Abstract
A model was built to examine the kinetics of regulatory cascades such as occur in developmental gene networks. The model relates occupancy of cis-regulatory target sites to transcriptional initiation rate, and thence to RNA and protein output. The model was used to simulate regulatory cascades in which genes encoding transcription factors are successively activated. Using realistic parameter ranges based on extensive earlier measurements in sea urchin embryos, we find that transitions of regulatory states occur sharply in these simulations, with respect to time or changing transcription factor concentrations. As is often observed in developing systems, the simulated regulatory cascades display a succession of gene activations separated by delays of some hours. The most important causes of this behavior are cooperativity in the assembly of cis-regulatory complexes and the high specificity of transcription factors for their target sites. Successive transitions in state occur long in advance of the approach to steady-state levels of the molecules that drive the process. The kinetics of such developmental systems thus depend mainly on the initial output rates of genes activated in response to the advent of new transcription factors.
Developmental processes are driven forward by spatial and temporal changes in regulatory state, that is, progression in the sets of transcription factors present in the cell nuclei. As these factors are produced by the regulatory genes active in the same nuclei, the underlying control process is often described as a transcriptional regulatory “cascade.” The steps of such a cascade are as follows: transcription factors present in certain nuclei in an embryo interact with the cis-regulatory elements of genes encoding other transcription factors; transcription of these previously quiescent genes is thereby induced, at a rate dependent on the levels of occupancy of their cis-regulatory elements; the new transcription factors accumulate, and together with other transcription factors, bind to different, previously inactive cisregulatory elements, and a yet again novel regulatory state is in this way generated. Of course, the basic requirement for the existence of the successive stages that make up a regulatory cascade is that each is used as a developmental control point at which multiple inputs from other regulatory genes and from external signal transduction systems are processed. Therefore developmental control systems have a network-like architecture, rather than acting as a linear chain of events (1–3); otherwise the initial regulatory state could just as well be used as the last, and there would be no cascade. But even though oversimplified, in that all collateral inputs are ignored, the idea of linear regulatory gene cascades is useful. They represent a commonly encountered fundamental of gene regulatory networks (GRNs), that is, processes in which the products of given regulatory genes are directly required for activation of downstream regulatory genes. The kinetics of regulatory state change cannot be faster than permitted by the kinetics with which the successive steps of the constituent regulatory gene cascades take place. Here, we address the question of how these kinetics operate, applying measured and estimated parameters from the developing sea urchin embryo: do the gene products at each stage attain a steady-state level required for activation of the succeeding stage, or do successive gene activations in a cascade follow one another without ever coming close to steady state?
Model
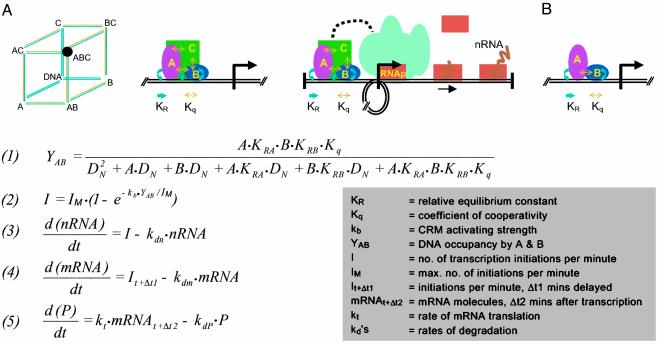
The mathematical model used for the simulations is summarized in Fig. 1. (Derivations are given in Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org.) The model is conveniently described in three parts: occupancy of cis-regulatory DNA by transcription factors with respect to transcription factor concentration, transcription initiation rate with respect to occupancy, and output of mRNA and proteins with respect to initiation rate.
Fig. 1.
Model for effect of cis-regulatory occupancy on transcriptional output. (A) Cartoon representing a three-factor cis-regulatory occupancy system. Factors A, B, and C can bind in any order (Left), although triple occupancy is needed for function. All factors interact energetically with one another when brought into proximity as well as with the DNA, i.e., bind cooperatively. Relative equilibrium constants (KR) for interaction with DNA are symbolized in blue, and cooperativity constants (Kq) are symbolized in yellow. The black circle highlights the functional state. (Center) Assembly of triple complex. (Right) DNA looping results in proximity of fully loaded CRM to the BTA (teal blob), activation thereof, and recruitment of polymerase (orange bricks); transcription ensues. (B) Two-factor CRM, model equations, and terms. For derivations, see Supporting Text. Both factors (A and B) must be bound to the CRM at once for activation of transcription to occur. Eq. 1 defines double occupancy of the CRM. Eq. 2 defines the relation between occupancy and initiation rate. Eqs. 3–5 define the kinetics of nRNA, mRNA, and protein output.
cis-Regulatory Module (CRM) Occupancy. Given CRMs are always engaged by multiple factors, the advent of which reflects, respectively, various prior developmental regulatory states (1). From this fundamental property derives critical features of the kinetic behavior of transcriptional regulatory systems, and the more components are required per CRM the more accentuated are these features. The cartoon in Fig. 1 A shows interactions with three DNA-binding transcription factors, but in the simulations, both for simplicity of calculation and to take the most conservative case, we consider only two transcription factors required to be bound in the CRM for it to be active (Fig. 1B). Eq. 1 of Fig. 1 calculates the double occupancy (YAB) of a CRM by factors A and B. Here we have followed the probabilistic solution of Ackers et al. (4) worked out originally for λ phage, but instead of using equilibrium constants, we have recast the calculations in terms of relative equilibrium constants (KR). For a given factor and target site, KR is the ratio of the equilibrium constant (KS) for interactions with the sequence-specific CRM sites to the equilibrium constant (KN) for the sequence-independent affinity of the factor for DNA in general. The use of KR to calculate DNA–protein complex formation was introduced by Felsenfeld and associates (5). In animal cells the amount of DNA is so large, and therefore there are so many nonspecific DNA target sites, that the accurate determinant of specific CRM site occupancy is KR, not KS (5, 6). An additional practical motivation is that many KR values for developmentally important sea urchin embryo transcription factors have been obtained (7), and with Eq. 1 we can now calculate occupancies by using these data. Note that Eq. 1 includes the term Kq for cooperative interactions between the transcription factors (4). As we show below, if there were no cooperativity (Kq = 1), the kinetic behavior of the transcription systems in our model would be quite different than if Kq is >1. However, highly cooperative binding behavior is usually observed whenever appropriate measurements are made on animal CRM–transcription factor complexes (e.g., refs. 8–16) and is undoubtedly the rule not the exception.
Occupancy and Initiation. The starting point is the principle that initiation rate, for any given basal transcription apparatus (BTA), is controlled by the level of occupancy of specific sites in the relevant CRM. This statement applies whether the CRM is a proximal element near the BTA or a distant enhancer. When the CRM is empty the gene is silent (or runs at a biologically insignificant “background” level). When the gene is being transcribed the rate should depend on the fraction of time the activators are in position on the relevant CRM. There is abundant quantitative evidence that CRM occupancy by the complex of proteins bound at the CRM target sites (plus other proteins bound in turn to these) determines transcription rate; see, for example, the precise regulatory control of output kinetics demonstrated for the sea urchin endo16 gene by mutation of individual CRM target sites (17, 18). Initiation rate in this discussion is taken to determine directly the ultimate transcription rate. Although small variations in the rate of polymerase translocation have been noted, the main part of the orders of magnitude dynamic range (from silence or basal rate to maximal rate) observed in regulated gene expression directly reflects frequency of initiation. This was first established unequivocally for in vivo animal systems by electron microscopy of transcription complexes at various levels of activity (19–21). The average rate at which the polymerase translocates along the gene during transcription is more or less constant for a given organism at a given temperature, and the transcriptional output depends on the rate at which the CRM causes polymerases to load into the BTA and begin transcribing.
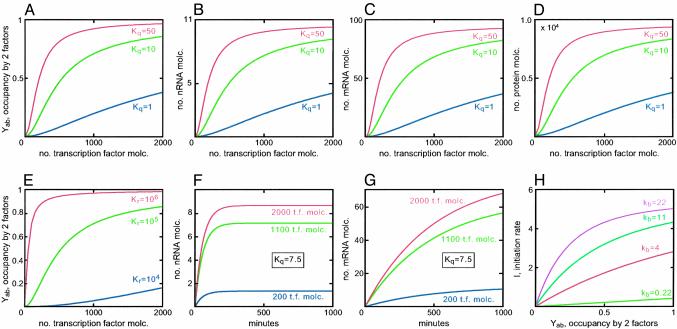
For sea urchin embryos at 15°C the translocation rate was measured at 6–9bp·s–1 (refs. 19 and 22; in the following we have used the upper limit, 9 bp·s–1). This sets the maximum initiation rate, because a new polymerase cannot enter the transcription complex at the initiation site and begin transcribing until the previous one has moved out of the way. At maximum initiation rate the polymerases are as closely packed as possible, with a center-to-center distance of ≈100 bp, and so the maximum rate is ≈11 transcripts initiated (and completed) per min, or approximately one transcript per 5.5 s (19). Activation of transcription by CRM transcription complexes occurs by several different (nonexclusive) pathways, e.g., by inducing nucleosome histone acetylation or by interaction with an activating mediator complex (23, 24). But whatever the mechanism, cis-regulatory activation of transcription requires an interaction between the CRM complex and the BTA (direct or otherwise). In our model it is assumed that these interaction events occur stochastically in time, at an average rate that is proportional to the occupancy of the CRM. As described in the derivation (see Supporting Text), we applied Poisson statistics to obtain Eq. 2 of Fig. 1. Eq. 2 has the form that at low levels of occupancy, the initiation rate is proportional to occupancy, but at very high occupancy, if prospective activating interactions occur too frequently to be useful because the maximal rate cannot be exceeded, they are excluded from function. In real life the proportionality constant (kb in Eq. 2) relating occupancy to initiation rate must vary, depending on the CRM and its constituents, for a given BTA. In our simulations we provisionally evaluated the average value of this constant by application of measured average KR and transcription factor concentration data to Eq. 1. The calculated occupancy was then used together with typical transcription rate measurements to find kb in Eq. 2 (see Fig. 2 legend and Supporting Text).
Fig. 2.
Simulations exploring effects of cooperativity of specific transcription factor binding affinity, transcription factor concentrations, and strength of activation function. Default settings were for Eq. 1, KRA = 105; Kq = 10; DN is taken as ≈20% of genome size. For molar calculations the volume of a sea urchin embryo cell nucleus was taken to be 4 × 10–15 liter. For 1.6 × 108 sites, DN is thus 0.07 M. For transcription factor concentration, A = B = 2,000 molecules (molc.) per nucleus. For Eq. 2, IM = 5.45 initiations per min–1 per gene; kb = 0.44 (see Supporting Text). For Eqs. 3–5, kdn = 0.02; kdm = 0.002; kdP = 0.02; Δt1 was taken as 20 min, and Δt2 as 7 min; kt = 2 molecules min–1 per mRNA. The values are all from Table 1, except that the default of kb was computed as follows: YAB was calculated from the default values (Eq. 1) and kb was obtained from Eq. 2, such that as YAB → 1, I → IM. The default for Kq is close to that for a –2 KCal cooperativity exchange (Kq = 7.5), as in the case considered in ref. 5. (A–D) Cooperativity effects. (A) Effects on occupancy of the CRM by both factors, as a function of number of transcription factor molecules per nucleus. (B–D) Effects on outputs of nRNA, mRNA, and protein, respectively, as transcription factor concentration is varied. (E) Effect of KR on CRM occupancy, as transcription factor concentration is varied (for examples of transcription factors for which these KR values apply, see Supporting Text). (F) Time kinetics of nRNA and mRNA accumulation, respectively, for different transcription factor concentrations; in E and FKq was set at 7.5. (H) Effect of varying kb on the relation between initiation rate (events per min) and occupancy (Eq. 2).
Transcription and Translation. Eqs. 3–5 of the model in Fig. 1 are standard descriptions of the overall processes of macromolecular synthesis and decay, assuming no translational regulation and average stochastic degradation processes. For the sea urchin embryo just such decay kinetics have been measured for individual mRNAs and mRNA populations (19, 25, 26). An important assumption in the model is that nuclear RNA (nRNA) processing is 100% efficient, so that in molecular terms transcription initiation rate equals the rate of mRNA entry into the cytoplasm (19, 25).
Parameters
Some relevant parameters from many measurements on sea urchin embryos are assembled in Table 1. The particular measurements on which Table 1 is based are listed in Table 2, which is published as supporting information on the PNAS web site. In the following simulations these are used to provide realistic constraints on the kinetic behavior of the model parameters. Note that whereas KS values depend very sharply on salt concentration, KR values are independent thereof (KR = KS/KN, where KS and KN are the equilibrium constants for site-specific and nonspecific interactions with DNA, respectively). The range in KRs in Table 1 will apply to any animal system. Numbers of mRNA and protein molecules per cell of course scale with cell size. For comparison with other systems the basic polymerase translocation and translation rates depend on temperature, as long ago observed, with a fold change in rate per 10°C (Q10) of ≈2–2.5 (19).
Table 1. Some transcription and prevalence parameters for sea urchin embryos.
| General rates* |
| PolII translocation rate: 9 bp·sec-1 |
| mRNA translation rate: Two molecules per mRNA-1·min-1 |
| Average t1/2 mRNA: 3-5 h |
| Average t1/2 nRNA: 20 min |
| Transcription rates† |
| Maximum: 11 molecules per min-1·gene-1 |
| Average for low prevalence species: 0.012 molecules per min-1·gene-1 |
| Average for moderate prevalence (20-40 h) species: 0.17 molecules per min-1·gene-1 |
| spec1: 0.9 molecules per min-1·gene-1 |
| mactin: 0.15 molecules per min-1·gene-1 (65 h) |
| Cyllla actin: 1.2-2.5 molecules per min-1·gene-1 (9-15 h) |
| Cyl actin: 0.16 molecules per min-1·gene-1 (65 h) |
| Parameters for transcription factors‡ |
| KR§ relative equilibrium constant (nine factors): 1.4 × 104 - 1.5 × 106 |
| P0¶, active protein molecules per nucleus (eight factors): 300-10,000 |
| KS∥, equilibrium constants (six factors): 2.8 × 107 M-1 to 1.7 × 108 M-1 |
| mRNAs per cell** (12 factors): av 37, range 5-150 |
For original sources and review, see Davidson (19). Data are for Strongylocentrotus purpuratus and/or Lytechinus pictus at 15°C.
See Table 2, from which these ranges are summarized.
Application of Parameters to Model: Transcriptional Control of a Single Gene
The simulations collated in Fig. 2 show the effects of the main biologically important parameters in cis-regulatory occupancy, and thence, output. Three of these parameters depend essentially on the intrinsic structure of the transcription factors involved, and the intrinsic sequence of their CRM target sites: the cooperativity constant Kq, the site-specific affinity constant KR, and kb, the constant representing the strength of the activation function that the occupied CRM generates. The fourth parameter, the concentration at any given time of the active transcription factors in the nucleus, is the variable on which gene regulation operates. This is of course controlled by upstream processes, namely transcriptional regulation of the genes encoding factors and signaling processes. In each of the simulations of Fig. 2 all of the parameters, except those in play, were set at default values taken or calculated from the measurements summarized in Table 1 (see Fig. 2 legend for values).
Fig. 2 A–D explores the kinetic consequences of cooperativity between transcription factors binding on the CRM and also shows that the outputs of the model are appropriate in absolute terms (compare Table 1). Fig. 2 A illustrates the sharp increase in occupancy as a function of transcription factor concentration, which even a modest amount of cooperativity produces. This steep occupancy function is reflected in the RNA and protein output curves (Fig. 2 B–D). Note that this is quantitatively a very conservative argument, because in real CRMs there are many more than two interacting factors, and the product of the Kqs will most likely far exceed the value 10, which implies only a little more than 2 Kcal of free energy per mole of complex. Much higher cooperativity values (red curves) only sharpen the relationship further. The important point is that, as expected (4), cooperativity in transcription factor binding produces a switch-like transcriptional output of nRNA, and hence mRNA and protein, with respect to input transcription factor concentrations. Fig. 2E demonstrates that the specific affinities of the factors for their target sites are similarly important: the higher the KRs the more switch-like the occupancy function (and similarly the mRNA and protein output functions, data not shown).
The dynamic output response to different transcription factor levels is shown in Fig. 2 F and G. Note that the response is very sensitive at low levels (i.e., compared with those levels typically observed; compare Table 1 and Supporting Text), and the steady-state nRNA outputs further increase only a little as factor concentration rises >1,000 molecules per nucleus. The slow approach to steady state of the output mRNA, still incomplete at 16 h, is caused by the typical half-lives of several hours, measured for sea urchin embryo mRNAs (19, 25, 26), and applied in the model simulations. The implication is that in the developing embryo, for transcription factor mRNAs that behave in this way, steady state will often never be attained, for given phases of transcription activity often last only for a few hours, usually <10.
Finally, the effect of differences in CRM activity constant kb in Eq. 2 is illustrated in Fig. 2H. We see that particularly weak or strong CRM activation complexes can very greatly alter the relation between CRM occupancy and initiation rate.
Cascade Kinetics
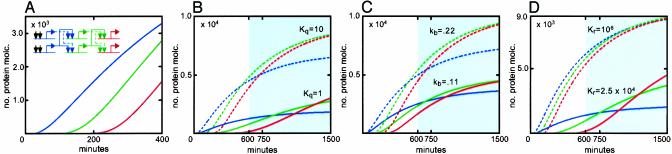
The kinetics of successive phases of expression in a three-gene model cascade are shown in Fig. 3. Again we consider a system in which the CRMs require binding of two factors for activity, here at each step of the cascade. The factor pairs required at each step in each CRM are produced by the preceding pair of regulatory genes (see cartoon in Fig. 3A). Initiation kinetics are illustrated in Fig. 3A, using the default settings indicated in the legend of Fig. 2. An interval of a couple of hours separates the successive accumulation curves. Their main feature is the character of the kinetics: successive phases of gene expression (i.e., activation of successive genes) occur while the preceding phase is still in its initial slope. That is, the steady-state values that would or could eventually result have virtually nothing to do with the essential initiation behavior of the regulatory cascade. This can be seen in the lower-resolution plots of Fig. 3 B–D. Here the kinetics are shown extending out to 25 h, but in life the last l5 h (or more) of the process would in most cases never occur (gray shading), because the pattern of gene expression would already have changed.
Fig. 3.
Simulations of cascade behavior. (A) Cartoon of three-step cascade and initiation rates. Double occupancy of each CRM is required as before. Only the first 400 min are shown; for default parameter values see Fig. 2. (B) Effects of cooperativity. (C) Effects of a 2-fold difference in kb, the activation efficiency (the default value calculated as in the legend of Fig. 2 is 0.44). (D) Effects of KR. In B–D, dashed lines represent upper parameter assumed; solid lines represent lower parameter; the gray areas represent the time after 10 h. These portions of the curves would probably never be observed in life because in the developing embryo given patterns of gene expression in given cells rarely extend into this time domain. Molc., molecules.
The effects of cooperativity are examined in Fig. 3B, and it is evident how important for the switch-like behavior of the gene activation cascade is this parameter. We again stress that the assumption in the model of a two-factor CRM is extremely conservative, and the all-or-none assembly of well studied CRM–transcription factor complexes (e.g., refs. 8, 10, and 16) suggests far higher cooperativity than the minimal default assumptions here; in life the activation functions will therefore be steeper. Variations in Kq and KR should and do behave similarly (Eq. 1): note in Fig. 3D that not only does low KR decrease output in the cascade, but like low Kq, it also greatly increases the interval separating activation phases. As noted earlier, Kq and KR are intrinsic parameters, dependent on the identity of the transcription factors, and so also is the activation efficiency of the CRM, which also may sharply affect cascade output (Fig. 3C). In life, the overall cascade kinetics, i.e., both the intervals between successive activations and the rates of transcript accumulation, depend immediately on these parameters. But ultimately the kinetic behavior of a given GRN transcription system depends on genomic regulatory programming. For it is the nucleotide sequence of the CRMs that determines the identities of the relevant factors that bind there, and the quality, organization, and number of their target sites in these CRMs that determine the cooperativity and DNA interaction energies generated when they bind.
Finally, the rapid response kintetics displayed by the model also suggest a possible answer to a conundrum. Because the turn on/off times for different regulatory genes (or the same gene in different cellular contexts) can be quite different, there can be no guarantee that regulatory inputs always change in a fixed order, resulting in potentially inappropriate spikes in gene expression. But the kinetic behavior of the model results in a smoothing out of the effects of input irregularities, essentially because the turnover rates are relatively slow, and the accumulation functions for the biosynthetic products of the transcriptional cascade are therefore insensitive to transient variations in regulatory input. The genes in the model thus act as “integrating amplifiers” of their regulatory inputs (a simulated illustration of this effect is shown in Fig. 4, which is published as supporting information on the PNAS web site).
Discussion
The model simulations in Figs. 2 and 3 show that genes are activated successively in a regulatory network cascade long before steady states are attained. Clearly an underlying cause of this behavior is the regulatory network architecture, which lacks adaptive level controls or other devices common in homeostatic systems. In early development the job is to move irreversibly forward, from one regulatory state to the next.
In considering the simulation results, an important question is how accurate are the parameters used likely to be? The data in Table 1 are based on solid measurements that are internally consistent with one another (ref. 19; Supporting Text), but there remain two unmeasured uncertainties not included in Table 1. The first is that absolute RNA synthesis and turnover measurements have not been directly made on transcription factor transcripts in sea urchin embryos. We have assumed that the same parameters hold for these as for other transcripts of similar prevalence (Supporting Text includes a relatively large number of regulatory factor mRNA prevalence measurements). Conceivably, though, these particular mRNAs turn over faster than the average, which would run the transcription rates up higher than we have assumed. However, this seems improbable: at least for several cases in the endomesoderm GRN (27) in which genes are turned off by a known repressor, the subsequent decay in mRNA prevalence indicates a several hour t1/2. The second unknown is protein turnover rate (kdp). Here there are almost no relevant data, and the assumptions we chose (t1/2 = 20 min) are supported only by a calculation based on rates of translation and factor prevalence, carried out for some factors that remain at similar levels throughout (Supporting Text; Table 1). Fortunately, this parameter appears only in Eq. 5. It affects none of the simulations in Fig. 2, or conclusions thereof, except the protein accumulation curves of Fig. 2D. If the turnover rates were lower than assumed, the behavior of the cascades in Fig. 3 would become even more accentuated, because at each point there would be more transcription factor for a given amount of mRNA. Furthermore, the prevalence data for the proteins (Table 2, P0 values) preclude a much higher turnover rate than we chose. In sum, we can regard the general conclusions with some confidence: first, regulatory gene cascades in sea urchin embryos are expected to act as kinetic systems that depend on initial rates, not on steady states; second, that these systems tend toward a mode of operation in which genes are switched on in succession. This characteristic depends on structural features, in particular the high specificity of the DNA-binding factors, and the organization of the CRMs. These contain clusters of target sites, which when occupied generate highly cooperative transcription factor complexes. We note, furthermore, that perturbation studies carried out on ftz and its target genes in Drosophila (28) produce sequential gene activation kinetics similar to those of Fig. 3A: although on a time scale of minutes rather than hours, the initiation of downstream gene expression in the cascade again occurs before the accumulation of the driver molecules ever approach steady state.
Aside from the obvious philosophical interest of these structure/function relationships, there is a practical import. The result tells us what kinds of kinetic treatments are reasonable for the analysis of developmental GRNs and what are not. It is also satisfying that the straightforward relationships of Fig. 1B make sense out of the absolute values of Table 1, in terms of the operation of the whole transcriptional system.
Early embryonic development is programmed at the genomic cis-regulatory level, essentially as a series of switches that are thrown given the appropriate inputs. The controlling events consist essentially of presentation in given nuclei at given times of active transcription factors, i.e., the CRM inputs. Although there are obviously tolerance ranges, many features lead one to doubt whether exquisite level control is essential. Among these are the transience and progressivity of regulatory patterns, the frequent occurrence of genomically mandated positive feedback loops (2, 3), and the Boolean behavior of repressive gene silencers. Activation of genes is controlled essentially by the products of the occupancy values, and this can result in sharp, Boolean-like changes in regulatory state. We think that the properties of developmental transcription kinetics are those required to execute the process of switching on genes progressively that is written in the genomic regulatory logic map. Given the sea urchin endomesoderm GRN (27), this idea can now be tested by comparison of model kinetic predictions against time-course measurements for expression of genes that are causally linked in cascade architecture.
Supplementary Material
Acknowledgments
We thank Professors David Galas, Richard A. Firtel, and Ellen Rothenberg for extremely helpful critical reviews of the manuscript. This work was supported by National Institutes of Health Grants GM-61005 and HD-37105.
Abbreviations: GRN, gene regulatory network; CRM, cis-regulatory module; BTA, basal transcription apparatus; nRNA, nuclear RNA.
References
- 1.Davidson, E. H. (2001) Genomic Regulatory Systems: Development and Evolution (Academic, San Diego).
- 2.Davidson, E. H., Rast, J. P., Oliveri, P., Ransick, A., Calestani, C., Yuh, C.-H., Minokawa, T., Amore, G., Hinman, V., Arenas-Mena, C., et al. (2002) Science 295, 1669–1678. [DOI] [PubMed] [Google Scholar]
- 3.Davidson, E. H., McClay, D. R. & Hood, L. (2003) Proc. Natl. Acad. Sci. USA 100, 1475–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ackers, G. K., Johnson, A. D. & Shea, M. A. (1982) Proc. Natl. Acad. Sci. USA 79, 1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emerson, B. M., Lewis, C. D. & Felsenfeld, G. (1985) Cell 41, 21–30. [DOI] [PubMed] [Google Scholar]
- 6.Lin, S. Y. & Riggs, A. D. (1975) Cell 4, 107–111. [DOI] [PubMed] [Google Scholar]
- 7.Calzone, F. J., Thézé, N., Thiebaud, P., Hill, R. L., Britten, R. J. & Davidson, E. H. (1988) Genes Dev. 2, 1074–1088. [DOI] [PubMed] [Google Scholar]
- 8.Garrity, P. A., Chen, D., Rothenberg, E. V. & Wold, B. J. (1994) Mol. Cell. Biol. 14, 2159–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Driever, W. & Nüsslein-Volhard, C. (1989) Nature 337, 138–143. [DOI] [PubMed] [Google Scholar]
- 10.Thanos, D. & Maniatis, T. (1995) Cell 83, 1091–1100. [DOI] [PubMed] [Google Scholar]
- 11.Sugawara M., Scholl, T., Mahanta, S. K., Ponath, P. D. & Strominger, J. L. (1995) J. Exp. Med. 182, 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sigvardsson, M., Clark, D. R., Fitzsimmons, D., Doyle, M., Åkerblad, P., Breslin, T., Bilke, S., Li, R., Yeamans, C., Zhang, G. & Hagman, J. (2002) Mol. Cell. Biol. 22, 8539–8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao, Z. X. & Nadal-Ginard, B. (1996) J. Biol. Chem. 271, 14371–14375. [DOI] [PubMed] [Google Scholar]
- 14.Gregory, R. C., Taxman, D. J., Seshasayee, D., Kensinger, M. H., Bieker, J. J. & Wojchowski, D. M. (1996) Blood 87, 1793–1801. [PubMed] [Google Scholar]
- 15.Braun, H. & Suske, G. (1998) J. Biol. Chem. 273, 9821–9828. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez-Munain, C., Roberts, J. L. & Krangel, M. S. (1998) Mol. Cell. Biol. 18, 3223–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuh, C.-H., Bolouri, H. & Davidson, E. H. (1998) Science 279, 1896–1902. [DOI] [PubMed] [Google Scholar]
- 18.Yuh, C.-H., Bolouri, H. & Davidson, E. H. (2001) Development (Cambridge, U.K.) 128, 617–628. [DOI] [PubMed] [Google Scholar]
- 19.Davidson, E. H. (1986) Gene Activity in Early Development (Academic, Orlando, FL), 3rd Ed.
- 20.Miller, O. L. & Bakken, A. H. (1972) Acta Endocrinol. 168, Suppl., 155–177. [DOI] [PubMed] [Google Scholar]
- 21.Busby, S. & Bakken, A. H. (1979) Chromosoma 79, 85–104. [DOI] [PubMed] [Google Scholar]
- 22.Aronson, A. I. & Chen, K. (1977) Dev. Biol. 59, 39–48. [DOI] [PubMed] [Google Scholar]
- 23.Malik, S. & Roeder, R. G. (2000) Trends Biochem. Sci. 25, 277–283. [DOI] [PubMed] [Google Scholar]
- 24.Ito, M. & Roeder, R. G. (2001) Trends Endocrinol. Metab. 12, 127–134. [DOI] [PubMed] [Google Scholar]
- 25.Cabrera, C. V., Lee, J. J., Ellison, J. W., Britten, R. J. & Davidson, E. H. (1984) J. Mol. Biol. 174, 85–111. [DOI] [PubMed] [Google Scholar]
- 26.Lee, J. J., Calzone, F. J. & Davidson, E. H. (1992) Dev. Biol. 149, 415–431. [DOI] [PubMed] [Google Scholar]
- 27.Davidson, E. H., Rast, J. P., Oliveri, P., Ransick, A., Calestani, C., Yuh, C.-H., Minokawa, T., Amore, G., Hinman, V., Arenas-Mena, C., et al. (2002) Dev. Biol. 246, 162–190. [DOI] [PubMed] [Google Scholar]
- 28.Nasiadka, A., Dietrich, B. H. & Krause, H. M. (2002) in Advances in Developmental Biology and Biochemistry, ed. DePamphilis, M. (Elsevier, Amsterdam), pp. 155–204.
- 29.Zeller, R. W., Britten, R. J. & Davidson, E. H. (1995) Dev. Biol. 170, 75–82. [DOI] [PubMed] [Google Scholar]
- 30.Höög, C., Calzone, F. J., Cutting, A. E., Britten, R. J. & Davidson, E. H. (1991) Development (Cambridge, U.K.) 112, 351–364. [DOI] [PubMed] [Google Scholar]
- 31.Calzone, F. J., Höög, C., Teplow, D. B., Cutting, A. E., Zeller, R. W., Britten, R. J. & Davidson, E. H. (1991) Development (Cambridge, U.K.) 112, 335–350. [DOI] [PubMed] [Google Scholar]
- 32.Cutting, A. E., Höög, C., Calzone, F. J., Britten, R. J. & Davidson, E. H. (1990) Proc. Natl. Acad. Sci. USA 87, 7953–7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coffman, J. A., Kirchhamer, C. V., Harrington, M. G. & Davidson, E. H. (1996) Dev. Biol. 174, 43–54. [DOI] [PubMed] [Google Scholar]
- 34.Martinez, P. & Davidson, E. H. (1997) Dev. Biol. 181, 213–222. [DOI] [PubMed] [Google Scholar]
- 35.Wang, D. G.-W., Kirchhamer, C. V., Britten, R. J. & Davidson, E. H. (1995) Development (Cambridge, U.K.) 121, 1111–1122. [DOI] [PubMed] [Google Scholar]
- 36.Arenas-Mena, C., Cameron, R. A. & Davidson, E. H. (2000) Development (Cambridge, U.K.) 127, 4631–4643. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.