Abstract
How RNA molecules fold into functional structures is a problem of great significance given the expanding list of essential cellular RNA enzymes and the increasing number of applications of RNA in biotechnology and medicine. A critical step toward solving the RNA folding problem is the characterization of the associated transition states. This is a challenging task in part because the rugged energy landscape of RNA often leads to the coexistence of multiple distinct structural transitions. Here, we exploit single-molecule fluorescence spectroscopy to follow in real time the equilibrium transitions between conformational states of a model RNA enzyme, the hairpin ribozyme. We clearly distinguish structural transitions between effectively noninterchanging sets of unfolded and folded states and characterize key factors defining the transition state of an elementary folding reaction where the hairpin ribozyme's two helical domains dock to make several tertiary contacts. Our single-molecule experiments in conjunction with site-specific mutations and metal ion titrations show that the two RNA domains are in a contact or close-to-contact configuration in the transition state even though the native tertiary contacts are at most partially formed. Such a compact transition state without well formed tertiary contacts may be a general property of elementary RNA folding reactions.
RNA is the key enzymatic component in a number of essential cellular processes, such as translation and splicing (1–4). Aside from these fundamental roles, RNA also finds important applications in modern biotechnology and medicine (5, 6). For example, recent developments in small interfering RNAs, protein-binding RNA aptamers, and target-specific catalytic RNAs suggest that these functional RNAs can serve as effective tools in functional genomics and proteomics and in gene therapy (5, 6). This increasing appreciation of RNA as a crucial biopolymer demands more than ever a clear picture of how RNA molecules fold into their native structures, which are vital to their functional properties. A fundamental understanding of RNA folding relies critically on the characterization of the associated folding transition states, i.e., the highest energy states along the reaction coordinates that dictate the transition kinetics. However, the characterization of the transition states of RNA folding lags far behind that of protein folding (7–11), in part because of a more rugged energy landscape for RNA that leads to multiple folding pathways and intermediate states (12–19), making it difficult to characterize elementary RNA folding transitions. Here, we demonstrate a solution to this problem by using single-molecule fluorescence spectroscopy (20, 21) on a model RNA enzyme, the hairpin ribozyme.
Our single-molecule time trajectories unambiguously identify multiple conformational states of the RNA and distinct structural transitions between effectively noninterchanging sets of unfolded and folded states. Using this technique, in conjunction with site-specific mutations, metal ion titrations, and electrostatic modeling, we have characterized key factors defining the transition state of an elementary RNA folding transition. Our mutational studies show that native contacts are at most partially formed in the transition state. Our metal ion titrations suggest that the folding transition state is compact and stabilized by electrostatic interactions in magnitude similar to those in the native state. We also observe slight shifting of compactness in the transition state as the solution conditions vary.
Materials and Methods
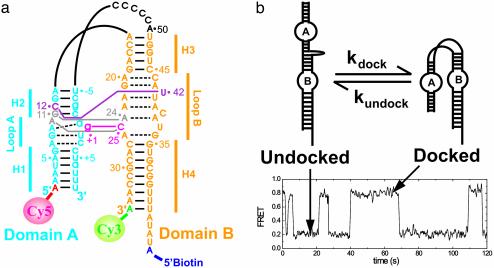
Preparation of Dye-Labeled RNA. As a simple model system for understanding RNA folding, we used a two-way junction hairpin ribozyme (22) in which two of the four-way junction arms of the original tobacco ringspot virus satellite RNA are deleted and the independently folding helical domains A and B are connected via a 6-nt bulge of sequence AC5 (Fig. 1a). In the experiment to test the effect of the g+1:C25 base pair on docking, a nicked WT hairpin ribozyme without the AC5 linker was used. In each case, catalysis was suppressed by a 2′ methoxy group at the cleavage site that minimally impacts folding (23). The Cy5-labeled RNA strand (RzA) and the biotin-labeled strand (RzB) were purchased from the Howard Hughes Medical Institute Biopolymer/Keck Foundation Biotechnology Resource Laboratory at the Yale University School of Medicine (New Haven, CT). Both strands were gel-purified and C8 reverse-phase HPLC-purified as described (23). The RzA strand was labeled postsynthetically with Cy3 dye and HPLC-purified as described (18, 22, 23), then annealed with the RzB strand by heating to 80°C for 45 s, followed by cooling to 37°C over 30 min.
Fig. 1.
Folding of the hairpin ribozyme. (a) The WT AC5 hairpin ribozyme used in this study. Watson–Crick and noncanonical base pairs of the docked conformer are indicated by solid and dashed lines, respectively. Tertiary interactions are represented as follows: pink, g+1:C25 Watson–Crick base pair; gray, ribose zipper; purple, U42 binding pocket (34). Biotin, Cy3, and Cy5 were attached as indicated (18). (b) Schematic of the docking and undocking transitions of the hairpin ribozyme with an experimental time trajectory showing the corresponding FRET changes at 37°C.
Single-Molecule Fluorescence Resonance Energy Transfer (FRET) Measurements. The annealed biotinylated ribozyme was bound in low concentration (50 pM) to a streptavidin-coated quartz slide surface via the biotin–streptavidin interaction, and the donor (ID) and acceptor (IA) fluorescence signals of optically resolved single molecules were detected on a total internal reflection fluorescence microscope as described (18, 24). The donor and acceptor fluorescence signals indeed photobleached in single steps, confirming single-molecule detection. The FRET ratio [defined as IA/(IA + ID)] was followed in real time for each individual molecule. Measurements were performed under a variety of cation conditions as indicated, but all solutions contained 50 mM Tris·HCl, pH 7.5, with an oxygen scavenging system consisting of 10% (wt/vol) glucose, 2% (vol/vol) 2-mercaptoethanol, 50 μg/ml glucose oxidase, and 10 μg/ml catalase to reduce photobleaching. Most of the experiments were performed at 37°C. However, because the rate constants for docking and undocking increase rapidly with increasing temperature (data not shown), we switched to 25°C in cases where the rate constant under consideration was too fast to be determined accurately at 37°C.
Nonlinear Poisson–Boltzmann Model Calculations. We devised a simplified model of the ribozyme in which its two domains are approximated by two negatively charged cylinders of A-type helical dimensions. Each cylinder is defined by a linear array of –2 charges with a radius of 11.5 Å, spaced 2.81 Å apart (25). One cylinder is composed of 24, the other of 14 of these –2 charges to mimic the actual size and charge of the hairpin ribozyme. The cylinders are treated as a low dielectric medium (dielectric constant of 2) immersed in a high dielectric aqueous solvent (dielectric constant of 80). Calculation of the electrostatic potentials and free energies with the nonlinear Poisson–Boltzmann equation in a mixed salt solution followed the numerical procedures of ref. 26.
Results and Discussion
The Hairpin Ribozyme as a Model System of RNA Folding. Our studies focus on the two-way junction form of the hairpin ribozyme, a model RNA enzyme derived from the autocatalytic negative strand of the tobacco ringspot virus satellite RNA (27). This ribozyme consists of two helix–loop–helix domains, A and B, that adopt either an extended (unfolded) state or specifically interact in a compact, enzymatically active docked (folded) state (Fig. 1) (23, 28). Such docking of two preformed elements of secondary structure is typical of the hierarchical assembly of RNA tertiary structure (15, 29).
To characterize the transition state of this elementary folding process, we use single-molecule FRET (14, 30–33) with a donor (Cy3) and acceptor fluorophore (Cy5) covalently linked to the ribozyme such that the docked state exhibits a higher FRET value than the undocked state (Fig. 1). FRET time trajectories of single ribozyme molecules (Fig. 1b) show stochastic transitions between the undocked (low FRET) and docked (high FRET) states. The dwell times of each docked and undocked events are calculated, and histograms of the dwell times are constructed, typically from several hundred molecules. The histograms of docked and undocked dwell times are then fit with single or multiple exponential decay functions to deduce the kundock and kdock values, respectively (18). The standard deviation of our kdock and kundock values from independent experiments is ≤8%. We previously showed that surface immobilization has a negligible effect on the docking and cleavage kinetics of the hairpin ribozyme (18), in accordance with the fact that the molecules behave consistently over the entire [Mg2+] range used in the present study.
Our measurements on the WT AC5 ribozyme at 12 mM Mg2+ and 25°C reveal a uniform docking rate constant (kdock = 0.018 s–1), but four distinct undocking rate constants (kundock,1 = 0.01 s–1, kundock,2 = 0.1 s–1, kundock,3 = 0.8 s–1, and kundock,4 = 6 s–1). Individual molecules are found to switch only very slowly (over several hours) between the four undocking behaviors. The fractions of molecules that display these four undocking rate constants are 59%, 11%, 14%, and 14%, respectively, whereas 2% seem not to dock at all. A total of 760 single-molecule FRET trajectories showing docking and undocking events were used to determine these fractions. These fractions do not change significantly with experimental conditions under all conditions tested. These results, in agreement with our previous measurements on the same construct with a nicked two-way junction (18), show that the hairpin ribozyme exhibits multiple, effectively noninterchanging populations with distinct undocking rates (18). A single-molecule approach is thus critical for distinguishing these populations so that the docking transition state of each population can be unambiguously characterized. In the following, we focus mainly on the population with the slowest undocking rate constant, in which all native tertiary interactions of the docked state are formed.
Tertiary Contact Formation in the Transition State. The crystal
structure (34) shows three
main tertiary contacts between domains A and B in the docked state of the
hairpin ribozyme (Fig.
1a): (i) the hydrogen bonding network of a
ribose zipper connects A10 and G11 in domain A with A24 and C25 in domain B;
(ii) U42 of domain B binds to a pocket formed by functional groups of
both domains; and (iii) g + 1 of domain A base pairs with C25 in a
G-binding pocket formed by domain B. In the following, we use Φ-value
analysis (35) to quantify the
tertiary contact formation in the folding transition state of the hairpin
ribozyme. We define
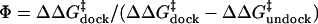 ,
with
,
with 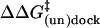 being the mutation-induced change in the free energy barrier for (un)docking.
Φ = 0 indicates that a mutation stabilizes the docked state relative to
the undocked state but does not destabilize the transition state, and thus
suggests that the tertiary contact disrupted by the mutation is not yet formed
in the transition state. By contrast, Φ = 1 indicates that the probed
tertiary contact is already formed in the transition state. A fractional
Φ-value indicates either a fractional tertiary contact in a single
transition state or the partitioning between parallel transition states, some
with fully formed tertiary contact, others with no contact at all
(35). Here, we refer to both
scenarios as partial tertiary contact formation.
being the mutation-induced change in the free energy barrier for (un)docking.
Φ = 0 indicates that a mutation stabilizes the docked state relative to
the undocked state but does not destabilize the transition state, and thus
suggests that the tertiary contact disrupted by the mutation is not yet formed
in the transition state. By contrast, Φ = 1 indicates that the probed
tertiary contact is already formed in the transition state. A fractional
Φ-value indicates either a fractional tertiary contact in a single
transition state or the partitioning between parallel transition states, some
with fully formed tertiary contact, others with no contact at all
(35). Here, we refer to both
scenarios as partial tertiary contact formation.
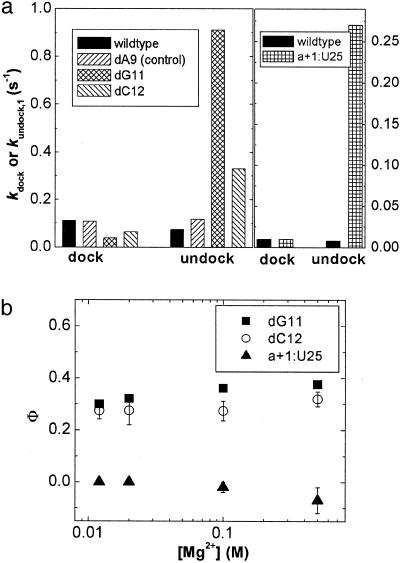
To test whether the ribose zipper is formed in the transition state, we replaced the 2′ hydroxyl group (2′ OH) of G11 with a hydrogen atom (2′ H) to disrupt two of the four hydrogen bonds in this motif (23, 27, 34). This dG11 mutation causes a 12.3-fold increase in kundock,1, but only a 2.9-fold decrease in kdock under standard conditions (at 12 mM Mg2+) (Fig. 2a), consistent with the previous observation of destabilized docking in bulk solution (23). As a control, the replacement of a 2′ OH not involved in tertiary contacts with a 2′ H (mutation dA9) changes kdock and kundock,1 only slightly (Fig. 2a). The small fractional Φ-values associated with the dG11 mutation throughout the entire accessible [Mg2+] range (12–500 mM) (Fig. 2b) suggest that the ribose zipper tertiary interaction is at most partially formed under these conditions (36).
Fig. 2.
Effect of mutations on the rate constants for docking and undocking. (a Left) kdock and kundock,1 of the WT and mutant ribozymes at 12 mM Mg2+ and 37°C. (a Right) kdock and kundock,1 of WT and mutant ribozymes at 12 mM Mg2+ and 25°C. (b) Dependence of the Φ-values on Mg2+ concentration. Values for the dG11 and dC12 mutations were obtained at 37°C, and those for the a+1/U25 mutant were obtained at 25°C. The error bars for the φ-values of dG11 are smaller than the symbols.
To test for the U42 binding pocket, the 2′ OH of C12 was replaced with a 2′ H (dC12 mutation), thus disrupting one of the five hydrogen bonds between U42 and its binding pocket (34). kundock,1 increases 4.4-fold (Fig. 2a), whereas kdock decreases by only 1.7-fold at 12 mM Mg2+. Again, the small fractional Φ-values at 12–500 mM Mg2+ (Fig. 2b) suggest that the U42 binding pocket is also at most partially formed in the transition state.
Finally, to test for the g+1:C25 base pair, we replaced it with a weaker a+1:U25 Watson–Crick base pair (18). Consistent with the previously observed sharp destabilization of docking (37), this mutation increases kundock,1 by ≈34-fold, while leaving kdock unchanged (Φ ≅ 0) (Fig. 2a), suggesting that the g+1:C25 base pair is not formed at all in the transition state. Significantly, the Φ-value for the a+1:U25 mutation remains very close to zero throughout the entire accessible [Mg2+] range (12–500 mM) (Fig. 2b).
This series of experiments provides us with a comprehensive picture of tertiary contact formation in the folding transition state of the hairpin ribozyme. Although the inter-domain Watson–Crick base pair g+1:C25 is not yet formed in the transition state, tertiary contacts involving backbone functional groups, i.e., the ribose zipper and U42 binding pocket, begin to form. Such nascent tertiary contact formation in the RNA folding transition state is different from previous observations for the Tetrahymena ribozyme, where tertiary contacts are essentially not formed (7, 8, 14).
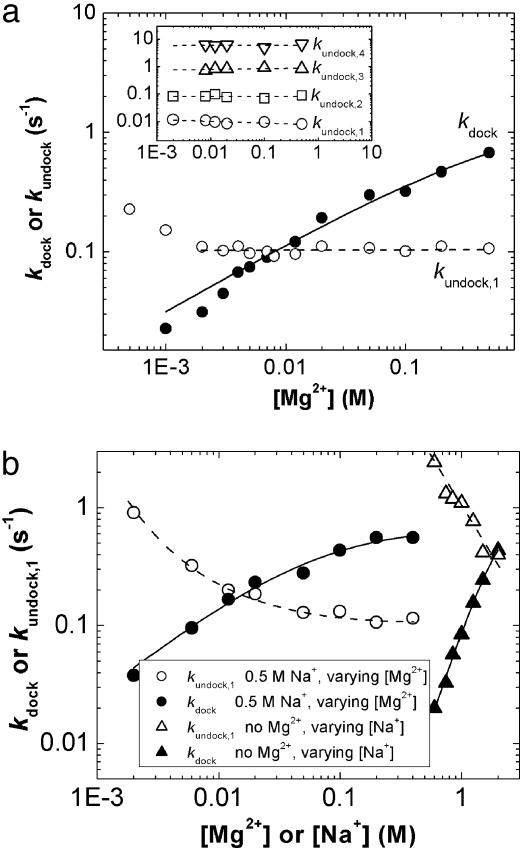
Metal Ion Dependence of the Docking and Undocking Rate Constants. Next, we probed directly the influence of metal ions on the folding process. Our single-molecule measurements reveal distinct Mg2+ dependencies of kdock and kundock (Fig. 3a), not previously observed in bulk-solution experiments (23, 28). kdock increases by >40-fold over a range from 1 to 500 mM Mg2+. The average slope ∂log(kdock)/∂log[Mg2+] can be interpreted to indicate an apparent uptake of 0.6 Mg2+ ions per RNA molecule on conversion from the undocked to the transition state, although it does not necessarily imply specific binding of Mg2+ (26). In stark contrast, kundock,1 is independent of [Mg2+] over nearly the entire range (2–500 mM), indicating no net Mg2+ uptake on conversion from the docked to the transition state (26). Interestingly, kundock,1 starts to increase from a constant value at very low [Mg2+] (≤1 mM), consistent with a gradual shift of the transition state along the reaction coordinate at the lowest ionic strength experimentally accessible at 37°C. At 25°C we are able to follow all four undocking rate constants (kundock,1–4) and find that they are all independent of [Mg2+] between 2 and 500 mM (Fig. 3a Inset).
Fig. 3.
Effect of metal ion concentration on the rate constant for docking and undocking. (a) Dependence of kdock (•) and kundock (○) of the WT ribozyme on [Mg2+] at 37°C. kdock values are fit to the Hill equation (solid line), kdock ≅ kmax [Mg2+]n/([Mg2+]n + (Mg1/2)n). Mg1/2 = 830 mM is the magnesium half-saturation point; n = 0.6 is the Hill constant, which approximates the average slope ∂log(kdock)/∂log[Mg2+]. (Inset) [Mg2+] dependence of all four undocking rate constants observed at 25°C. At this temperature, kdock increases continuously with [Mg2+] in a similar fashion to the kdock values at 37°C (data not shown). (b) The [Na+] dependence of kdock and kundock,1 (triangles) in the absence of Mg2+ and the [Mg2+] dependence of kdock and kundock,1 in the presence of 500 mM Na+ (circles). kdock values are fit to the Hill equation (solid line), yielding Mg1/2 = 60 mM, n = 0.8 and Na1/2 = 2.1 M, n = 3, respectively.
Notably, the effects of monovalent ions (Na+) on kdock and kundock are clearly distinct from those of divalent ions (Fig. 3b). The increase of kdock with [Na+] suggests that the transition state is also stabilized by Na+ ions, which are less likely to interact with RNA by specific site binding (38, 39). The slopes ∂log(kdock)/∂log[Na·] and ∂log(kundock,1)/∂log[Na·] suggest a significant uptake of Na+ ions on conversion from the transition to the docked state and from the undocked to the transition state (26). The comparable slopes indicate that about half of the total Na+ ion uptake on docking already occurs in the transition state.
In mixed ion experiments, monovalents and divalents are expected to compete for nonspecific interactions with the RNA, and this competition is indeed observed in Mg2+ titration experiments in the absence and presence of Na+ (compare Fig. 3 a and b). In the absence of Na+, kundock starts to increase from a constant value (≈0.1 s–1) only below 1 mM Mg2+ (Fig. 3a), suggesting that the binding of Mg2+ ions to the transition state is saturated above that [Mg2+]. In contrast, in the presence of a background of 500 mM Na+, kundock starts to increase from a constant value below 50 mM Mg2+ (Fig. 3b), suggesting that Mg2+ binding is not saturated up to this much higher [Mg2+] because of the competition of Na+ for nonspecific binding sites.
A Compact RNA Folding Transition State. The fact that the hairpin ribozyme folds and functions in both divalent and monovalent salt solutions (Fig. 3) (27, 40) suggests that specific metal site binding is not obligatory for folding, whereas nonspecific electrostatic interactions (diffuse binding modes) are certainly crucial to the folding process of the hairpin ribozyme, as they are for RNA in general (26, 39). Although we cannot formally rule out effects of specific metal binding in folding the hairpin ribozyme, we propose a simple model based solely on electrostatic considerations to understand our experimental observations: the docked, transition, and undocked states have different charge densities and thus attract metal ions with different affinities. The dramatic increase of kdock with [Mg2+] and lack of a [Mg2+] dependence of kundock (between 2 and 500 mM Mg2+ in the absence of monovalents) suggest a significant Mg2+ uptake by the RNA on conversion from undocked to transition state and no significant uptake on conversion from docked to transition state, respectively, which in turn suggests that the charge density of the transition state must be significantly higher than that of the undocked state, but similar to that of the docked state.
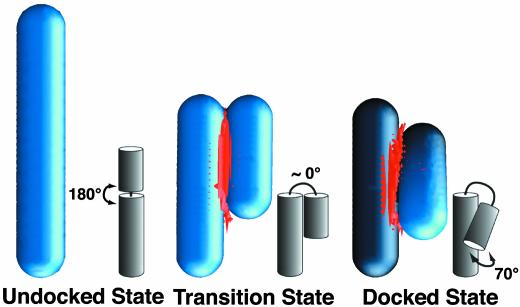
To further understand the structural implications of the metal ion dependence of docking and undocking, we have used the nonlinear Poisson–Boltzmann equation to calculate the electrostatic interactions in generalized cylinder models of the ribozyme (Fig. 4; Table 1) (26). The four major configurations of the ribozyme considered are: (i) an extended configuration that represents the undocked state (U), (ii) a docked configuration where the two domains are in contact and at a 70° angle as in the crystal structure (34) (D), (iii) a contact model of the transition state where the two domains are in direct contact at a parallel 0° angle (C), and (iv) a noncontact model of the transition state where the two domains surfaces are 5 Å apart at a 0° angle (NC), allowing for the insertion of hydrated metal ions (Fig. 4). Our calculations show a large accumulation of divalent metal ions (M2+) at the interface of the two RNA domains on conversion from U to C states (Fig. 4) (the uptake of M2+ ions per RNA is ΔΓU→C = 1.3; Table 1), whereas the reorientation of the two domains on conversion from C to D states is accompanied by only an insignificant M2+ loss (Fig. 4) (ΔΓC→D = –0.01; Table 1). In contrast, the M2+ binding properties of the NC state are distinct from those of both the U and D states, with significant and comparable uptake and loss of M2+ in moving from the U and D states, respectively, toward the NC state (ΔΓU→NC = 0.68, ΔΓNC→D = 0.59; Table 1).
Fig. 4.
A theoretical model describing the electrostatic interactions of the hairpin ribozyme with metal ions. Domains A and B of the ribozyme were modeled as two connected cylinders (blue) with their relative orientations depicted in gray. The nonlinear Poisson–Boltzmann equation was used to determine the divalent metal ion distribution around the RNA in solvent containing 25 mM monovalent and 10 mM divalent salt. The 3D isoconcentration contour (red) at 3.0 M shows the accumulation of divalent metal ions at the domain interface in the docked state and the contact model of the transition state. On titration with Mg2+ alone, the most probable transition state is represented by the contact model, where domains A and B are in contact as shown. However, at <1 mM Mg2+ in the absence of Na+, on titration with Na+ alone, or on Mg2+ titration in a background of 500 mM Na+, the more appropriate transition state is a noncontact model where the two domains are only slightly separated (see text). In all cases, docking likely occurs via an ensemble of transitions states that all satisfy these restrictions. The figure was rendered by using the program grasp (50).
Table 1. Calculated electrostatic contribution to domain docking at 25 mM M+X-, 10 mM M2+ (X-)2.
| Transition | ΔGcoulombic, kT | ΔGsolvation, kT | ΔG(M+X-), kT | ΔG(M2+(X-)2), kT | ΔΓ |
|---|---|---|---|---|---|
| U → C | 2,450 | -3,140 | -68 | -16 | 1.3 |
| C → D | 0.02 | -133 | -21 | 1.9 | -0.01 |
| U → NC | 895 | -1,900 | -86 | -6.8 | 0.68 |
| NC → D | 1,560 | -1,370 | -3.5 | -7.4 | 0.59 |
ΔGcoulombic, coulombic interaction free energy among all charges in a uniform dielectric constant of 2. ΔGsolvation, stabilization conferred by pure water on transfer into an aqueous, salt-free solution (dielectric constant of 80). ΔG(M+X-), stabilization conferred by monovalent salt (M+X-) on transfer from an aqueous (salt-free) solution to a solution containing 25 mM M+X-. ΔG(M2+(X-)2), stabilization conferred by divalent salt (M2+X2-) on transfer from a monovalent salt solution to a solution containing both 25 mM M+X- and 10 mM M2+X2-. ΔΓ, change in preferential interaction coefficient or net M2+ ion uptake on conversion of one RNA structure into another. ΔΓ is equivalent to the slope of the metal ion dependence of the interconversion rate constant, i.e., ΔΓ = ∂log(k)/∂log [M2+] (26). In our electrostatic calculations, this term describes the change in number of thermodynamically “bound” counterions and includes both M2+ uptake and X- release (26). This term is directly related to the electrostatic free energies of the initial and end structures (26). U, undocked; C, contact; D, docked; NC, noncontact.
In the context of our experimental results in the absence of monovalent salt, which show a significant Mg2+ uptake on conversion from undocked to transition state and no significant uptake on conversion from transition to docked state (Fig. 3a), the above calculations strongly suggest that domains A and B are in a contact configuration in the folding transition state, whereas their relative orientation is not necessarily native-like. It is plausible that docking occurs via an ensemble of such contact transition states with different relative domain orientations. Only at very low Mg2+ concentrations (≤1 mM) does Mg2+ uptake on conversion from transition to docked state become significant, indicating perhaps a slight separation of domains A and B in the transition state, similar to the NC state.
In contrast, on titration with Na+ alone, metal ions are equally taken up on conversion from undocked to transition state and from transition to docked state (Fig. 3b). Similar behavior is found in the Mg2+ titration in a background of 500 mM Na+ (Fig. 3b). These results suggest that the location of the transition state shifts along the reaction coordinate dependent on the solvent ionic strength. In the latter two cases, a suitable model for the transition state is in fact our NC model discussed above in which the two RNA domain surfaces are 5 Å apart, leading to comparable uptake of metal ions from undocked to transition state and from transition to docked state. Moving the domains further apart (for example to adopt an L-shaped configuration) leads to a transition state with electrostatic properties similar to the undocked state, predicting little Mg2+ dependence in the docking rate constant (ΔΓU→L = 0.25, ΔΓL→D = 1.03), which is inconsistent with our experimental data. Again, under these latter conditions (<1 mM Mg2+ in the absence of Na+, on titration with Na+ alone, or on Mg2+ titration in the presence of 500 mM Na+) docking possibly occurs via an ensemble of transition states. These states may comprise ones with different relative orientations between domains A and B as well as with slightly varying domains distances; however, the distances cannot be significantly larger than in our NC model (5 Å).
Thus all our metal ion titration data are consistent with compact transition states in which domains A and B of the hairpin ribozyme are either in a contact configuration (as in the case of our Mg2+ titration) or in a close-to-contact configuration (as in the cases of our Na+ titration or our Mg2+ titration in the presence of Na+). The ubiquity of domain docking events in RNA folding (15, 29) and the generality of the electrostatic behavior observed in our generalized cylinder models suggest that our conclusions may be generally valid for tertiary structure assembly events in RNA folding.
What Constitutes the RNA Folding Free Energy Barrier? Another finding from our calculations is that not only metal ions but also the water solvent enormously stabilizes the compact transition and docked states against the extended undocked state, completely alleviating any coulombic repulsion (compare ΔGsolvation with ΔGcoulombic in Table 1). This effect arises from the increased polarization of the water molecules around the RNA by the stronger electrostatic fields surrounding more compact states. Our calculations also show that the addition of monovalent and divalent salt further stabilizes the compact transition and docked states against the undocked state, as indicated by ΔG(M+X–) and ΔG(M2+(X–)2), respectively, in Table 1. Thus we propose that the unfavorable free energy barrier to folding must involve additional, nonelectrostatic factors. As suggested before (41, 42), an analogy can be made to protein folding where transition states are often also stabilized by general, nonspecific interactions, i.e., the hydrophobic interactions of nonpolar amino acid side chains (9–11, 43).
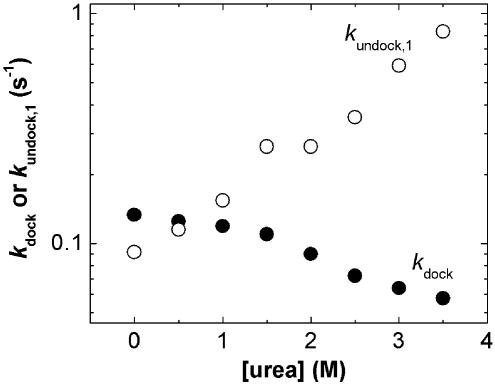
Among potential factors contributing to the free energy barrier of RNA folding, breaking of the helical stack between domains A and B in the undocked state (Fig. 1) is an obvious candidate. If such unstacking were the major cause of the folding barrier and/or presented a kinetic trap, the addition of a denaturant such as urea may be expected to significantly increase kdock (44, 45). In contrast, kdock decreases with increasing urea concentration (Fig. 5), indicating that unstacking is unlikely to be the major contributor to the folding free energy barrier. Consistent with this finding, disfavoring domain stacking by introducing either a three- or a nine-atom spacer between A14 and A15 of the 5′ ribozyme segment at the domain junction (Fig. 1a) does not accelerate kdock in the WT AC5 ribozyme construct used in the study (data not shown).
Fig. 5.
Effect of urea concentration on the rate constants for docking (•) and undocking (○).
Even under the most favorable experimental conditions, docking of the two small RNA domains of the hairpin ribozyme is slow (≈1 s–1, at 37°C and 500 mM Mg2+, Fig. 3a) compared with the analogous docking of α-helices in proteins (up to 106 s–1) (46). Based on our finding that the transition state is compact, we propose that the associated decrease in chain conformational entropy may contribute significantly to the folding free energy barrier. In addition, RNA secondary structure is very stable compared with protein secondary structure, resulting in strongly hierarchical folding pathways with tertiary structure forming after secondary structure (15, 29). The necessary breaking and rearrangement of secondary structure in the loop regions of domains A and B on docking (34, 47) may therefore also contribute significantly to the folding free energy barrier of the hairpin ribozyme.
Conclusions
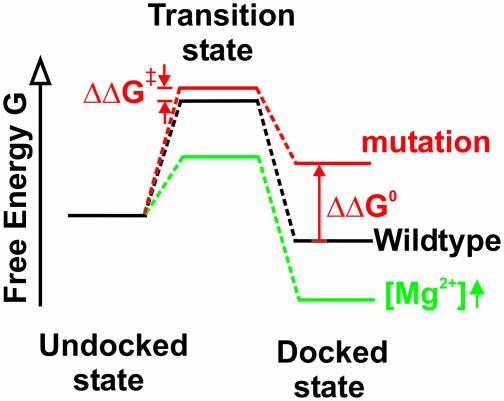
In summary, we have demonstrated that single-molecule FRET is a powerful tool for probing the transition state of an RNA folding reaction. Using this technique in combination with site-specific mutagenesis, metal ion titrations, and electrostatic modeling we have obtained an in-depth characterization of the transition state of a model two-state folding reaction, where two RNA helical domains dock to make specific tertiary contacts. Our mutational studies have shown that, although the inter-domain base pair is not formed yet in the transition state, the tertiary contacts involving backbone functional groups begin to form. We have shown, using metal ion titrations, that the transition state for folding is compact and electrostatically stabilized by metal ions and water solvent to a similar extent as the native state (Fig. 6). The favorable electrostatic stabilization of the transition state relative to the unfolded state leads us to propose that the unfavorable barrier to RNA folding is at least partly nonelectrostatic in nature, potentially caused by the decrease in conformational entropy and/or the breaking (rearrangement) of secondary structure. The compact transition states without well formed tertiary contacts observed here may be a general phenomenon in elementary RNA folding reactions. In large, multidomain RNA molecules such compact states are further stabilized and observed as folding intermediates, or metastable local minima, in contrast to the transition state we observe here for a small catalytic RNA (15, 19, 48, 49).
Fig. 6.
Free energy diagram of the docking and undocking transitions in dependence
of either a mutation that disrupts a tertiary interaction or an increase in
Mg2+ concentration, as concluded from our
single-molecule studies. The symbols are defined as
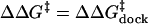 and
and
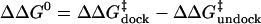 .
.
Acknowledgments
We thank Carol Fierke, David Engelke, David Draper, Sunney Xie, and Charles Lieber for helpful suggestions and comments on the manuscript. This work was supported in part by grants from the Office of Naval Research (to X.Z.), the National Institute of General Medical Sciences and the American Chemical Society (to N.G.W.), a National Institutes of Health training grant in the Molecular, Cellular, and Chemical Biology Program (to G.B.), a postdoctoral fellowship from the Swiss National Funds (to D.R.), a predoctoral fellowship from the National Science Foundation (to M.M.R.), and a National Institutes of Health training grant in Biophysics (to A.G.).
Abbreviation: FRET, fluorescence resonance energy transfer.
References
- 1.Doudna, J. A. & Cech, T. R. (2002) Nature 418, 222–228. [DOI] [PubMed] [Google Scholar]
- 2.Moore, P. B. & Steitz, T. A. (2002) Nature 418, 229–235. [DOI] [PubMed] [Google Scholar]
- 3.Collins, C. A. & Guthrie, C. (2000) Nat. Struct. Biol. 7, 850–854. [DOI] [PubMed] [Google Scholar]
- 4.Valadkhan, S. & Manley, J. L. (2002) Nat. Struct. Biol. 9, 498–499. [DOI] [PubMed] [Google Scholar]
- 5.Famulok, M. & Verma, S. (2002) Trends Biotechnol. 20, 462–466. [DOI] [PubMed] [Google Scholar]
- 6.Sullenger, B. A. & Gilboa, E. (2002) Nature 418, 252–258. [DOI] [PubMed] [Google Scholar]
- 7.Bartley, L. E., Zhuang, X., Das, R., Chu, S. & Herschlag, D. (2003) J. Mol. Biol. 328, 1011–1026. [DOI] [PubMed] [Google Scholar]
- 8.Silverman, S. K. & Cech, T. R. (2001) RNA 7, 161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bilsel, O. & Matthews, C. R. (2000) Adv. Protein Chem. 53, 153–207. [DOI] [PubMed] [Google Scholar]
- 10.Myers, J. K. & Oas, T. G. (2002) Annu. Rev. Biochem. 71, 783–815. [DOI] [PubMed] [Google Scholar]
- 11.Gruebele, M. (2002) Curr. Opin. Struct. Biol. 12, 161–168. [DOI] [PubMed] [Google Scholar]
- 12.Sclavi, B., Sullivan, M., Chance, M. R., Brenowitz, M. & Woodson, S. A. (1998) Science 279, 1940–1943. [DOI] [PubMed] [Google Scholar]
- 13.Treiber, D. K., Rook, M. S., Zarrinkar, P. P. & Williamson, J. R. (1998) Science 279, 1943–1946. [DOI] [PubMed] [Google Scholar]
- 14.Zhuang, X., Bartley, L. E., Babcock, H. P., Russell, R., Ha, T., Herschlag, D. & Chu, S. (2000) Science 288, 2048–2051. [DOI] [PubMed] [Google Scholar]
- 15.Thirumalai, D., Lee, N., Woodson, S. A. & Klimov, D. (2001) Annu. Rev. Phys. Chem. 52, 751–762. [DOI] [PubMed] [Google Scholar]
- 16.Liphardt, J., Onoa, B., Smith, S. B., Tinoco, I. J. & Bustamante, C. (2001) Science 292, 733–737. [DOI] [PubMed] [Google Scholar]
- 17.Russell, R., Zhuang, X., Babcock, H. P., Millett, I. S., Doniach, S., Chu, S. & Herschlag, D. (2002) Proc. Natl. Acad. Sci. USA 99, 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhuang, X., Kim, H., Pereira, M. J., Babcock, H. P., Walter, N. G. & Chu, S. (2002) Science 296, 1473–1476. [DOI] [PubMed] [Google Scholar]
- 19.Fang, X. W., Thiyagarajan, P., Sosnick, T. R. & Pan, T. (2002) Proc. Natl. Acad. Sci. USA 99, 8518–8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie, X. S. & Lu, H. P. (1999) J. Biol. Chem. 274, 15967–15970. [DOI] [PubMed] [Google Scholar]
- 21.Moerner, W. E. & Orrit, M. (1999) Science 283, 1670–1676. [DOI] [PubMed] [Google Scholar]
- 22.Walter, N. G. (2001) Methods 25, 19–30. [DOI] [PubMed] [Google Scholar]
- 23.Walter, N. G., Hampel, K. J., Brown, K. M. & Burke, J. M. (1998) EMBO J. 17, 2378–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ha, T. (2001) Methods 25, 78–86. [DOI] [PubMed] [Google Scholar]
- 25.Sharp, K. A., Friedman, R. A., Misra, V., Hecht, J. & Honig, B. (1995) Biopolymers 36, 245–262. [DOI] [PubMed] [Google Scholar]
- 26.Misra, V. K. & Draper, D. E. (2002) J. Mol. Biol. 317, 507–521. [DOI] [PubMed] [Google Scholar]
- 27.Walter, N. G. & Burke, J. M. (1998) Curr. Opin. Chem. Biol. 2, 24–30. [DOI] [PubMed] [Google Scholar]
- 28.Walter, N. G., Burke, J. M. & Millar, D. P. (1999) Nat. Struct. Biol. 6, 544–549. [DOI] [PubMed] [Google Scholar]
- 29.Brion, P. & Westhof, E. (1997) Annu. Rev. Biophys. Biomol. Struct. 26, 113–137. [DOI] [PubMed] [Google Scholar]
- 30.Ha, T., Enderle, T., Ogletree, D. F., Chemla, D. S., Selvin, P. R. & Weiss, S. (1996) Proc. Natl. Acad. Sci. USA 93, 6264–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deniz, A. A., Laurence, T. A., Beligere, G. S., Dahan, M., Martin, A. B., Chemla, D. S., Dawson, P. E., Schultz, P. G. & Weiss, S. (2000) Proc. Natl. Acad. Sci. USA 97, 5179–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Talaga, D. S., Lau, W. L., Roder, H., Tang, J., Jia, Y., DeGrado, W. F. & Hochstrasser, R. M. (2000) Proc. Natl. Acad. Sci. USA 97, 13021–13026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuler, B., Lipman, E. A. & Eaton, W. A. (2002) Nature 419, 743–747. [DOI] [PubMed] [Google Scholar]
- 34.Rupert, P. B. & Ferre-D'Amare, A. R. (2001) Nature 410, 780–786. [DOI] [PubMed] [Google Scholar]
- 35.Fersht, A. R., Itzhaki, L. S., ElMasry, N. F., Matthews, J. M. & Otzen, D. E. (1994) Proc. Natl. Acad. Sci. USA 91, 10426–10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fersht, A. R., Matouschek, A. & Serrano, L. (1992) J. Mol. Biol. 224, 771–782. [DOI] [PubMed] [Google Scholar]
- 37.Pinard, R., Lambert, D., Walter, N. G., Heckman, J. E., Major, F. & Burke, J. M. (1999) Biochemistry 38, 16035–16039. [DOI] [PubMed] [Google Scholar]
- 38.Murray, J. B., Seyhan, A. A., Walter, N. G., Burke, J. M. & Scott, W. G. (1998) Chem. Biol. 5, 587–595. [DOI] [PubMed] [Google Scholar]
- 39.Pyle, A. M. (2002) J. Biol. Inorg. Chem. 7, 679–690. [DOI] [PubMed] [Google Scholar]
- 40.Fedor, M. J. (2000) J. Mol. Biol. 297, 269–291. [DOI] [PubMed] [Google Scholar]
- 41.Cate, J. H., Hanna, R. L. & Doudna, J. A. (1997) Nat. Struct. Biol. 4, 553–558. [DOI] [PubMed] [Google Scholar]
- 42.Murthy, V. L. & Rose, G. D. (2000) Biochemistry 39, 14365–14370. [DOI] [PubMed] [Google Scholar]
- 43.Daggett, V. & Fersht, A. R. (2003) Trends Biochem. Sci. 28, 18–25. [DOI] [PubMed] [Google Scholar]
- 44.Rook, M. S., Treiber, D. K. & Williamson, J. R. (1998) J. Mol. Biol. 281, 609–620. [DOI] [PubMed] [Google Scholar]
- 45.Fang, X. W., Pan, T. & Sosnick, T. R. (1999) Nat. Struct. Biol. 6, 1091–1095. [DOI] [PubMed] [Google Scholar]
- 46.Mayor, U., Guydosh, N. R., Johnson, C. M., Grossmann, J. G., Sato, S., Jas, G. S., Freund, S. M., Alonso, D. O., Daggett, V. & Fersht, A. R. (2003) Nature 421, 863–867. [DOI] [PubMed] [Google Scholar]
- 47.Walter, N. G., Harris, D. A., Pereira, M. J. & Rueda, D. (2001) Biopolymers 61, 224–242. [DOI] [PubMed] [Google Scholar]
- 48.Buchmueller, K. L., Webb, A. E., Richardson, D. A. & Weeks, K. M. (2000) Nat. Struct. Biol. 7, 362–366. [DOI] [PubMed] [Google Scholar]
- 49.Russell, R., Millett, I. S., Tate, M. W., Kwok, L. W., Nakatani, B., Gruner, S. M., Mochrie, S. G., Pande, V., Doniach, S., Herschlag, D. & Pollack, L. (2002) Proc. Natl. Acad. Sci. USA 99, 4266–4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicholls, A., Sharp, K. A. & Honig, B. (1991) Proteins 11, 281–296. [DOI] [PubMed] [Google Scholar]