Abstract
We report here the identification of an angiopoietin-related growth factor (AGF). To examine the biological function of AGF in vivo, we created transgenic mice expressing AGF in epidermal keratinocytes (K14-AGF). K14-AGF mice exhibited swollen and reddish ears, nose and eyelids. Histological analyses of K14-AGF mice revealed significantly thickened epidermis and a marked increase in proliferating epidermal cells as well as vascular cells in the skin compared with nontransgenic controls. In addition, we found rapid wound closure in the healing process and an unusual closure of holes punched in the ears of K14-AGF mice. Furthermore, we observed that AGF is expressed in platelets and mast cells, and detected at wounded skin, whereas there was no expression of AGF detected in normal skin tissues, suggesting that AGF derived from these infiltrated cells affects epidermal proliferation and thereby plays a role in the wound healing process. These findings demonstrate that biological functions of AGF in epidermal keratinocytes could lead to novel therapeutic strategies for wound care and epidermal regenerative medicine.
Skin tissues, especially epidermis, are a barrier against the environment, which becomes accessible in wounds and various infections. An important goal in wound management is to achieve rapid wound closure. Analysis of gene activation in skin tissues shows that transforming growth factor-α (TGF-α) (1) produced by keratinocytes and keratinocyte growth factor (KGF) (2, 3) made by dermal fibroblasts are both powerful growth factors for epidermal keratinocytes, indicating an important role for the interaction between dermis–epidermis in skin development. Although these two growth factors play critical roles in wound healing, gene inactivation studies show that these factors are not essential for epidermal growth or regeneration and suggest that regulation of epidermal growth is more complex than has been previously appreciated (4, 5).
Angiopoietin-1 (Ang-1) (6) is a ligand for the receptor tyrosine kinase TIE2 (7, 8), which contributes to signaling in angiogenesis (9). Ang-1 is characterized structurally by two domains, a coiled-coil domain and a fibrinogen-like domain (10). Recently, members of the angiopoietins (Angs) family have been identified by a domain homology-based molecular cloning strategy. One of these proteins, angiopoietin-related protein 2 (ARP2), was also reported as an angiogenic factor (11). Several recent reports demonstrate that angiopoietin-related proteins (ARPs) show pleiotropic effects not only on vascular cells but on cells of other lineages. For example, it has been shown that ANGPTL3 (12, 13) and FIAF/PGAR/ARP4 (14) may play central roles in lipid/adipocyte metabolism as well as in angiogenesis. Here we identify, by screening EST databases, a previously undescribed angiopoietin-related growth factor (AGF), which is abundantly expressed in hepatocytes. To determine whether AGF promotes in vivo angiogenesis as does Ang-1, we created transgenic (TG) mice overexpressing mouse AGF in epidermal basal keratinocytes of skin (K14-AGF) and investigated vascularization of their dermal layer. K14-AGF mice show not only increased numbers of microvessels in the skin but thickened epidermal layers, resulting in rapid wound closure in the healing process and unusually rapid closure of holes punched in the ears of K14-AGF mice. Here we focus on the role of AGF in skin and show that AGF promotes the proliferation of epidermal keratinocytes and its biological functions could lead to novel therapeutic strategies for wound care and epidermal regenerative medicine.
Materials and Methods
Generation of TG Mice. The pK14-AGF-pA plasmid was generated by inserting the coding region of mouse AGF cDNA into the pK14-pA plasmid (15). We subsequently generated K14-AGF mice according to standard methods (16). We identified transgenic offspring by PCR of tail genomic DNA using forward (5′-GCTCCTGGGCAACGTGCTGG-3′) and reverse (5′-CTGCTGTCTCAAGCTCTGC-3′) primers. Three independent K14-AGF TG lines were backcrossed with wild-type BALB/c mice (purchased from SLC, Shizuoka, Japan). Mice were housed in environmentally controlled rooms of the Laboratory Animal Research Center under the guidelines of Keio University for animal and recombinant DNA experiments.
Preparation of cDNA from Hematopoietic Cells and RT-PCR Analysis. A cell suspension from femur bone marrow of C57BL/6 mice (SLC) was prepared. For preparation of bone marrow-derived mast cells (BMMCs), total bone marrow cells were cultured as described elsewhere (17). For purification of various hematopoietic cells, total bone marrow cells were analyzed and sorted by FACSVantage (BD Biosciences, Palo Alto, CA). The mAbs used in immunofluorescence staining and procedures for flow cytometry were as described (18). Procedures for RT-PCR analysis were as described (19). For AGF the forward primer was 5′-CATGGAGGGATTGTGCAGAG-3′ and the reverse was 5′-AGCCGGGTCAACATAACAGC-3′ For GAPDH the forward primer was 5′-AATCCCATCACCATCTTCCA-3′ and the reverse was 5′-CCAGGGGTCTTACTCCTTG-3′. Each PCR cycle consisted of a 1-min denaturation at 94°C, 1 min of annealing at 64°C, and 1 min of extension at 72°C.
Immunohistochemical Analysis. To detect AGF protein in sections and by Western blotting, we prepared anti-mouse AGF polyclonal antibodies that were produced by immunizing rabbits with a synthetic peptide corresponding to amino acids 202–217 of mouse AGF (NTSRRLDQTPEHQREQ). Fixed sections from liver, back skin, and ears of K14-AGF mice and controls were stained with 1:500 diluted anti-mouse AGF antibody, antiphospho-histone H3 antibody (Upstate Biotechnology, Lake Placid, NY), anti-phospho-Akt for Ser-473 antibody (Cell Signaling, Beverly, MA), anti-phospho-p44/42 MAPK (Thr-202/Tyr-204) antibody (Cell Signaling), or 1:1,000 diluted anti-mouse keratin 1, 5, and 14 antibodies (Covance, Berkeley, CA). For BrdUrd staining, we injected BrdUrd i.p. into 3-month-old K14-AGF mice and controls and obtained epidermal sections from ears 1 h after injection. Staining was performed by using a BrdUrd staining kit (Oncogene, Boston). Sections were washed and counterstained with Mayer's haematoxylin.
Wound-Healing Experiments. A 2-mm hole was made in the center of both ears of K-14 AGF mice and controls by using a metal ear punch (Natsume, Tokyo) as described elsewhere (20). Macroscopic observation of the morphological alteration of holes was followed for wound closure. In another case, all ear and tail were amputated at 5 mm site from the tip with a single stroke of a scalpel blade in anesthetized mice. In a similar fashion, 1, 2, 3, 5, or 8 days later, an additional 5 mm of ear edge or tail end was removed from each of the animals. These additional amputated segments were used for both histological examination and Northern blotting analysis.
Northern Blot Analysis. We prepared total RNA from skin and liver from K14-AGF mice and controls, and excisional wounded skin from wild-type BALB/c mice with TRIzol (GIBCO/BRL, Gaithersburg, MD) according to the manufacturer's instructions. We performed Northern blotting analysis with the probes for AGF or KGF cDNA. Ten micrograms of total RNA were size-fractionated by electrophoresis on a 1.0% agaroseformaldehyde gel and transferred to nylon membranes (Amersham Pharmacia). The membranes were hybridized with 32P-labeled probes in ExpressHyb hybridization solution (BD Biosciences) at 65°C for 20 h. The membranes were washed serially, with the final wash in 0.1× SSC, 0.1% SDS at 65°C. The exposure time was 24 h, and the signals were detected by using Fujix BAS2500 image analyzer (Tokyo).
Statistics. Data are expressed as mean ± SD. Statistical analysis was conducted by using the Student's t test. Statistical significance was defined as P < 0.05.
Results
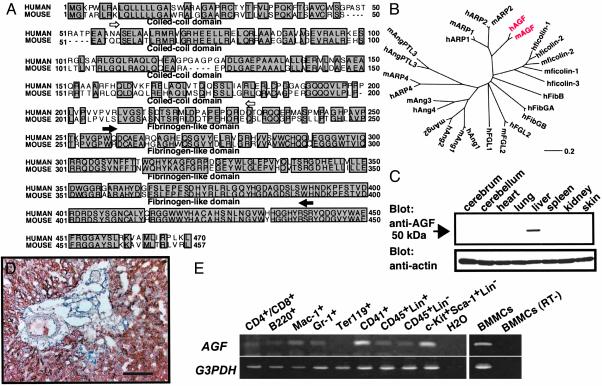
Cloning of Human and Mouse AGF. To identify novel genes belonging to the Ang family (6, 10), we searched EST databases for sequences homologous to Angs. One of the highest scores was a single EST (AA919135). Based on its sequence, we designed nested oligonucleotides and performed 5′ RACE on RNA isolated from the human fetus (purchased from BD Biosciences), yielding the complete coding sequence and 5′ untranslated region of the gene. The extended cDNA sequence encodes a previously undescribed 470-aa protein (Fig. 1A) with a coiled-coil domain in the NH2-terminal portion and a fibrinogen-like domain in the C-terminal region, both of which are conserved in Angs. We next identified a single mouse EST (GenBank accession no. AI194628) similar to human AGF (GenBank accession no. AB054064) and obtained a cDNA sequence of mouse AGF (GenBank accession no. AB054065) by using a RACE strategy. The 457-aa mouse protein shares 74.0% amino acid identity with human AGF (Fig. 1 A). The probable evolutionary relationships of the angiopoietin superfamily were derived using the DNASIS for Windows Ver 2.1 (Hitachi Software, Tokyo), indicating that AGF is evolutionarily closer to the recently reported angiopoietin-related proteins ARP1 and ARP2 (11) than it is to Angs and ARP4 (Fig. 1B). Northern blot analysis revealed 4.0- and 1.8-kb transcripts expressed abundantly in liver and weakly in heart, brain, lung, kidney, and testis (data not shown). Western blotting analysis revealed that the mouse AGF product was 50-kDa and that it was also abundantly expressed in liver (Fig. 1C). Immunohistochemical analysis revealed that hepatocytes specifically express AGF (Fig. 1D). This expression pattern is similar to that of Ang-1. Therefore, we next examined expression of AGF in hematopoietic cells, because Ang-1 expression is restricted to hematopoietic stem cells (HSCs). RT-PCR analysis showed that AGF was broadly expressed in hematopoietic cells, and abundantly expressed in CD41-positive megakaryocytes/platelets, HSCs and mast cells (BMMCs) (Fig. 1E). We found AGF in granules in mast cells by in situ hybridization and immunohistochemical staining, suggesting that mast cells secrete AGF (Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org).
Fig. 1.
Sequence and expression analyses of AGF. (A) Deduced amino acid sequences of human and mouse AGF. Open and filled arrows indicate the limits of the coiled-coil and fibrinogen-like domains, respectively. (B) The evolutionary relationship of AGF (red) to the angiopoietin superfamily was derived by using dnasis for windows v. 2.1 (Hitachi Software, Tokyo). The length of each horizontal line is proportional to the degree of amino acid sequence divergence. (C) Western blot analysis of various mouse tissues by using the anti-AGF (Upper) and anti-actin (Lower) antibodies. (D) Immunoreaction using the anti-AGF antibody shows AGF specifically expressed in hepatic parenchymal cells, not in the Glisson region. (Scale bar = 100 μm.) (E) Analysis of AGF mRNA expression in hematopoietic cells from adult bone marrow or BMMCs by RT-PCR. CD4+/CD8+, T cell; B220, B cell; Mac-1, macrophage and monocyte; Gr-1, granulocyte; Ter119, erythrocyte; CD41, megakaryocyte/platelet; CD45+Lin+, mature HCs; CD45+Lin-, immature HCs; c-Kit+Sca-1+Lin-, hematopoietic stem cell-enriched population. A mixture of anti-Mac-1, -Gr-1, -B220, -CD4, -CD8, and -Ly-6 antibodies was used as a lineage marker (Lin). GAPDH mRNA served as a loading control. All RNA without RT treatment (RT-) show no transcript by PCR. BMMCs (RT-) is one representative data.
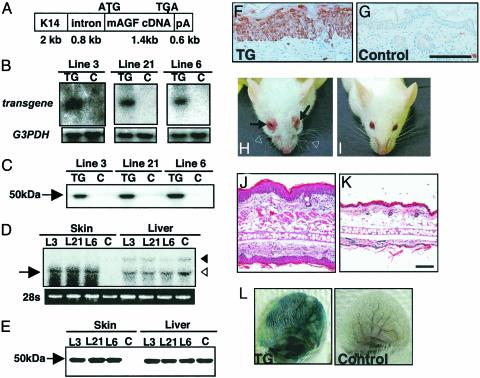
Epidermal Phenotype of K14-AGF TG Mice. AGF is structurally similar to Ang-1, suggesting that it may be involved in angiogenesis. Therefore to investigate whether AGF functions in angiogenesis, transgenic mice expressing AGF in the epidermis under control of the keratinocyte specific promoter K14 (15, 21–24) were generated (Fig. 2A). We selected three independent K14-AGF mouse lines expressing AGF abundantly for analysis based on Northern and Western blotting analysis of whole neonatal skin tissues from 16 TG mice carrying the transgene (Fig. 2 B, C, and D). We next characterized that protein level of transgene expression in skin tissues was equivalent to the endogeneous AGF expression in liver (Fig. 2E). Immunohistochemical analysis of ear tissues from selected transgenic mice lines revealed that AGF was detected in the epidermal layer (Fig. 2F), whereas there was no AGF expression in either the epidermis or dermis of non-TG controls (Fig. 2G). These three lines showed essentially equivalent phenotypes. K14-AGF mice were grossly red and showed swollen and reddish nose and eyelids (Fig. 2 H and I). We initially found significantly increased numbers of microvessels in the dermis, suggesting that AGF promotes angiogenesis (Fig. 7, which is published as supporting information on the PNAS web site). Surprisingly, we found a markedly thickened epidermis and edematous thickened ears in K14-AGF mice compared with controls (Fig. 2 J and K). Therefore, we examined vascular leakiness after injection of Evans blue dye into the tail vein as described elsewhere (ref. 24 and Supporting Materials and Methods, which is published as supporting information on the PNAS web site). The ears of K14-AGF mice become significantly and broadly blue compared with controls (Fig. 2L), and spectrophotometric analysis revealed a 2.2-fold increase in vascular leakiness, indicating that leakiness induces edematous thickened ears. We next compared the epidermis of 7-week-old K14-AGF mice to that of same-aged K14-VEGF mice, in which marked vascular leakiness has been shown (22, 23). No thickened epidermis was detected in K14-VEGF mice, whereas marked edematous alterations appeared in the ear, indicating that vascular leakiness cannot induce epidermal proliferation (Fig. 8A, which is published as supporting information on the PNAS web site).
Fig. 2.
Markedly thickened epidermal layers in K14-AGF mice. (A) Schematic representation of the transgene used to generate K14-AGF mice. K14, intron, and pA indicate the human K14 promoter, rabbit β-globin intron, and a polyadenylation signal derived from the K14 gene, respectively. (B and C) Expression of the transgene was detected in the whole skin of F1 mice (TG) 3 days after birth by Northern (B) and Western (C) blotting analysis. No expression of the transgene was detected in controls (C). Blotting analysis for GAPDH was performed as an internal control experiment. (D and E) Comparison of the mRNA (D) and protein (E) level of AGF from skin and liver between K14-AGF mice and controls. Arrow in D indicates the transcription of the transgene. Open and filled arrowheads in D indicate 1.8- and 4.0-kb endogeneous AGF transcripts, respectively. Five micrograms of protein was loaded in each lane in E.(F and G) Immunohistochemical analysis of AGF detects expression of the transgene in the epidermis of skin from the ears of F1 K14-AGF mice (F) and their controls (G). (Scale bar = 100 μm.) (H and I) Front view of the K14-AGF mouse and controls. Swelling of the eyelid (arrows in H), ears, and nose, and wavy whiskers (open arrowheads in H) were detected in K14-AGF mice. (J and K) Hematoxylin/eosin histology of swollen ear of the K14-AGF mouse (J) and control (K). (Scale bar = 100 μm.) (L) Photographs of ears injected intravenously with Evans blue dye to visualize plasma leakage. The ear of a K14-AGF mouse was strongly blue, whereas the control ear was not changed. One representative experiment is shown.
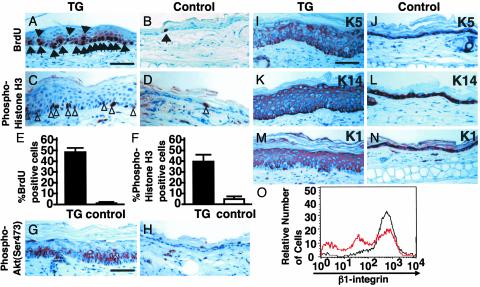
Actively Cycling and Differentiated Epidermal Cells in K14-AGF Mice. To determine whether there was an increase in the number of mitotic cells in the epidermis of K14-AGF mice, we undertook bromo-deoxyuridine (BrdUrd) labeling of skin tissues from 4 week-old K14-AGF mice. An increased number of BrdUrd-positive keratinocytes in the epidermis of K14-AGF mice revealed a marked increase in proliferating cells in the epidermis compared with controls (Fig. 3 A, B, and E). Immunohistochemical analysis using an anti-phospho-histone H3 antibody also revealed an increase in mitotic cells in the epidermis of K14-AGF mice compared with controls (Fig. 3 C, D, and F). Furthermore, we found AGF derived from keratinocytes directly promotes keratinocyte growth in an autocrine manner in K14-AGF mice using in vitro culture analysis (Fig. 9, which is published as supporting information on the PNAS web site). These findings indicate that AGF directly enhances the growth of keratinocytes. Significant immunoreactivity was also detected in the epidermis of K14-AGF mice by using an antibody directed against phospho-Ser-473 of Akt compared with that of controls (Fig. 3 G and H). No expression of phospho-p44/42 mitogen-activated protein kinase (MAPK) was detected in epidermis of either K14-AGF mice or controls (data not shown), suggesting that Akt signaling is involved in epidermal proliferation induced by AGF.
Fig. 3.
Actively cycling expression of keratin proteins in epidermal cells of K14-AGF mice. (A–F) Comparison of levels of DNA synthesis in the epidermis from K14-AGF mice and controls. Skin sections from both mice were stained immunohistochemically for BrdUrd (A and B) and anti-phospho-histone H3 (C and D)by using peroxidase-based detection. Sections were counterstained with hematoxylin. Arrows and arrowheads indicate examples (brown-stained nuclei) of BrdUrd-positive and phospho-histone H3-positive cells, respectively. (E and F) The average numbers of labeled cells with BrdUrd and anti-phospho-histone H3 immunoreactivity from five sections each from three mice, respectively. (G and H) Immunoreactivity against anti-phospho-Akt antibody was seen in the thickened epidermis from the K14-AGF mouse (G), whereas no immunoreactivity was seen in epidermis from controls (H). (I–N)K5(J) and K14 (L) were detected in the basal layer of the epidermis in controls. Similar sections obtained from K14-AGF mice show positive staining in the suprabasal layer of the epidermis as well(I and K). K1 staining was similar for both the K14-AGF mouse (M) and controls (N). (Scale bar = 50 μm.) (O) Surface levels of β1-integrins in basal keratinocytes of K14-AGF (red line) and controls (black line). Three peaks for intensity of β1-integrin expression are detected in K14-AGF, whereas one peak with high intensity is seen in controls.
When we used keratin expression as a marker of epidermal differentiation, we found strong immunoreactivity against anti-K14 and -K5 antibodies in both basal and suprabasal layers of K14-AGF mice (Fig. 3 I and K) compared with control skin, where the basal layer was predominantly stained (Fig. 3 J and L). Anti-K1 staining of epidermal cells revealed increased numbers of keratinocytes in the suprabasal layer of K14-AGF mice (Fig. 3 M and N). This keratin pattern exhibited by K14-AGF mice was similar to that seen in various hyperproliferative epidermal diseases (1, 25, 26). FACS analysis revealed that β1-integrinnegative and β1-integrinlow fractions are prominent in the epidermis of 5-old-day K14-AGF mice, indicating that the majority of thickened keratinocytes of K14-AGF mice were differentiated (27–30) (Fig. 3O).
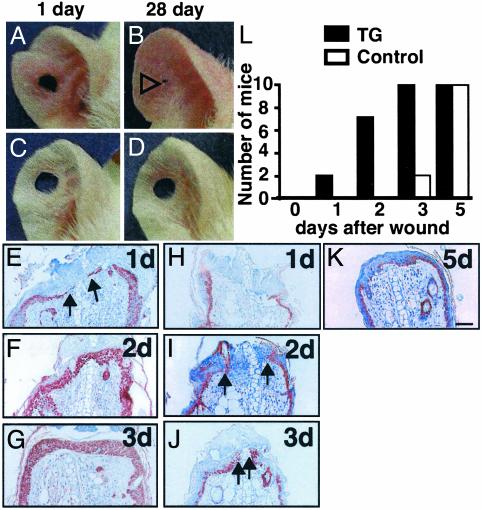
Proliferative Activity of AGF for Regeneration in Skin Tissues. To investigate whether AGF plays a role in remodeling and regeneration in skin tissues, we undertook a wound-healing assay using K14-AGF mice. We detected unusual closure of holes punched in the ears (20) of K14-AGF mice relative to wild type controls (Fig. 4 A–D). Histological examination for closed ear punches of K14-AGF mice revealed that wounds show marked progress toward full closure due to extensive epidermal proliferation and were well supplied with blood vessels, connective tissues, and cartilage in the new growth zone (data not shown). We next compared the wound-healing process after tail amputation and ear excision of K14-AGF mice and controls. One day after the initial excision of ears, migrating and proliferating K14-positive epidermal cells were already detected in K14-AGF mice (Fig. 4E), whereas no K14-positive epidermal cells were detected at wounding sites in control mice (Fig. 4H). Angiogenesis was observed in the dermis of controls 2 days after the initial wound, whereas microvessels at the wounding region in the dermis of K14-AGF mice had already increased on day 1. Between 3 and 5 days were required for overlapping of epidermal cells at the wounding site in controls (Fig. 4 F, G, and L), whereas keratinocytes in K14-AGF mice completely covered the wound site before 3 days (Fig. 4 I, J, and L). We hypothesize that AGF derived from K14-positive keratinocytes in K14-AGF mice plays an important role in the wound-healing process through promoting proliferation of epidermal cells and angiogenesis. Similar findings were detected in tail amputation experiments (data not shown). These findings show the biological functions of AGF in remodeling and regeneration in skin tissues. Immunohistochemical analysis of wounded skin from controls revealed that no expression of AGF in either the epidermis or dermis was detected, whereas AGF was detected in the regenerated epidermal layer in addition to nonwounded keratinocytes in K14-AGF mice (data not shown).
Fig. 4.
Reepithelialization of wounds in K14-AGF mice. (A–D) Representative photograph ear wound healing. K14-AGF and control ears were punched in the center creating a 2-mm open hole and followed for 28 days. (A and B) One can see the progression of hole closure from day 1 (A) to day 28 (B). Open arrowhead in B indicates shortened hole in K14-AGF. (E–K) Wounding was accomplished by ear segment excisions. Shown are representative data of skin sections with staining for anti-K14 antibody from the exposed portion of the remaining ear of K14-AGF mice (E–G) and control littermates (H–K)at1,2,3, and 5 days after the initial wounds. All sections were photographed at the same magnification. (Scale bar = 100 μm.) Arrowheads indicate migrating and proliferating epidermal keratinocytes, indicating that keratinocytes overlapped the injury site rapidly in K14-AGF mice. (L) Analysis of frequencies of wound closure by histological examination at 1, 2, 3, and 5 days after the initial wounding. Filled (controls) and open (K14-AGF mice) columns represent the number of mice in which the wound was completely covered with keratinocytes. Ten mice were examined on each day of the experiment.
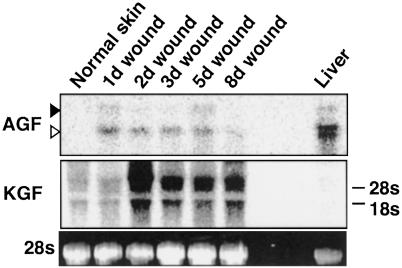
mRNA Expression of AGF in Wounded Skin. To investigate the mRNA expression of AGF in wounded tail and ear skin, we isolated RNA from excisional wounds at different intervals after wounding and performed Northern blotting analysis. For each time point, wounded segments from five mice were excised and used for RNA isolation. Normal skin from nonwounded mice was used as a control. We found that expression of AGF was induced at day 1 after wounding, whereas no expression of AGF was detected in normal skin (Fig. 5). Consistent with previous findings (31), KGF was detected in normal skin, and was considerably increased during wound healing. Relative to KGF expression, much lower levels of AGF were found in wounded skin and it decreased to the basal value within 8 days after wounding in contrast to the prolonged induction of KGF expression (Fig. 5).
Fig. 5.
Expression of AGF mRNA in normal and wounded skin. The total RNA (10 μg) from normal and wounded ear skin was analyzed by Northern blotting analysis with cDNA probes for AGF and KGF. The relative amount of each mRNA was quantified with normalization to 28S rRNA levels. The time after injury is indicated on top of each lane: 1, 2, 3, 5, and 8 days.
Discussion
Here we report the identification of a novel angiopoietin-related growth factor, AGF. K14-AGF transgenic mice in which expression of AGF was forced in epidermal cells show keratinocyte proliferation and increased cutaneous wound healing, suggesting that AGF promotes keratinocyte growth. In addition, we found that K14-AGF mice exhibit increased vascular permeability and increased numbers of blood vessels, suggesting that AGF has also angiogenic activity like other Angs and ARPs, such as Ang-1, ARP2, and ANGPTL3. These findings indicate that AGF has pleiotropic effects not only in vascular cells but in other cell lineages. Based on the assumption that angiogenesis and proliferation of keratinocytes are critical components in wound healing (32, 33), the biological functions of AGF in both keratinocytes and vascular cells could potentially lead to novel therapeutic strategies for wound care and epidermal regenerative medicine.
Several lines of evidence suggest that regulation of epidermal growth is far more complex than was appreciated previously and likely involves several factors. Here, we found that forced expression of AGF in K14-AGF mice has potential to accelerate the wound healing process through its activity in promoting keratinocyte proliferation. These findings led us to ask whether endogenous AGF plays a physiological role in pathological settings. In this study, we have provided evidence that mRNA of AGF from wounded skin tissues was induced, whereas no immunoreactivity for anti-AGF antibody was detected in either the epidermis or dermis. Wound healing results from the interplay of different tissue structures and several resident and infiltrating cell types. The latter consist mainly of leukocyte subsets, such as neutrophils, macrophages, mast cells, platelets, and lymphocytes (33–35). These cells sequentially infiltrate the wound site and serve as sources of various growth factors and cytokines, which promote angiogenesis (32) and/or proliferation of keratinocytes in the wound healing process. Based on our findings that AGF is expressed abundantly in various blood cells, we speculate that AGF from these infiltrated blood cells may pathologically and/or physiologically contribute to repair of wounding sites through promoting proliferation of keratinocytes and angiogenesis. To investigate the physiological role of endocrine AGF from hepatocytes and paracrine AGF from infiltrated cells, analysis of AGF-deficient mice is required.
Critical to understanding the function of a ligand is the identification and characterization of its cognate receptor. The fibrinogen-like domain at the C terminus (36) of Angs is a binding site for the TIE2 receptor (7, 8). AGF also contains this domain, suggesting that AGF could be a ligand for either TIE1 or TIE2. However, binding analyses show that AGF does not bind to either immobilized TIE1-Fc or TIE2-Fc protein (data not shown). This is consistent with our findings that neither receptor is expressed in epidermis and there are significant phenotypic differences in the epidermis between K14-AGF and K14-Ang1 mice (22–24) (Fig. 8B). Identification of an AGF receptor is essential to identify the precise role of AGF.
Supplementary Material
Acknowledgments
We thank Dr. George D. Yancopoulos for providing us with K14-Ang1 and K14-VEGF mice. This work was supported by grants-in-aid for Scientific Research on Priority Areas from the Ministry of Education, Science, and Culture of Japan, by the Yamanouchi Foundation for Research on Metabolic Disorders, and by a grant-in-aid from the Tokyo Biochemical Research Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AGF, angiopoietin-related growth factor; ARP, angiopoietin-related protein; Ang, angiopoietin; TG, transgenic; KGF, keratinocyte growth factor; BMMC, bone marrow-derived mast cell.
References
- 1.Vassar, R. & Fuchs, E. (1991) Genes Dev. 5, 714-727. [DOI] [PubMed] [Google Scholar]
- 2.Guo, L., Yu, Q-C. & Fuchs, E. (1993) EMBO J. 12, 973-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Werner, S., Smola, H., Liao, X., Longaker, M. T., Krieg, T., Hofschneider, P. H. & Williams, L. T. (1994) Science 266, 819-822. [DOI] [PubMed] [Google Scholar]
- 4.Mann, G. B., Fowler, K. J., Gabriel, A., Nice, E. C., Williams, R. L. & Dunn, A. R. (1993) Cell 73, 249-261. [DOI] [PubMed] [Google Scholar]
- 5.Guo, L., Degensteim, L. & Fuchs, E. (1996) Genes Dev. 10, 165-175. [DOI] [PubMed] [Google Scholar]
- 6.Suri, C., Jones, P. F., Patan, S., Bartunkova, S., Maisonpierre, P., Davis, S., Sato, T. N. & Yancopoulos, G. D. (1996) Cell 87, 1171-1180. [DOI] [PubMed] [Google Scholar]
- 7.Iwama, A., Hamaguchi, I., Hashiyama, M., Murayama, M., Yasunaga, K. & Suda, T. (1993) Biochem. Biophys. Res. Commun. 195, 301-309. [DOI] [PubMed] [Google Scholar]
- 8.Dumont, D. J., Yamaguchi, T. P., Conlon, R. A. Rossant, J. & Breitman, M. L. (1992) Oncogene 7, 1471-1480. [PubMed] [Google Scholar]
- 9.Sato, T. N., Tozawa, Y., Deutsch, U., Wolburg-Buchholz, K., Fujiwara, Y., Gendron-Maguire, M., Grigley, T., Wolburg, H., Risau, W. & Qin, Y. (1995) Nature 376, 70-74. [DOI] [PubMed] [Google Scholar]
- 10.Gale, N. W. & Yancopoulos, G. D. (1999) Genes Dev. 13, 1055-1066. [DOI] [PubMed] [Google Scholar]
- 11.Kim, I., Moon, S. O., Koh, K. N., Kim, H., Uhm, C. S., Kwak, H. J., Kim, N. G. & Koh, G. Y. (1999) J. Biol. Chem. 274, 26523-26528. [DOI] [PubMed] [Google Scholar]
- 12.Koishi, R., Ando, Y., Ono, M., Shimamura, M., Yasumo, H., Fujiwara, T., Horikoshi, H. & Furukawa, H. (2002) Nat. Genet. 30, 151-157. [DOI] [PubMed] [Google Scholar]
- 13.Camenisch, G., Pisabarro, M. T., Sherman, D., Kowalski, J., Nagel, M., Hass, P., Xie, M. H. & Gurney, A. (2002) J. Biol. Chem. 277, 17281-17290. [DOI] [PubMed] [Google Scholar]
- 14.Yoon, J. C., Chickering, T. W., Rosen, E. D., Dussault, B., Qin, Y., Soukas, A., Friedman, J. M., Holmes, W. E. & Spiegelman, B. M. (2000) Mol. Cell. Biol. 20, 5343-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunisada, T., Yamazaki, H., Hirobe, T., Kamei, S., Omoteno, M., Tagaya, H., Hemmi, H., Koshimizu, U., Nakamura, T.& Hayashi, S. I. (2000) Mech. Dev. 94, 67-78. [DOI] [PubMed] [Google Scholar]
- 16.Hogan, B., Beddington, R., Costantini, F. & Lacy, E. (1994) Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 17.Boesiger, J., Tsai, M., Maurer, M., Yamaguchi, M., Brown, L. F., Claffey, K. P., Dvorak, H. F. & Galli, S. J. (1998) J. Exp. Med. 188, 1135-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takakura, N., Watanabe, T., Suenobu, S., Yamada, Y., Noda, T., Ito, Y., Satake, M. & Suda, T. (2000) Cell 102, 199-209. [DOI] [PubMed] [Google Scholar]
- 19.Hamada, K. Oike, Y., Ito, Y., Maekawa, H., Miyata, K., Shimomura, T. & Suda, T. (2003) Arterioscler. Thromb. Vasc. Biol. 23, 190-197. [DOI] [PubMed] [Google Scholar]
- 20.Clark, L. D., Clark, R. K. & Heber-Katz, E. (1998) Clin. Immunol. Immunopathol. 88, 35-45. [DOI] [PubMed] [Google Scholar]
- 21.Suri, C., McClain, J., Thurston, G., McDonald, D. M., Zhou, H., Oldmixon, E. H., Sato, T. N. & Yancopoulos, G. D. (1998) Science 282, 468-471. [DOI] [PubMed] [Google Scholar]
- 22.Detmar, M., Brown, L. F., Schon, M. P., Elicker, B. M., Velasco, P., Richard, L., Fukumura, D., Monsky, W., Claffey, K. P. & Jain, R. K. (1998) J. Invest. Dermatol. 113, 1047-1053. [DOI] [PubMed] [Google Scholar]
- 23.Thurston, G., Suri, C., Smith, K., McClain, J., Sato, T. N., Yancopoulos, G. D. & McDonald, D. M. (1999) Science 286, 2511-2514. [DOI] [PubMed] [Google Scholar]
- 24.Elson, D. A., Thurston, G., Huang, L. E., Ginzinger, D. G., McDonald, D. M., Johnson, R. S. & Arbeit, J. M. (2001) Genes Dev. 15, 2520-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss, R. A., Eichner, R. & Sun, T. T. (1984) J. Cell Biol. 98, 1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stoler, A., Kopan, R., Duvic, M. & Fuchs, E. (1988) J. Cell Biol. 107, 427-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, P. H. & Watt, F. M. (1993) Cell 73, 713-724. [DOI] [PubMed] [Google Scholar]
- 28.Jones, P. H., Harper, S. & Watt, F. M. (1995) Cell 80, 83-93. [DOI] [PubMed] [Google Scholar]
- 29.Kaur, P. & Li, A. (2000) J. Invest. Dermatol. 114, 413-420. [DOI] [PubMed] [Google Scholar]
- 30.Watt, F. M. (2002) EMBO J. 21, 3919-3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Werner, S., Peters, K. G., Longaker, M. T., Fuller-Pace, F., Banda, M. J. & Williams, L. T. (1992) Proc. Natl. Acad. Sci. USA 89, 6896-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tonnesen, M. G., Feng, X. & Clark, R. A. F. (2000) J. Invest. Dermatol. 5, 40-46. [Google Scholar]
- 33.Singer, A. J. & Clerk, R. A. F. (1999) N. Engl. J. Med. 341, 738-746. [DOI] [PubMed] [Google Scholar]
- 34.Artuc, M., Hermes, B., Steckelings, U. M., Grutzkau, A. & Henz, B. M. (1999) Exp. Dermatol. 8, 1-16. [DOI] [PubMed] [Google Scholar]
- 35.Trabucchi, E., Radaelli, E., Marazzi, M., Foschi, D., Musazzi, M., Veronesi, A. M. & Montorsi, W. (1988) Int. J. Tissue React. 10, 367-372. [PubMed] [Google Scholar]
- 36.Procopio, W. N., Pelavin, P. I., Lee, W. M. F. & Yeilding, N. M. (1999) J. Biol. Chem. 274, 30196-30201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.