added and is caused entirely by the contaminant external DNA background; i.e., no DNA is ejected. The top curve is for a sample where LamB is present but no PEG8000 has been added; here, the measured absorbance corresponds to DNA nucleotides from fully ejected DNA. This maximum intensity minus the background is a measure of the amount of DNA ejected from the phage. All of the other curves correspond to samples with LamB and different PEG8000 weight fractions. At 29% PEG8000, the ejection of DNA from the phage is essentially completely suppressed.
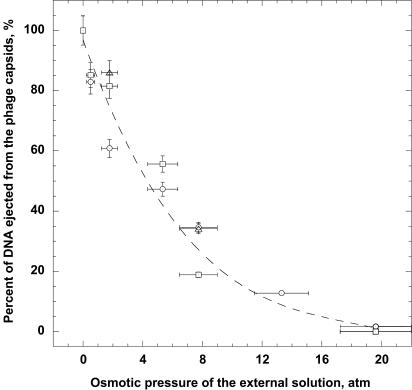
The relation between the osmotic pressure and the polymer concentration and the temperature is well established for PEG8000 (20), which allows us to deduce the fraction of DNA ejection as a function of osmotic pressure. The ejection fraction at a PEG weight percent is calculated as the ratio of the difference [260 absorbance (% PEG) – 260 absorbance (background)] divided by the difference [260 absorbance (0% PEG) – 260 absorbance (background)]. Fig. 3 shows that the DNA ejection has been completely suppressed when the external solution pressure reaches ≈20 atm. The results presented in Fig. 3 include data from sets of aliquot samples from three different phage stocks. The horizontal error bars were obtained by propagating the error in the PEG concentration, caused primarily by the high viscosity of the PEG solutions, which makes pipeting less accurate. The vertical bars reflect differences between repeated measurements of UV absorbance for the same sample with all conditions kept constant.
Fig. 3.
Fraction of DNA ejected as a function of the osmotic pressure (calculated from the concentration of PEG8000). Data are presented for samples taken from three different phage stocks. Horizontal and vertical error bars represent one standard deviation and were determined by propagation of errors in the PEG concentration and the absorbance.
Discussion
The progressive inhibition of DNA ejection with increasing osmotic pressure can be understood most simply in terms of the excluded volume interactions between the DNA and PEG. More explicitly, for the DNA to be ejected from the capsid, it needs to be inserted into the PEG solution. This inhibition in turn requires that a cavity must be formed, devoid of PEG molecules, of sufficient size to accommodate the DNA chain. The work associated with forming this cavity arises from both pressure-volume and surface contributions, and the work per unit length of inserted chain corresponds directly to the force resisting entry of the chain into the colloidal suspension (8). Now, the force driving the chain out of the capsid (because of its strong confinement) has been shown (1–4) to decrease monotonically as ejection proceeds because of the progressive relief of stress inside the capsid. It follows that ejection will proceed only to the point where the ejection force, feject, has dropped to a value equal to the resisting force, fresist, associated with the work of insertion into the PEG solution. What we have shown in the present work is that a PEG concentration of ≈30% by weight is sufficient to give rise to a resisting force that balances the maximum ejection force, i.e., to an fresist that prevents the ejection process from even beginning.
Whereas the inhibition of ejection arises from a force balance being achieved, it does not correspond to equality of the pressures (i.e., forces per unit area). This is because the relationship between force and pressure is quite different inside as compared with outside. These respective relationships have been investigated theoretically and, using the presently reported data, we can quote them here to make a rough estimate of the pressure inside the capsid. More explicitly, Castelnovo et al. (8) have estimated that a PEG pressure of 20 atm corresponds to a resisting force of ≈20 pN. In turn, Tzlil and coworkers (2, 3) have estimated that an ejection force of this magnitude corresponds to a pressure inside the capsid of ≈40 atm. Thus, the pressure in the fully packaged capsid is estimated to be larger than (but comparable to) the osmotic pressure found sufficient to suppress the ejection completely. Inferring more quantitative estimates from the present measurements will require a more detailed theoretical analysis of the salt conditions and polymer molecular weight, etc. (A.E., M. Castelnovo, C.M.K., and W.M.G., unpublished results). We note, however, that the 41.5-kb-long DNA that we have used in these experiments is 7-kb shorter than that in the WT phage. Hence, the pressure inside the WT capsids will be substantially higher.
Our interpretation of the experimental results is based on several assumptions that can be checked. We have assumed that the PEG8000 does not enter the capsid. Experiments in which the osmotic pressure is produced with PEG4000 give results essentially identical to those with higher molecular weight polymer. If, however, glycerol is used as the osmotic stress agent, DNA ejection is complete even at glycerol concentrations corresponding to osmotic pressures >50 atm (see http://aqueous.labs.brocku.ca/data/glycerol). We interpret these results as demonstrating that the glycerol, which has a diameter of a few tenths of a nanometer, permeates the capsid, and therefore cannot cause an osmotic pressure difference. As a result, the effective force resisting the ejection is identically zero. On the other hand, the radii of gyration of the PEG8000 and PEG4000 polymers (3.5 and 2.3 nm, respectively) (21) are sufficiently large that they cannot permeate the phage capsid. Note that possible effects of osmotic pressure differences on the efficiency of DNA packaging had been pointed out earlier in the context of in vitro phage formation in the presence of dextran, sugars, and polyols (22). We have also assumed that the fraction of DNA ejected is the same for all opened capsids, as opposed to some ejecting completely, and others not at all. Measurements made by extracting DNA from the capsids and determining its length by analytical ultracentrifugation and pulsed-field electrophoresis have independently shown this assumption to be correct (unpublished results).
Also, when the excess of LamB was increased by a factor of 2, there was no change in 260-nm absorbance; an increase would have been expected if there had been an equilibrium involving LamB and PEG. Finally, we have performed experiments in which a concentration of 1 mM spermine chloride is present in the incubating samples. We find that ejection is completely suppressed at a pressure of 4 atm rather than 20 atm. This finding suggests further that inhibition of ejection is not caused by PEG interfering with the action of the LamB receptor. In the absence of spermine, >50% of the genome is released at 4 atm PEG. Indeed, the spermine results confirm that a significantly smaller resisting force is sufficient to stop the ejection, consistent with our interpretation of the mechanism of inhibition. The tetravalent spermine cation permeates the capsid and mediates the repulsions between neighboring portions of the packaged DNA, thereby reducing the pressure and forces within the capsid (23, 24). It follows that the osmotic pressure and forces necessary for inhibition will be correspondingly smaller. We believe that our experimental approach can be systematically used in this way to determine the forces and pressures inside viral capsids as a function of packaged genome lengths and ambient salt conditions.
Acknowledgments
We thank Alexandra Graff for making available the purified LamB, octyl-polyoxyethylene detergent, and detailed protocols for purification of LamB. Emir Berkane kindly provided pop154 E. coli strain for LamB purification. We acknowledge many stimulating discussions of these experiments with Martin Castelnovo, Francoise Livolant, Lucienne Letellier, Marta de Frutos, Paulo Tavares, Bengt Jönsson, and Ulf Olsson. We have also benefited from the generous technical help and expert advice of James W. Gober, Sabeeha Merchant, Rainer Figge, Jeffery L. Moseley, Maria Kaczor-Grzeskowiak, Robert Mencel, Martin Phillips, and Mari Gingery (University of California, Los Angeles). Richard E. Dickerson kindly provided laboratory space. We acknowledge the use of the UCLA-DOE facilities. A.E. has received financial support from The Swedish Foundation for International Cooperation in Research and Higher Education. This work was supported by National Science Foundation Grants CHE00-7931 and CHE99-88651 (to C.M.K. and W.M.G.).
Abbreviation: PEG, polyethylene glycol.
References
- 1.Smith, D. E., Tans, S. J., Smith, S. B., Grimes, S., Anderson, D. L. & Bustamante, C. (2001) Nature 413, 748–752. [DOI] [PubMed] [Google Scholar]
- 2.Tzlil, S., Kindt, J. T., Gelbart, W. M. & Ben-Shaul, A. (2002) Biophys. J. 84, 1616–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kindt, J., Tzlil, S., Ben-Shaul, A. & Gelbart, W. M. (2001) Proc. Natl. Acad. Sci. USA 98, 13671–13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purohit, P. K., Phillips, R. & Kondev, J. (2003) Proc. Natl. Acad. Sci. USA 100, 3173–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Earnshaw, W. C. & Harrison, S. C. (1997) Nature 268, 598–602. [DOI] [PubMed] [Google Scholar]
- 6.Novick, S. L. & Baldeschwieler, J. D. (1988) Biochemistry 27, 7919–7924. [DOI] [PubMed] [Google Scholar]
- 7.Anderson, T. F., Rappaport, C. & Muscatine, N. A. (1953) Ann. L'Institut Pasteur 84, 5–14. [PubMed] [Google Scholar]
- 8.Castelnovo, M., Bowles, R. K., Reiss, H. & Gelbart, W. M. (2003) Eur. Phys. J. 10, 191–197. [DOI] [PubMed] [Google Scholar]
- 9.Goldschmidtclermont, M. & Rahire, M. (1986) J. Mol. Biol. 191, 421–432. [DOI] [PubMed] [Google Scholar]
- 10.Moseley, J. L., Page, M. D., Alder, N. P., Eriksson, M., Quinn, J., Soto, F., Theg, S. M., Hippler, M. & Merchant, S. (2002) Plant Cell 14, 673–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maniatis, T., Fritsch, E. F. & Sambrook, J. (1983) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 12.Silhavy, T. J., Berman, M. L. & Enquist, L. W. (1984) Experiments with Gene Fusions (Cold Spring Harbor Lab. Press, Plainview, NY).
- 13.Graff, A., Sauer, M., Van Gelder, P. & Meier, W. (2002) Proc. Natl. Acad. Sci. USA 99, 5064–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roa, M. & Scandella, D. (1976) Virology 72, 182–194. [DOI] [PubMed] [Google Scholar]
- 15.Randall-Hazelbauer, L. & Schwartz, M. (1973) J. Bacteriol. 116, 1436–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roa, M. (1981) FEMS Microbiol. Lett. 11, 257–262. [Google Scholar]
- 17.Berrier, C., Bonhivers, M., Letellier, L. & Ghazi, A. (2000) FEBS Lett. 476, 129–133. [DOI] [PubMed] [Google Scholar]
- 18.Prilipov, A., Phale, P. S., Van Gelder, P., Rosenbusch, J. P. & Koebnik, R. (1998) FEMS Microbiol. Lett. 163, 65–72. [DOI] [PubMed] [Google Scholar]
- 19.Pollard, E. C. & Solosko, W. (1971) Biophys. J. 11, 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parsegian, V. A., Rand, R. P., Fuller, N. L. & Rau, D. C. (1986) Methods Enzymol. 127, 400–416. [DOI] [PubMed] [Google Scholar]
- 21.Abbott, N. L., Blankschtein, D. & Hatton, T. A. (1992) Macromolecules 25, 5192–5200. [Google Scholar]
- 22.Serwer, P., Masker, W. E. & Allen, J. L. (1983) J. Virol. 45, 665–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rau, D. C. & Parsegian, V. A. (1992) Biophys. J. 61, 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raspaud, E., de la Cruz, M. O., Sikorav, J. L. & Livolant, F. (1998) Biophys. J. 74, 381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]