Abstract
The microtubule (MT)-associated protein tau is important in neuronal development and in Alzheimer's and other neurodegenerative diseases. Genetic analyses have established a cause-and-effect relationship between tau dysfunction/misregulation and neuronal cell death and dementia in frontotemporal dementia and parkinsonism associated with chromosome 17; several mutations causing this dementia lead to increased ratios of four-repeat (4R) to three-repeat (3R) wild-type tau, and an attractive hypothesis is that the abnormally high ratio of 4R to 3R tau might lead to neuronal cell death by altering normal tau functions in adult neurons. Thus, we tested whether 3R and 4R tau might differentially modulate the dynamic instability of MTs in vitro using video microscopy. Although both isoforms promoted MT polymerization and decreased the tubulin critical subunit concentration to approximately similar extents, 4R tau stabilized MTs significantly more strongly that 3R tau. For example, 4R tau suppressed the shortening rate, whereas 3R tau had little or no detectable effect. Similarly, 3R tau had no effect on the length shortened during a shortening event, whereas 4R tau strongly reduced this parameter. Further, when MTs were diluted into buffer containing 4R tau, the MTs were stabilized and shortened slowly. In contrast, when diluted into 3R tau, the MTs were unstable and shortened rapidly. Thus, 4R tau stabilizes MTs differently and significantly more strongly than 3R tau. We suggest a “dosage effect” or haploinsufficiency model in which both tau alleles must be active and properly regulated to produce appropriate amounts of each tau isoform to maintain MT dynamics within a tolerable window of activity.
Many neurodegenerative diseases exhibit abnormal pathological fibers composed primarily of the microtubule (MT)-associated protein, tau (for a recent review, see ref. 1). These disorders, termed “tauopathies,” include Alzheimer's disease, frontotemporal dementia and parkinsonism associated with chromosome 17 (FTDP-17), Pick's disease, and progressive supranuclear palsy. In 1998, several groups reported a direct genetic linkage between mutations in the tau gene and FTDP-17 (2–5). Although some tau mutations are structural and others are regulatory, all exhibit dominant phenotypes. Thus, both dysfunction and misregulation of tau are causally related to neuronal cell death, neurodegenerative disease, and dementia.
Tau is normally present predominantly in the cell bodies and axons of neuronal cells and is necessary for the establishment of neuronal cell polarity and axon outgrowth, axonal transport, and maintenance of axonal morphology (6–10). Tau is also expressed in glial cells (11). Mechanistically, tau stimulates MT polymerization, stabilizes MTs, and suppresses MT dynamics (12–15). Because MT dynamics must be tightly regulated for cells to function and remain viable (e.g., ref. 16), it follows that the action of tau must also be finely regulated.
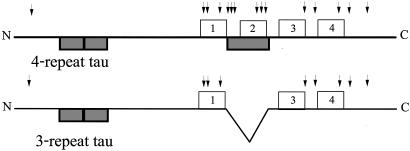
Although there is only a single tau gene, alternative splicing of tau mRNA produces six different isoforms in the CNS (17). The isoforms fall into two groups (17, 18), one of which contains four 18-aa imperfect repeats near the C termini and the other, three such repeats (Fig. 1). In both cases, the repeats are separated by 13- to 14-aa-long interrepeats. Previous work has shown that four-repeat (4R) tau binds to MTs ≈3-fold more strongly than three-repeat (3R) tau (19–21) and also assembles MTs more effectively than 3R tau (22, 23).
Fig. 1.
Schematic of 4R and 3R tau. Boxes above each line correspond to the 18-aa imperfect repeats. Dark boxes below each line correspond to regions encoded by alternatively spliced exons. The inclusion or excision of sequences encoded by the exon in the repeat region leads to the synthesis of 4R or 3R tau, respectively. Arrows above the line mark the positions of amino acid substitutions that lead to FTDP-17.
The biological significance of tau RNA alternative splicing is highlighted by two observations. First, tau isoform expression is tightly regulated during development: whereas only the shortest 3R isoform is expressed in fetal neurons, all six tau isoforms are expressed in adult human brain, with 3R and 4R tau being expressed at approximately equal levels (5, 17, 18, 24–26). Transition from the simple fetal to the complex adult expression pattern coincides more or less temporally with the arrival of growth cones at their targets, leading to the model that the developmental transition of tau isoform expression reflects different functional requirements for MTs in fetal vs. adult neurons. The second observation highlighting the importance of tau RNA splicing is that many FTDP-17 tau mutations are regulatory mutations that alter the pattern of tau RNA splicing without altering the primary sequence of the encoded tau protein (2–4, 27). These mutations result in an increase in the expression ratio of 4R to 3R tau in adult neurons, an alteration sufficient to cause neuronal cell death and dementia. Thus, there must be fundamentally important functional differences between 3R and 4R tau.
Although the genetic linkage between tau mutations and FTDP-17 establishes a clear cause-and-effect relationship between tau dysfunction/misregulation and neuronal cell death and dementia, the underlying molecular mechanism leading to dementia and neuronal cell death is not understood. In line with the growing number of dominantly inherited degenerative diseases involving abnormal protein folding (28), several investigators have suggested a “gain-of-a-toxic-function” mechanism in which the mutations confer on tau an increased probability of forming abnormal cytotoxic neurofibrillary tangles (5, 29). Although this is a plausible model for FTDP-17 mutations that alter the tau amino acid sequence, it is less obvious what toxic function might be acquired by the tau RNA splicing mutations in which only the ratio of otherwise wild-type proteins is affected.
To obtain a more mechanistic understanding of normal and pathological tau action, we have analyzed the abilities of 3R and 4R tau to regulate the growing and shortening dynamics of individual MTs in vitro using video microscopy. We find that the abilities of 3R and 4R tau to suppress the rate and extent of MT shortening and to stabilize MTs are different. On the basis of these data, we propose an alternative to the gain-of-a-toxic-function model of tau action. We suggest a “dosage effect” or haploinsufficiency model in which both tau alleles must be active and properly regulated to produce appropriate amounts of each tau isoform to maintain MT dynamics within a tolerable window of activity. In this model, migration outside the tolerable window of dynamic activity leads to cell death. The model provides insights into tau action during neurodegeneration resulting from either tau dysfunction or misregulation, as well as during the normal development and maintenance of neuronal function.
Materials and Methods
Purification of 3R and 4R Tau Isoforms and of Tubulin. Recombinant full-length adult rat 4R and 3R tau were synthesized in Escherichia coli by using the pET vector expression system (Novagen) (15). The tau concentration was determined by subjecting an aliquot of a large tau sample to acid hydrolysis followed by MS determination of the mass of each amino acid, by using known amino acid standards for comparison. This assay revealed a major discrepancy with the commonly used colorimetric or light spectroscopic methods. Specifically, the MS assay revealed that previously used colorimetric assays (and the extinction coefficient determined based on those assays) overestimated the concentration of tau by 2.7-fold. Thus, a 1 mg/ml solution of highly purified tau has an absorbance at 278 nm of 0.78 rather than 0.29, as determined (30). Purified tau was stored at –70°C. Bovine brain MT protein (≈70% tubulin and 30% MT-associated proteins) was prepared by three cycles of assembly and disassembly (31). Tubulin was purified from the MT protein by elution through a Whatman P-11 phosphocellulose column (32) and equilibrated in 50 mM Pipes/1 mM MgSO4/1 mM EGTA/0.1 mM GTP. Purified tubulin (>99% pure) was drop frozen in liquid nitrogen and stored at –70°C.
Analysis of Dynamic Instability of Individual MTs by Video Microscopy. Purified tubulin (10 or 12 μM) was polymerized at the ends of sea urchin (Strongylocentrotus purpuratus) axonemal seeds at 37°C in the presence or absence of the desired tau isoform in 87 mM Pipes/36 mM Mes/1.4 mM MgCl2/1 mM EDTA, pH 6.8 (PMME buffer). The dynamic instability behavior at the plus ends of individual MTs was recorded at 37°C. The ends were designated as plus or minus on the basis of the growth rate, the number of MTs that grew at opposite ends of the seeds, and the relative lengths of the MTs (15, 33). Dynamic instability was analyzed both during the early elongation phase of polymerization (at 10 μM tubulin) and when the MT suspension was approaching but not yet at steady state (12 μM tubulin). For analysis of dynamics at near steady state, usually 30 μl of tubulin solution was assembled onto the ends of the seeds for 30 min at 37°C. For analysis, 3 μl of the suspension was applied to a coverslip and prepared for video microscopy as described (15, 32). For analysis of dynamics during initial polymerization, 3 μl of a 10 μM tubulin solution was mixed with axonemal seeds at 37°C and placed on a coverslip. The dynamics of individual MTs were analyzed between ≈2 min and 7 or 8 min and not longer than 10 min after initiation of polymerization. Data points were collected at 3- to 5-s intervals. MT length changes with time were determined as described (15). We considered a MT to be in a growing phase if it increased in length by >0.2 μm at a rate >0.3 μm/min. MTs showing length changes ≤0.2 μm over the duration of six data points were considered to be in an attenuated state. Twenty to 30 MTs were analyzed for each experimental condition.
Results
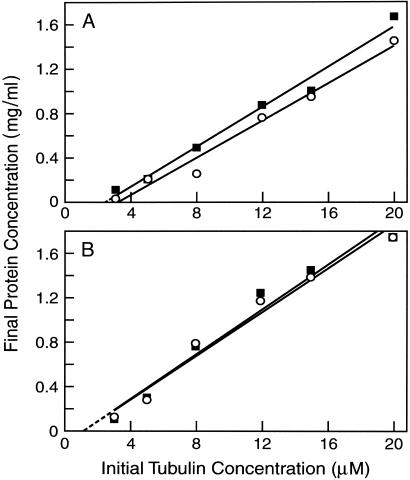
Effects of 3R and 4R Tau on the Tubulin Critical Subunit Concentration. Before examining the effects of 3R and 4R tau on individual dynamic instability parameters, we first compared their respective abilities to reduce the critical subunit concentration for MT assembly using a sedimentation assay. Three- or 4R tau was incubated with different concentrations of tubulin, and the MTs were allowed to assemble for 30 min at 37°C. The MTs were then separated from unassembled tubulin by sedimentation and the protein content of the pellets determined. The critical concentration for tubulin in the absence of added tau was >10 μM (data not shown). At the lower concentration of tau analyzed (0.09 μM, Fig. 2A), both 4R and 3R tau strongly reduced the critical concentration, with 4R tau doing so slightly more strongly than 3R tau (to 3.3 and 2.5 μM tubulin for 3R and 4R tau, respectively). At 0.74 μM tau (Fig. 2B), 4R and 3R tau reduced the critical concentration to the same extent, to 1.25 μM tubulin. We also compared the relative abilities of 3R and 4R tau to promote MT polymerization. Whereas half-maximal MT assembly was achieved by 4R tau at 0.11–0.15 μM, half-maximal MT assembly was achieved by 3R tau at 0.19–0.22 μM (data not shown). Thus, consistent with earlier work (e.g., refs. 22 and 23), both 3R and 4R tau promote MT assembly, with 4R tau doing so somewhat more effectively than 3R tau.
Fig. 2.
The effects of 3R (circles) and 4R tau (squares) on the tubulin critical subunit concentration. MTs were assembled at 37°C in the presence of 0.09 μM tau isoforms (A) or 0.74 μM isoforms (B). After 30 min, the MTs were sedimented at 150,000 × g in a Beckman TL100 centrifuge (Beckman Coulter). The quantity of polymer was determined after dissolving the MT pellets in 100 μl of 100 mM Pipes/1 mM MgS04/1 mM EGTA, pH 6.8, at 0°C for 1 h.
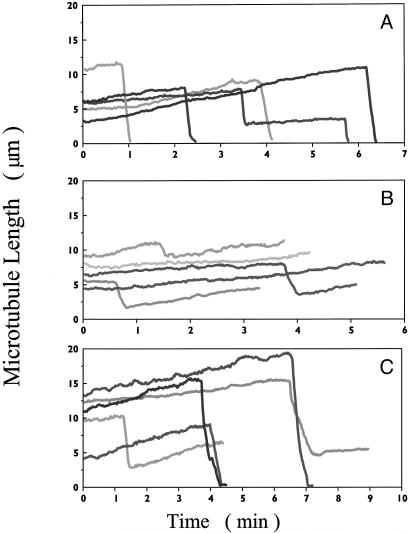
3R and 4R Tau Differentially Modulate MT Dynamics both Near Steady State and During the Initial Phase of Polymerization. The effects of subsaturating concentrations of 3R and 4R tau on the in vitro dynamic instability parameters at the plus ends of individual MTs were first analyzed near steady state (Fig. 3). Similar to work carried out at steady state (15), 0.74 μM 4R tau (tau/tubulin molar ratio of 1:16) strongly suppressed the dynamic instability behavior of the MTs. Importantly, 4R tau strongly reduced the rate and extent of shortening compared with control MTs (Fig. 3, compare A and B), whereas 3R tau did not appear to affect these parameters (Fig. 3 A and C).
Fig. 3.
Life history traces; the effects of 0.74 μM 3R and 4R tau on the dynamics of individual MTs at 37°C near steady state. The growing and shortening dynamic of individual MTs was determined by video microscopy (see Materials and Methods). (A) Control (no added tau). (B) 4R tau. (C) 3R tau. Each trace represents a single MT.
The differential effects of 3R and 4R tau on several of the dynamic instability parameters near steady state are shown quantitatively in Table 1. 3R and 4R tau promoted the rate of MT growth similarly. Specifically, 0.74 μM 4R tau increased the growth rate 46% from 0.96 μm per min to 1.4 μm per min, whereas 0.74 μM 3R tau increased the growth rate by 42% to 1.36 μm per min. In addition, both 3R and 4R tau reduced the overall dynamicity of the MTs to approximately similar extents. In marked contrast, 3R and 4R tau affected the rate of shortening very differently. 4R tau (0.74 μM) reduced the shortening rate by 54% from 36.2 to 16.7 μm per min, whereas 0.74 μM 3R tau reduced the shortening rate by only 19%, to 28.8 μm per min. Similarly, the length the average MT shortened during a shortening event was unaffected by 0.74 μM 3R tau, whereas it was reduced 43% by 4R tau.
Table 1. Effects of 3R and 4R tau on several dynamic instability parameters at microtubule plus ends in vitro near steady state.
|
3R tau, μm
|
4R tau, μm
|
|||
|---|---|---|---|---|
| Control | 0.37 | 0.74 | 0.74 | |
| Rate, μm/min | ||||
| Growing | 0.96 ± 0.1 | 1.1 ± 0.12 | 1.36 ± 0.24 | 1.4 ± 0.14 |
| Shortening | 36.2 ± 3.0 | 29.5 ± 3.1 | 28.8 ± 5.9 | 16.7 ± 3 |
| Length excursion, μm per event | ||||
| Growing | 1.8 ± 0.24 | 2.0 ± 0.22 | 1.9 ± 0.2 | 1.4 ± 0.14 |
| Shortening | 6 ± 0.6 | 8.4 ± 0.95 | 7.2 ± 1.2 | 3.4 ± 0.5 |
| Dynamicity, μm/min | 2.45 | 2.38 | 1.26 | 2 |
Data are ±SEM except for transition frequencies, which are ±SD
The differential effects of 3R and 4R tau on the rate and extent of shortening were even more pronounced when their effects on dynamics were analyzed during the early phase of polymerization (Table 2). The effects of 3R tau were analyzed at tau/tubulin ratios between 1:110 (0.09 μM 3R tau) and 1:27 (0.37 μM 3R tau) and of 4R tau at tau/tubulin ratios of 1:54 (0.19 μM 4R tau) and 1:27 (0.37 μM 4R tau). Similar to what occurred near steady state, 3R and 4R tau promoted the rate of MT growth to similar extents. Both 3R and 4R tau also increased the length grown during a growth event to the same extent. In contrast, 3R and 4R tau exerted very different effects on the rate and extent of shortening. Specifically, 0.19 μM 4R tau suppressed the shortening rate by 37% from 26.8 to 17 μm/min, and 0.37 μM 4R tau suppressed this rate by 53%. In contrast, 3R tau had no detectable effect on the shortening rate at any concentration. Similarly, 0.37 μM 3R tau had no apparent effect on the length shortened during shortening events, whereas 0.37 μM 4R tau strongly reduced this parameter (Table 2).
Table 2. Effects of 3R and 4R tau on dynamic instability at plus ends of individual microtubules in vitro during the early elongation phase of polymerization.
|
3R tau, μM
|
4R tau, μM
|
|||||
|---|---|---|---|---|---|---|
| Parameter | 0 (control) | 0.09 | 0.19 | 0.37 | 0.19 | 0.37 |
| Rate, μm/min | ||||||
| Growing | 1.2 ± 0.2 | 1.4 ± 0.3 | 1.7 ± 0.3 | 1.8 ± 0.3 | 1.4 ± 0.4 | 1.9 ± 0.5 |
| Shortening | 26.8 ± 6.1 | 26.4 ± 8.0 | 31 ± 16.8 | 26 ± 9 | 17.0 ± 6.2 | 12.5 ± 5.5 |
| Length excursion, μm per event | ||||||
| Growing | 5.3 ± 3.7 | 6.2 ± 3 | 7.1 ± 3.5 | 8.5 ± 5.7 | 7.6 ± 4.5 | 8.5 ± 3 |
| Shortening | 8.8 ± 4 | 8.8 ± 3.3 | 8.1 ± 2.5 | 10 ± 9 | 7.1 ± 2.9 | 1.9 ± 1 |
| Percent time in phase | ||||||
| Growing | 94 | 97.9 | 95 | 98 | 96.6 | >99 |
| Shortening | 6 | 2.1 | 5 | 2 | 3.4 | <1 |
| Dynamicity, μm/min | 2.7 | 2.6 | 2.2 | 1.9 | 1.7 | 2 |
Data are ±SEM. The effects of 3R and 4R tau were analyzed on the dynamic instability parameters at MT plus ends during the growth (elongation) phase of polymerization, between 2 and 10 min after initiation of polymerization
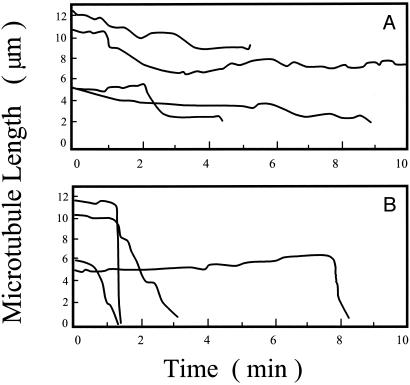
Differential Ability of 3R and 4R Tau to Stabilize MTs. To independently confirm the differential actions of 3R and 4R tau on MT stability, we assembled MTs to steady state at 12 μM tubulin in the absence of tau and then diluted the MT suspensions 4-fold, either into a solution of 0.74 μM 3R tau or 0.74 μM 4R tau or into buffer containing no tau. When no tau was present, the MTs disassembled immediately, because the 4-fold dilution drops the tubulin concentration well below the critical concentration for assembly (data not shown). When the MTs were diluted into 4R tau, they were quite stable, and when they shortened, they did so slowly (Fig. 4A). In contrast, when diluted into 3R tau, the MTs were relatively unstable and shortened rapidly (Fig. 4B). Thus, 3R tau exerted a significantly weaker ability to stabilize MTs than 4R tau.
Fig. 4.
The effects of 3R and 4R tau on the stability of individual MTs after dilution-induced disassembly. MTs were polymerized in PMME buffer at 37°C (12 μM tubulin) and diluted 4-fold into PMME buffer containing 0.74 μM 4R (A)or3Rtau(B). The diluted MT suspensions were analyzed between 2 and 10 min after dilution. Each trace represents a single MT.
Discussion
The most important finding described here is that, under conditions in which 4R and 3R tau promote the rate and extent of MT growth similarly, 4R tau strongly suppresses the rate and extent of MT shortening, whereas 3R tau has minimal effect on these parameters. These isoform-specific mechanistic differences are consistent with earlier competition (34) and structure–function (21) studies suggesting that 4R and 3R tau interact with MTs with at least some distinct molecular mechanisms. The data indicate that 4R tau stabilizes MTs differently and substantially more strongly than 3R tau. Ongoing work in our laboratories has established that these same differences between the actions of microinjected 4R and 3R tau on the growth and shortening dynamics of MTs also occur in living cells (J. Bunker, M. A. Jordan, L.W., and S.C.F., unpublished results). The increased ability of 4R tau to stabilize neuronal MTs as compared with 3R tau may play a role in neuronal cell death and dementia in FTDP-17 individuals expressing altered 4R/3R tau ratios.
There have been few detailed studies on the effects of tau on MT dynamics. Initially, Drechsel et al. (13) found that 4R tau at high concentrations close to those saturating the tau-binding sites on the MT surface strongly promotes the rate and extent of polymerization, decreases the transition frequency from the growing to the shortening state, and inhibits the rate of depolymerization. Trinczek et al. (14) examined the effects high concentrations of 3R and 4R tau on various dynamic instability parameters during the initial stages of MT polymerization and observed similar but not identical effects. Especially relevant to the work described here, 4R tau exerted a markedly stronger ability to slow the tubulin dissociation rate at MT ends than 3R tau.
In a previous report, we found that very low concentrations of 4R tau powerfully suppress MT dynamics (15). The ratios of 4R tau to tubulin in the MTs used in that work, here corrected for the tau concentration as determined by MS, varied between 1:473 at 0.028 μM 4R tau (previously reported as 0.075 μM tau) and 1:27 at 0.44 μM tau (previously reported as 1.2 μM tau). These extremely low ratios reduced the rate and extent of shortening and, because the MT suspension was at steady state, they also reduced the rate and extent of growth. 4R tau also markedly increased the percentage of total time that the MTs spent in an attenuated (paused) state, neither growing nor shortening detectably.
Of considerable interest, the suppressing action of low tau concentrations on MT dynamics resembles the action on dynamics of the MT-stabilizing drug taxol. Like tau, taxol also binds strongly to MTs, possibly to the same site at which tau binds (35), and suppresses dynamics at very low ratios of taxol bound per molecule of tubulin in the MTs (36). Such actions of low microtubule-associated protein concentrations (MAPs) support the idea that drugs such as taxol and other drugs that modulate MT dynamics are mimicking the action of regulatory MAPs (36, 37).
Mechanistic Implications for Tau-Mediated Neuronal Cell Death and Disease: Gain of a Toxic Function or a Dosage Effect? Tau dysfunction (e.g., accumulation of “neurofibrillary tangles”) is correlated with a number of neurodegenerative disorders (1), and the recent identification of tau mutations linked to FTDP-17 (2–4) demonstrates that errors in tau action or regulation lead to neuronal cell death and dementia. There are two classes of FTDP-17 mutations. One involves alteration of tau RNA splicing that results in an increase in the ratio of wild-type 4R to 3R tau (2–4, 27). The second are point mutations resulting in amino acid substitutions. The majority of the point mutations map to the repeat and interrepeat region of tau or lie immediately adjacent to it (2–4), in sequences known to mediate or regulate tau's MT binding and assembly activities (Fig. 1; see also refs. 19–21, 34, 38, and 39). The initial in vitro analyses of point-mutated tau proteins revealed relatively subtle reductions in MT-binding and assembly activities (5, 40–42). Many point-mutated proteins also increase tau fiber formation activity in vitro (29, 41, 43, 44). The final important clue to tau action provided by the FTDP-17 mutations is that all of the mutations exhibit dominant rather than recessive phenotypes.
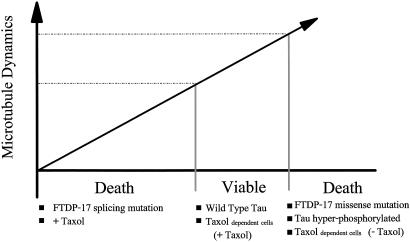
A number of investigators have suggested that the molecular mechanism underlying FTDP-17 tau-mediated neuronal cell death might involve the gain of a toxic function, consistent with the dominance of the phenotype. One idea is that the mutated tau polypeptides may have acquired an increased capability to form toxic fibers (5, 29). Although a plausible model to explain the substitution mutations, it is less obvious what new activity would account for cell death in the RNA splicing mutations, because the tau proteins remain wild type. On the basis of the data presented here, we propose an alternative model to explain FTDP-17 mutant phenotypes based on dosage effects. We suggest that the properly regulated activities of both tau alleles are necessary to produce appropriate amounts of each isoform to maintain MT dynamics within the narrow range required for cell function and viability (Fig. 5). If MT dynamics in neurons migrate outside this range, becoming too rapid or too suppressed, cell functions dependent on the dynamics will be impaired and, in time, cell death could occur.
Fig. 5.
Dosage effect model for normal and pathogenic tau action. Cell function and viability require properly regulated MT dynamics. Overly suppressed dynamics, such as those that occur in the presence of taxol and, as we propose, might occur in the FTDP-17 RNA splicing mutations, result in cell death. Similarly, overly dynamic MTs, such as those that occur in taxol-resistant and -dependent cells in the absence of taxol (16) and those that might occur in the FTDP-17 missense mutations and in cells expressing hyperphosphorylated tau, also result in cell death.
This dosage model can readily account for both the RNA splicing and amino acid substitution classes of FTDP-17 mutations. With the RNA splicing mutations, increasing the ratio of 4R to 3R tau will lead to less MT shortening and increased MT stabilization. With the mutations causing amino acid substitutions, most of the mutated tau molecules possess defects in MT-binding and/or assembly activities (e.g., see refs. 5, 40, and 42), and cells expressing such mutated tau proteins are likely to have overly dynamic MTs. Our model predicts that cell death should occur in both classes of mutations, because the pattern of MT dynamics would migrate outside the window of viability, one way or the other.
Evidence for the need to maintain MT dynamics within a narrow window has recently been obtained in mitotic cells. Specifically, low concentrations of taxol powerfully suppress MT dynamics in mitotic spindles of dividing cells, resulting in the inability of the cells to progress normally from metaphase to anaphase (45, 46). Low concentrations of taxol that suppress spindle MT dynamics by only ≈15–25% are sufficient to impair mitotic progression and induce apoptosis (for example, refs. 45 and 46). Moreover, in lung tumor A549 cells that are both resistant to taxol and dependent on the presence of taxol for viability, removal of taxol results in a significant increase in MT dynamics (16). Importantly, on removal of taxol, the dependent cells become blocked in mitosis and die, as do their wild-type counterparts treated with normal taxol concentrations. Thus, during mitosis, there appears to be a narrow window of tolerated MT dynamics, and excessively rapid dynamics as well as suppressed dynamics lead to mitotic inhibition and cell death (16).
A dosage effect model has additional important features. First, it can accommodate the longstanding correlation between tau hyperphosphorylation and neuronal cell death. Because most tau phosphorylation events reduce tau's binding affinity for MTs, they are also likely to reduce the ability of tau to regulate MT dynamics. Indeed, phosphorylation of tau by MAP2 kinase (13), MARK, or cdk5 (14) reduces tau's ability to influence MT dynamics. If the reduction of dynamics-regulating activity is sufficient to move the cell outside the dynamics window required for viability, the model predicts cell death. A second feature of the model is that abnormal tau fiber formation is relegated to a downstream consequence of the primary cause of cell death, which is abnormal tau effects on MT dynamics. In this regard, Drosophila-overexpressing human tau exhibit many features of Alzheimer's disease, including early onset and progressive neurodegeneration, but without abnormal tau fiber formation (47). Thus, abnormal tau fiber formation is not an obligatory feature of tau-induced neuronal cell death.
Finally, the dosage effect and gain-of-toxic-function models are not mutually exclusive. In addition to the possibility that the fibers may themselves be toxic under some circumstances, fiber formation would be expected to contribute to deregulation of MT dynamics by virtue of sequestering the pool of tau, which should in turn lead to overly dynamic MTs and subsequent cell death.
How Might Altered MT Dynamics Cause Cell Death? In nonneuronal dividing cells, drugs that alter MT dynamics can lead to aberrant cell movement, cell morphology, chromosome segregation, and intracellular transport mechanisms (48). Although chromosome segregation and major cell movements are not highly relevant for postmitotic neurons, neurons should be very sensitive to perturbations affecting axonal transport or the maintenance of cell morphology. Hence, if altered neuronal MT dynamics underlie tau-mediated neuronal cell death in FTDP-17 disease, the actual cause of cell death might be the inability to properly transport cargo along the axon and/or to maintain the elongated cell morphology necessary to acquire and transport target-derived trophic factors. Indeed, overexpression of tau in cultured neuronal cells leads to aberrant axonal transport (10). Further, compromising axonal transport via overexpression of dynactin in mice causes late-onset death in motor neurons (49).
Implications for Normal Neuronal Development and Maintenance. The transition from the simple fetal 3R tau expression pattern to the more complex adult expression pattern (approximately equal amounts of 3R and 4R tau) coincides approximately with the arrival of growth cones at targets. This correlation has led to the model that the developmental transition in tau isoform expression confers on MTs the different functional capabilities required in growing immature neurons vs. mature neurons. However, the mechanistic rationale for the developmental regulation of tau RNA splicing has remained elusive. On the basis of the data presented here, the MTs in fetal neurons may be significantly more dynamic that those in adult neurons. Integrating this notion with the model schematized in Fig. 5 leads to the conclusion that the window of acceptable dynamic behavior shifts as development proceeds. This possibility is not without precedent, because the level of MT dynamics can vary widely in different cell types, within the same cell at different times in the cell cycle, and even in different regions of the same cell (50, 51).
Acknowledgments
We thank Dr. Herb Waite for help with the MS analysis and Mr. Herb Miller for technical assistance. This research was supported by U.S. Public Health Service Grants NS13560 (to L.W.) and NS35010 (to S.C.F.).
Abbreviations: MT, microtubule; FTDP-17, frontotemporal dementia and parkinsonism associated with chromosome 17; 4R, four repeat; 3R, three repeat.
References
- 1.Lee, V. M., Goedert, M. & Trojanowski, J. Q. (2001) Annu. Rev. Neurosci. 24, 1121-1159. [DOI] [PubMed] [Google Scholar]
- 2.Hutton, M., Lendon, C. L., Rizzu, P., Baker, M., Froelich, S., Houlden, H., Pickering-Brown, S., Chakraverty, S., Isaacs, A., Grover, A., et al. (1998) Nature 393, 702-705. [DOI] [PubMed] [Google Scholar]
- 3.Spillantini, M. G., Nurrell, J. R., Goedert, M., Farlow, M. R., Klug, A. & Ghetti, B. (1998) Proc. Natl. Acad. Sci. USA 95, 7737-7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark, L. N., Poorkaj, P., Wszolek, Z., Geschwind, D. H., Nasreddine, Z. S., Miller, B., Li, D., Payami, H., Awert, F., Markopoulou, K., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 13103-13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong, M., Zhukareva, V., Vogelsberg-Ragaglia, V., Wszolek, Z., Reed, L., Miller, B. I., Geschwind, D. H., Bird, T. D., McKeel, D., Goate, A., et al. (1998) Science 282, 1914-1917. [DOI] [PubMed] [Google Scholar]
- 6.Drubin, D. G., Feinstein, S. C., Shooter, E. M. & Kirschner, M. W. (1985) J. Cell Biol. 101, 1799-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caceres, A. & Kosik, K. (1990) Nature 343, 461-463. [DOI] [PubMed] [Google Scholar]
- 8.Esmaeli-Azad, B., McCarty, J. & Feinstein, S. C. (1994) J. Cell Sci. 107, 869-879. [DOI] [PubMed] [Google Scholar]
- 9.Liu, C., Lee, G. & Jay, D. G. (1999) Cell Motil. Cytoskeleton 43, 232-242. [DOI] [PubMed] [Google Scholar]
- 10.Trinczek, B., Ebneth, A., Mandelkow, E. M. & Mandelkow, E. (2002) J. Cell Biol. 156, 1051-1063.11901170 [Google Scholar]
- 11.LoPresti, P., Szuchet, S., Papasozomenos, S. C., Zinkowski, R. P. & Binder, L. I. (1995) Proc. Natl. Acad. Sci. USA 92, 10369-10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fellous, A., Francon, J., Lennon, A.-M. & Nunez, J. (1977) Eur. J. Biochem. 78, 167-174. [DOI] [PubMed] [Google Scholar]
- 13.Drechsel, D. N., Hyman, A. A., Cobb, M. H. & Kirschner, M. W. (1992) Mol. Biol. Cell 3, 1141-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trinczek, B., Biernat, J., Baumann, K., Mandelkow, E.-M. & Mandelkow, E. (1995) Mol. Biol. Cell 6, 1887-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panda, D., Goode, B. L., Feinstein, S. C. & Wilson, L. (1995) Biochemistry 34, 11117-11127. [DOI] [PubMed] [Google Scholar]
- 16.Goncalves, A., Braguer, D., Kamath, K., Martello, L., Briand, C., Horwitz, S., Wilson, L. & Jordan, M. A. (2001) Proc. Natl. Acad. Sci. USA 98, 11737-11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Himmler, A. (1989) Mol. Biol. Cell 9, 1389-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Himmler, A., Drechsel, D., Kirschner, M. W. & Martin, D. W. (1989) Mol. Cell. Biol. 9, 1381-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butner, K. A. & Kirschner, M. W. (1991) J. Cell Biol. 115, 717-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gustke, N., Trinczek, B., Biernat, J., Mandelkow, E.-M. & Mandelkow, E. (1994) Biochemistry 33, 9511-9522. [DOI] [PubMed] [Google Scholar]
- 21.Goode, B. L., Chau, M., Denis, P. E. & Feinstein, S. C. (2000) J. Biol. Chem. 275, 38182-38189. [DOI] [PubMed] [Google Scholar]
- 22.Lee, G., Neve, R. L. & Kosik, K. S. (1989) Neuron 2, 1615-1624. [DOI] [PubMed] [Google Scholar]
- 23.Goedert, M. & Jakes, R. (1990) EMBO J. 9, 4225-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goedert, M., Spillantini, M. G., Jakes, R., Rutherford, D. & Crowther, R. A. (1989) Neuron 3, 519-526. [DOI] [PubMed] [Google Scholar]
- 25.Goedert, M., Spillantini, M. G., Potier, M. C., Ulrich, J. & Crowther, R. A. (1989) EMBO J. 8, 393-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kosik, K. S., Orecchio, L. D., Bakalis, S. & Neve, R. L. (1989) Neuron 2, 1389-1397. [DOI] [PubMed] [Google Scholar]
- 27.D'Souza, I., Poorkaj, P., Hong, M., Nochlin, D., Lee, V. M., Bird, T. D. & Schellenberg, G. D. (1999) Proc. Natl. Acad. Sci. USA 96, 5598-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor, J. P., Hardy, J. & Fischbeck, K. H. (2002) Science 296, 1991-1994. [DOI] [PubMed] [Google Scholar]
- 29.Gamblin, T. C., King, M. E., Dawson, H., Vitek, M. P., Kuret, J., Berry, R. W. & Binder, L. I. (2000) Biochemistry 39, 6136-6144. [DOI] [PubMed] [Google Scholar]
- 30.Cleveland, D. W., Hwo, S. W. & Kirschner, M. W. (1977) J. Mol. Biol. 116, 227-247. [DOI] [PubMed] [Google Scholar]
- 31.Toso, R. J., Jordan, M. A., Farrell, K. W., Matsumoto, B. & Wilson L. (1993) Biochemistry 32, 1285-1293. [DOI] [PubMed] [Google Scholar]
- 32.Panda, D., Miller, H. P. & Wilson, L. (1999) Proc. Natl. Acad. Sci. USA 96, 12459-12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker, R. A., O'Brien, E. T., Pryer, N. K., Soboeiro, M. F., Voter, W. A., Erickson, H. P. & Salmon, E. D. (1988) J. Cell Biol. 107, 1437-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goode, B. L. & Feinstein, S. C. (1994) J. Cell Biol. 124, 769-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kar, S., Fan, J., Smith, M. J., Goedert, M. & Amos, L. A. (2003) EMBO J. 22, 70-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Derry, W. B., Wilson, L. & Jordan, M. A. (1995) Biochemistry 34, 2203-2211. [DOI] [PubMed] [Google Scholar]
- 37.Wilson, L., Panda, D. & Jordan, M. A. (1999) Cell Struct. Funct. 24, 329-335. [DOI] [PubMed] [Google Scholar]
- 38.Brandt, R. & Lee, G. (1993) J. Biol. Chem. 268, 3414-3419. [PubMed] [Google Scholar]
- 39.Goode, B., Denis, P., Panda, D., Miller, H., Radeke, M. J., Wilson, L. & Feinstein, S. C. (1997) Mol. Biol. Cell 8, 353-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hasegawa, M., Smith, M. & Goedert, M. (1998) FEBS Lett. 437, 207-210. [DOI] [PubMed] [Google Scholar]
- 41.Yen, S. H., Hutton, M., DeTure, M., Ko, L. W. & Nacharaju, P. (1999) Brain Pathol. 9, 695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeTure, M., Ko, L. W., Yen, S., Nacharaju, P., Easson, C., Lewis, J., van Slegtenhorst, M., Hutton, M. & Yen, S. H. (2000) Brain Res. 853, 5-14. [DOI] [PubMed] [Google Scholar]
- 43.von Bergen, M., Barghorn, S., Li, L., Marx, A., Biernat, J., Mandelkow, E. M. & Mandelkow, E. (2001) J. Biol. Chem. 276, 48165-48174. [DOI] [PubMed] [Google Scholar]
- 44.Li, L., von Bergen, M., Mandelkow, E. M. & Mandelkow, E. (2002) J. Biol. Chem. 277, 41390-41400. [DOI] [PubMed] [Google Scholar]
- 45.Jordan, M. A., Wendell, K., Gardiner, S., Derry, W. B., Copp, H. & Wilson, L. (1996) Cancer Res. 56, 816-825. [PubMed] [Google Scholar]
- 46.Yvon, A. M. C., Wadsworth, P. & Jordan, M. A. (1999) Mol. Biol. Cell 10, 947-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wittmann, C. W., Wszolek, M. F., Shulman, J. M., Salvaterra, P. M., Lewis, J., Hutton, M. & Feany, M. B. (2001) Science 293, 711-714. [DOI] [PubMed] [Google Scholar]
- 48.Dustin, P. (1984) Microtubules (Springer, Berlin), 2nd Ed.
- 49.LaMonte, B. H., Wallace, K. E., Holloway, B. A., Shelly, S. S., Ascano, J., Tokito, M., Van Winkle, T., Howland, D. S. & Holzbaur, E. L. (2002) Neuron 34, 715-727. [DOI] [PubMed] [Google Scholar]
- 50.Bass, P. W. & Black, M. M. (1990) J. Cell Biol. 111, 495-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wordemann, L. & Mitchison, T. J. (1994) in Microtubules, eds. Hyams, J. & Lloyd, C. (Wiley–Liss, New York), pp. 287-301.