Abstract
Assays of drug action typically evaluate biochemical activity. However, accurately matching therapeutic efficacy with biochemical activity is a challenge. High-content cellular assays seek to bridge this gap by capturing broad information about the cellular physiology of drug action. Here, we present a method of predicting the general therapeutic classes into which various psychoactive drugs fall, based on high-content statistical categorization of gene expression profiles induced by these drugs. When we used the classification tree and random forest supervised classification algorithms to analyze microarray data, we derived general “efficacy profiles” of biomarker gene expression that correlate with anti-depressant, antipsychotic and opioid drug action on primary human neurons in vitro. These profiles were used as predictive models to classify naïve in vitro drug treatments with 83.3% (random forest) and 88.9% (classification tree) accuracy. Thus, the detailed information contained in genomic expression data is sufficient to match the physiological effect of a novel drug at the cellular level with its clinical relevance. This capacity to identify therapeutic efficacy on the basis of gene expression signatures in vitro has potential utility in drug discovery and drug target validation.
Keywords: pharmacogenomics, predictive efficacy, drug screening
Microarray-based gene expression patterns can be used as fingerprints of cellular physiology. The variety of cellular physiologies discernable by gene expression profile fingerprinting is expanding as an increasing range of cell types and cellular manipulations are investigated, and statistical methods of expression profile classification are refined. In yeast, distinctive profiles of genomic expression have been used to characterize cellular responses to diverse environmental transitions (1), functionally classify genetic manipulations, and discover a novel target for a drug of partially characterized function (2). In cancer studies, microarray data has been used to classify solid tumors (3), correlate tumor characteristics to clinical outcome (4), and cluster cell lines on the basis of their tissue of origin and response to drugs (5–9). In the area of toxicogenomics, large-scale gene expression analysis of toxin-treated cells and animals has yielded a highly accurate capacity to recognize the toxic potential of novel drug candidates (10–14), resulting in an increase in the efficiency of drug triage in the pharmaceutical development pipeline.
Multiple statistical methods have been applied to classification and recognition of expression profiles. Supervised classification analysis methods, which can classify patterns of novel data based on prior knowledge of sample classes, include linear discriminant analysis and genetic algorithm/K-nearest neighbors (11, 15), Fisher discriminant analysis (16), support vector machines (17), neural networks (18), and tree-based analysis (19). Here, we use human primary neurons treated with multiple members of multiple classes of antidepressant drugs, antipsychotic drugs, and opioid receptor agonists to generate DNA microarray gene expression data representative of these classes of treatment. We investigate whether example gene expression profiles from these drug treatments can be used to construct statistical models capable of predicting drug efficacy. We find that the classification tree (CT) and random forest (RF) supervised classification schemes can be used to predict the functional category of members of each of these drug classes with good accuracy, based on analysis of the gene expression profile induced by a drug.
Materials and Methods
Cell Cultures. Primary human neuronal precursor cells (Clonex-press, Gaithersburg, MD) were cultured for 7 days in growth media (GM) (50:50 DMEM/F12, 5% FBS, 10 ng/ml basic fibroblast growth factor, 10 ng/ml epidermal growth factor, 1:100 Clonexpress neuronal cell supplement, penicillin/streptomycin), and differentiated for 7 days in six-well plates at 900,000 cells per well in GM plus 100 μM dibutyrtyl cAMP, 20 ng/ml nerve growth factor, 1:100 matrigel, with 72-h media changes. Morphologically neuronal cells comprised ≈80% of the cultures.
Drug Treatments. Drugs were dissolved in DMSO and added to cultures at a final DMSO concentration of 0.04%. Drug concentrations represented pharmaceutically relevent doses: 2.0 μM amoxepine, 2.0 μM clomipramine, 2.0 μM desipramine, 1.0 μM doxepin, 2.0 μM imipramine, 1.0 μM maprotiline, 0.7 μM nortryptyline, 0.4 μM protriptyline, 1.5 μM trimipramine, 0.3 μM citalopram, 0.3 μM paroxetine, 1.4 μM sertraline, 0.4 μM tranylcypromine, 0.8 μM phenelzine, 0.6 μM iproniazid, 2.0 μM trazadone, 1.0 μM amitriptyline, 0.5 μM fluoxetine, 1.5 μM fluvoxamine, 2.3 μM bupropion, 1.0 μM chlorpromazine, 1.0 μM trifluperazine, 0.8 μM triflupromazine, 0.05 μM pimozide, 4.0 μM clozapine, 0.2 μM haloperidol, 0.04 μM risperidone, 0.5 μM loxapine, 0.1 nM BW373U86, 1.0 μM Enkephalin, 0.1 nM U50488, 1.0 μM U62066, 1.0 μM Endomorphin, 0.1 μM Tyr-d-Arg-Phe-Lys-NH2 (DALDA), 0.1 μM Tyr-d-Ala-Gly-NMe-Phe-Gly-ol (DAMGO), 0.1 μM Dynorphin A. Three control cultures were treated with DMSO only; two were treated with 5.0 μM phencyclidine (PCP) or 5.0 μM amphetamine. All treatments were 24 h in duration and conducted simultaneously. Drugs were purchased from Sigma or Tocris Cookson (Ellisville, MO).
Sample Processing. Cells were lysed in Trizol. Biotin-labeled cDNA was made by using 15 μg of total RNA with poly(T) primers. Gene expression was evaluated by hybridization to the proprietary CuraChip microarray (CuraGen, New Haven, CT) of ≈11,000 oligonucleotide probes. Slides were hybridized for 15 h at 30°C with constant rotation, washed for 30 min at room temperature (RT), incubated in streptavidin solution (4°C, 30 min), washed three times for 15 min at RT, incubated in Cy3-conjugated detection buffer (4°C, 30 min), and washed three times for 15 min at RT. Slides were scanned (GMS 418 Scanner, Genetic Microsystems, Woburn, MA) and analyzed by using imagene software (BioDiscovery, Marina Del Rey, CA). Of 11,000 genes on the microarray, ≈4,700 were found to be expressed at least 3× background.
Data Analysis. All genes detectable at least 3× background after signal normalization were included in the data sets for analysis. Data were prefiltered by using a generous Kruskal–Wallis filter (P < 0.001, ≈4,700 genes over 36 samples). Both CT and RF models were constructed by using the data sets from the 36 drug-treated samples, and calculated within a leave-one-out cross-validation loop to minimize the influence of marker prefiltering on model accuracy. The two highest-count markers selected by the CT decision tree were then removed from the data set and this process was repeated, then repeated again with the two top markers from the second iteration removed as well. For RF and each iteration of the CT algorithm, the samples were weighted such that an unknown would have an equal probability of falling within any class, and not default to the over-represented class (antidepressants). The RF algorithm was also calculated with 1,000 trees grown and two random inputs attempted at each split. Two additional RF models were generated with the entire selective serotonin reuptake inhibitor (SSRI) or tricyclic class removed in a leave-one-subclass-out cross-validation loop. All statistical algorithms were performed by using the “R” statistical software system (www.cran.r-project.org).
Three-dimensional graphs were generated with decisionsite (Spotfire, Somerville, MA). For CT iteration-one results, class separation provided by the four identified biomarkers was graphed for the four possible three-way combinations (leaving each gene out once). For RF results, class separation provided by the three markers with greatest confidence measures was graphed. The volumes occupied by the respective sample classes were delineated by lines interconnecting all of the correctly classified members in each group.
Results
Supervised Classification of Drug-Treated Samples. The supervised classification methods of RF and CT were used to analyze the gene expression profiles of drug-treated neurons. These methods have the advantage that examples of known classes can be used to build models of salient features that provide categorical distinction between the data sets used to build the models. These models can then be tested empirically with data sets of known class that were not used in model construction (cross validation). The ability of the model to correctly classify naïve samples serves as a measure of model quality. With both CT and RF methods, a “leave-one-out” training and testing series was conducted for all 36 drug-treated samples. Thus, 36 individual models were constructed, each trained with 35 example gene expression profiles, with one profile withheld from training for evaluation. After construction of each model, the profile excluded from the training set was tested by the model for assignment to one of the three drug treatment categories. Overall effectiveness of each method was calculated as the percent correct classifications out of the total 36 training and testing events conducted by the method.
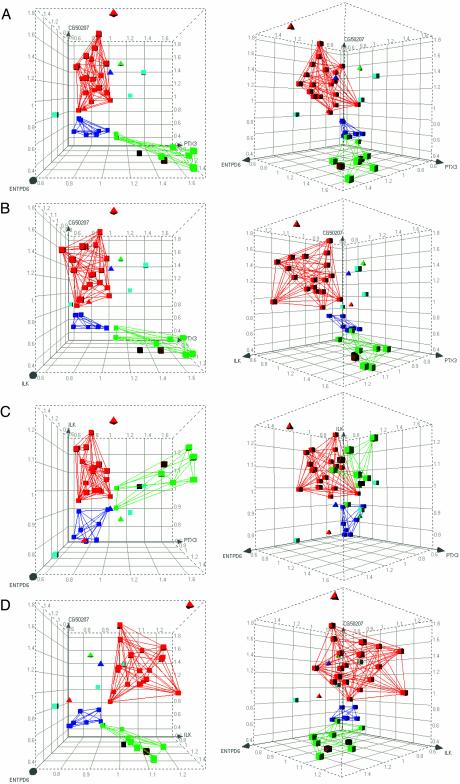
Classification Tree. The CT method classified 32 of 36 expression profiles in the category corresponding to the therapeutic application of the drug used to treat the cells (88.9% correct classification) (Table 1). Of the 20 antidepressants, 18 were correctly classified. Of the eight antipsychotics, seven were correctly classified. Of the eight opioid receptor agonists, seven were correctly classified. Interestingly, four gene markers were sufficient to provide this level of resolution accuracy among these expression profiles, with pentaxin 3 (PTX3) and integrin-linked kinase (ILK) being sufficient to provide the majority of resolution between classes, as indicated by the high marker count for these genes (Table 2). Three-dimensional graphical representation of all possible three-way combinations of these four biomarkers illustrates the robust class separation provided by expression level comparison (Fig. 1).
Table 1. Classification of gene expression profiles induced by different drug treatments.
| Compound | True class | CT pred | RF pred | Subclass |
|---|---|---|---|---|
| Amoxepine | AD | AD | AD | Tricyclic |
| Clomipramine | AD | AD | AD | Tricyclic |
| Desipramine | AD | OP | AD | Tricyclic |
| Doxepin | AD | AD | AD | Tricyclic |
| Imipramine | AD | AD | AD | Tricyclic |
| Maprotiline | AD | AD | AD | Tricyclic |
| Nortryptyline | AD | AD | AD | Tricyclic |
| Protriptyline | AD | AD | AD | Tricyclic |
| Trimipramine | AD | AD | AD | Tricyclic |
| Amitriptyline | AD | AD | AD | Tricyclic |
| Citalopram | AD | AD | AD | SSRI |
| Paroxetine | AD | AD | AD | SSRI |
| Sertraline | AD | AD | AD | SSRI |
| Fluoxetine | AD | AD | AD | SSRI |
| Fluvoxamine | AD | AD | AD | SSRI |
| Tranylcypromine | AD | AD | AD | MAOI |
| Phenelzine | AD | AP | AP | MAOI |
| Iproniazid | AD | AD | AP | MAOI |
| Trazadone | AD | AD | AD | Atypical |
| Buproprion | AD | AD | AD | Atypical |
| Chlorpromazine | AP | AP | AP | Classic |
| Trifluperazine | AP | AP | AP | Classic |
| Triflupromazine | AP | AP | AP | Classic |
| Pimozide | AP | AP | AP | Classic |
| Clozapine | AP | AD | AD | Atypical |
| Haloperidol | AP | AP | AP | Atypical |
| Risperidone | AP | AP | AP | Atypical |
| Loxapine | AP | AP | AD | Atypical |
| BW373U86 | OP | OP | AD | δ OPR |
| Enkephalin | OP | AD | AD | δ OPR |
| U50488 | OP | OP | OP | κ OPR |
| U62066 | OP | OP | OP | κ OPR |
| Dynorphin A | OP | OP | AD | κ/μ OPR |
| DALDA | OP | OP | OP | μ OPR |
| DAMGO | OP | OP | OP | μ OPR |
| Endomorphin | OP | OP | OP | μ OPR |
| Percent ”correct” | 88.9 | 83.3 |
True class, known therapeutic utility; AD, antidepressant; AP, antipsychotic; OP, opioid receptor agonist. CT pred and RF pred, classes predicted by those methods for the expression profiles elicited by the drugs listed on the left; boldface designations, predicted therapeutic classes different from the ”true” class. Subclass, the common pharmacological sub-classification; MAOI, monoamine oxidase inhibitor; δ, κ, μOPR, δ, κ, μ opioid receptor agonist, respectively
Table 2. Marker sets resulting from three sequential iterations of the CT analysis method.
| Marker identity | Marker count |
|---|---|
| CT iteration 1: 88.9% accuracy | |
| PTX3, pentaxin 3 | 35 |
| ILK, integrin linked kinase | 34 |
| ENTPD6, ectonucleoside triphosphate diphosphohydrolase 6 | 1 |
| GPCR CG50207 | 1 |
| CT iteration 2: 72.2% accuracy | |
| SFRS7, splicing factor, arginine/serine-rich 7 | 34 |
| ENTPD6, ectonucleoside triphosphate diphosphohydrolase 6 | 24 |
| CBRC7TM_424 GPCR | 1 |
| APAF-1, apoptotic protease activating factor 1 | 1 |
| ERMAP, erythroblast membrane-associated protein | 8 |
| CGFLC_31120 | 1 |
| GPCR CG50207 | 1 |
| LDHA, lactate dehydrogenase A | 1 |
| CT iteration 3: 80.6% accuracy | |
| LYPLA1, lysophospholipase 1 | 34 |
| GPCR CG50207 | 26 |
| CBRC7TM_424, GPCR | 1 |
| APAF-1, apoptotic protease activating factor 1 | 8 |
| CGFLC_31120 | 1 |
| LDHA, lactate dehydrogenase A | 1 |
For each iteration, the two most frequent markers from the previous iteration were deleted from the expression data training set. Percent accuracy, the proportion of drug treatments correctly predicted by the respective marker set. G protein-coupled receptor (GPCR) CG50207 and CGFLC_31120 are novel sequences of GPCR and unknown function, respectively. Marker count, number of times out of the 36 model building episodes a particular gene was selected as a marker
Fig. 1.
Three-dimensional representation of class discrimination on the basis of biomarker expression: classification tree. All possible three-way combinations of the four-gene marker set from CT iteration-one are displayed: (CG50207 vs. ENTPD6 vs. PTX3) (A), (CG50207 vs. ILK vs. PTX3) (B), (ILK vs. ENTPD6 vs. PTX3) (C), and (CG50207 vs. ENTPD6 vs. ILK) (D). Axes represent relative expression levels of marker genes, with means set to 1.0. Each graph is shown perpendicular to the XY plane (Left), and from 45° rotation around the y axis (Right). Red, antidepressant; dark blue, antipsychotic; green, opioid; brown, PCP; black, amphetamine; light blue, vehicle control; squares, correctly predicted treatments; triangles, misclassified treatments. Lines connecting correctly predicted treatments delineate the volume occupied by each accurately defined sample class. Note distinct class separation and placement of untreated control samples outside the treatment classes.
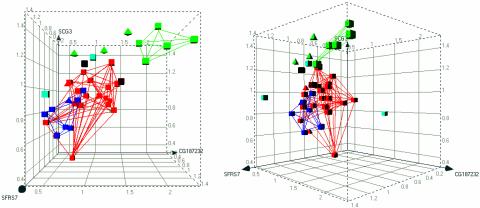
Random Forest. The RF method classified 30 of 36 expression profiles in the category corresponding to the therapeutic application of the drug used to treat the cells (83.3% correct classification) (Table 1). Of the 20 antidepressants, 18 were correctly classified. Of the eight antipsychotics, seven were correctly classified. Of the eight opioid receptor agonists, five were correctly classified. The RF analysis identified 326 markers (not shown) used to construct the predictive models. The markers assumed an importance measure between zero and one. A large importance measure indicates that random permutation of that gene causes samples to be misclassified more often (hence that gene is important). Thirty-two markers had an importance measure >0.35. Three markers had an importance measure >0.75: SFRS7 (splicing factor, arginine/serine-rich 7), SCG3 (secretogranin III), and hypothetical protein CG187232-01. Class separation provided by only these three top biomarkers (Fig. 2) is less distinct than the separation yielded by the biomarkers identified by the CT. This is probably because of the relative proficiency of the CT and RF algorithms with data sets containing a few strong markers, or a large number of weak markers, respectively.
Fig. 2.
Three-dimensional representation of class discrimination on the basis of biomarker expression: random forest. The three biomarkers with importance measures >0.75 are depicted: (SFRS7 vs. SCG3 vs. CG187232–01). Axes represent relative expression levels of marker genes, with means set to 1.0. The graph is shown perpendicular to the XY plane (Left), and from 45° rotation around the y axis (Right). Red, antidepressant; dark blue, antipsychotic; green, opioid; brown, PCP; black, amphetamine; light blue, vehicle control; squares, correctly predicted treatments; triangles, misclassified treatments.
Novel Subclass Prediction. The ability to accurately predict the functional class of an unrepresented drug type was tested by constructing models with an entire subclass omitted from the training data set. Two independent iterations of RF were conducted, with all expression data from SSRI-treated samples or tricyclic-treated samples withheld from the training data, respectively. Each model was then used to classify members of the antidepressant drug subclass withheld from training. The SSRI-naïve model correctly identified five of five (100%) SSRI treatments as antidepressants, and the tricyclic-naïve model correctly predicted 10 of 10 (100%) tricyclics as antidepressants, even though neither model was constructed with explicit examples of these respective subclasses.
Outgroup Identification by Graphical Depiction. A measure of model strength is the ability to distinguish inactive treatments from active drugs of various classes. In the most stringent case, a model should be able to exclude outgroup treatments from the target categories, even with outgroups not used to build the model. To determine whether the constructed models are capable of identifying novel outgroups as separate from established drug classes, we withheld vehicle-treated and nontarget class drug-treated control culture microarray data from the CT and RF training sets, to serve as unfamiliar outgroups. After the genes that distinguish the drug classes were identified, the top biomarkers from the CT and RF methods were depicted graphically, incorporating all of the data from each drug class as well as control samples (Figs. 1 and 2). A minimum volume encompassing the correctly classified samples was depicted by linearly connecting all members of each drug class. Vehicle-treated samples within these views were clearly excluded from the minimum volumes of each of the three drug classes, thereby illustrating a means by which outgroups may be effectively distinguished from therapeutic treatments, even when those outgroups are not included in the training data sets. PCP- and amphetamine-treated samples were effectively separated from the three main drug classes by RF, and fell very close to but outside the opioid category in the CT visualization.
Multiple Iterations of Classification Tree. Because relatively few genes were identified as drug class markers by the CT, we investigated the robustness of the approach in the absence of these markers by successively performing a second and third iteration of the analysis, excluding from the data set the predominant gene markers identified by the first and second iterations, respectively. The second iteration resulted in the identification of eight gene markers sufficient to provide 72.2% resolution accuracy. Misclassified as antidepressants in this second iteration were trifluperazine, clozapine, enkephalin, DALDA, and dynorphin. Misclassified as antipsychotics were clomipramine, phenelzine, iproniazid, and buproprion. Paroxetine was misclassified as opioid. Interestingly, the third iteration resulted in the identification of fewer gene markers (six) than the second iteration (Table 2), sufficient to provide greater resolution accuracy (80.6%) between treatment classes, even though the strongest markers from the second iteration were removed from the data set. Misclassfications made by the third iteration were identical to the second iteration, except paroxetine, buproprion and DALDA were correctly classified. Because CT and RF are based on similar recursive partitioning algorithms, the two approaches would be expected to identify many of the same markers. Indeed, both methodologies identified SFRS7 as a top marker. Overall, 55% of the markers from CT iterations 1–3, and six of the eight CT markers with counts >1, are among the 326 RF markers.
Discussion
The goal of the present study was to determine whether in vitro gene expression profiles of drug efficacy could be generated and used to predict the therapeutic classes of compounds administered to cell cultures. Recognition of toxin-induced expression profiles has proven possible because of the characteristic transcriptional responses to various types of cellular insult. Whether different classes of drugs applied at nontoxic doses elicit discernable signature expression profiles has not been investigated. In particular, we were interested in whether drugs distinguished by therapeutic indication at the patient level induce distinct expression profiles at the cellular level that correlate with clinical efficacy. To test this hypothesis, we selected three general classes of psychotropic compounds that might be expected to elicit transcriptional responses in human neurons: two that are used clinically (antidepressants and antipsychotics) and one that acts on a therapeutically relevant target class (opioid receptor agonists). Within each of these three categories, multiple subclasses were represented in order to capture general transcriptional features of each drug class.
CT and RF supervised classification schemes proved effective at predicting the physiological consequence of drug treatments on the basis of induced gene expression profile. The progressive leave-one-out strategy of model building and testing allowed us to comprehensively assess the ability of these schemes to accurately classify profiles induced by “novel” drugs (drugs not used to build the statistical predictive model). Because each of the three drug categories were weighted equally in our analyses, the accuracy of categorization based simply on random chance would be expected to be 33.3%. The 83.3% and 88.9% success rates of RF and CT, respectively, indicate that salient features of the distinct cellular physiologies induced by these drug classes are being identified by these methods, and that they can be used to correctly predict the clinical activity of unfamiliar compounds. In principle, this approach can be expanded beyond the three classes resolved here to encompass a large number of drug classes and cellular physiologies. Such high-content output is principally limited only by the number of pharmaceutical classes to which a given cell type is responsive.
The difference in accuracy between the CT and RF is likely caused by relative emphases on individual marker strength. Both methods partition feature-space to classify samples (20). CT performs this split once, to identify a few genes that each explain a large portion of the differences among classes. RF calculates many trees to identify many genes that each account for a small amount of the differences among classes. Although the CT method has several advantages over more traditional learning algorithms, the hierarchical nature of the data partition is especially sensitive to outliers, manifesting in high model variance and misclassification rates (20, 21). By virtue of its iterative character, RF improves model variance, generally yielding classification accuracies equal to or better than CT augmented by variance-reduction techniques such as boosting or bagging (20, 22, 23). Our finding that the first iteration of CT has a lower misclassification rate than RF (Table 1) suggests that these CT markers are especially important in distinguishing among the drug groups. This is supported by the subsequent two iterations of CT which each result in classification accuracies lower than the RF (Table 2).
How the specific biomarkers identified here relate systematically to the medical processes with which they correlate, and whether these biomarkers are also functional intervention points for further pharmaceutical development, are questions under investigation. Subclasses of drugs within each broad class are medicinally similar, but mechanistically heterogeneous [e.g., SSRIs, monoamine oxidase (MAO) inhibitors, and norepinephrine-uptake inhibitors act on distinct drug targets within multiple neurotransmitter systems, yet can exert similar effects on clinical depression]. This suggests that these drugs may share a common mechanism of action downstream from their proximal biochemical activities. Detected by our microarray were many genes involved in the transmitter systems targeted by the three drug classes, including MAO-a, MAO-b, multiple serotonin receptors, D2, and other dopamine receptors. Several expected targets, such as the serotonin transporter, norepinephrine transporter, and the δ, κ, and μ opioid receptors were not detected by the microarray, indicating they may be expressed below the level of detection, or that other, unconventional targets are the primary locus of action in this culture system. The presence of gene expression biomarkers that robustly predict drug class is consistent with the presence of convergent mechanisms of action among the respective subclasses of the three broad drug classes studied here.
Although the practical emphasis in expression profile finger-printing to date has been to discern drug-toxicity profiles, drug-efficacy profiles may have utility in several important areas. One apparent application is to prioritize lead compounds early in drug development. The absence of a clear understanding of the mechanisms of psychosis and depression has prevented the development of a general cellular assay for drugs treating these disorders. For example, there is no one cellular assay for “antidepression.” Apart from direct binding of specific drugs to different molecular targets, antidepressant activity can only be tested in vivo at the present time, presenting throughput limitations. Pharmaceutical development has thus been largely confined to “me-too” molecules of specific activity similar to existing compounds. Our discovery of efficacy profiles common to drugs of disparate activity but within a common therapeutic class provides a simple in vitro tool for finding functionally valuable drugs that act via a range of targets. Moreover, this method works whether the mechanism of action is known or novel. In our study, even when entire drug subclasses were left out of the RF training protocol, their medical utility was correctly identified upon testing of the models. The accurate classification of SSRI and tricyclic antidepressants by models naïve to these important pharmaceutical classes illustrates the prospect of discovering therapeutically valuable drugs that function by unknown mechanisms. Had SSRIs not been discovered before this study, for example, they could still have been accurately identified in our assay as having antidepressant activity.
In future refinements, it will be necessary to recognize false positives by including outgroups into the modeling protocol or establishing an alternative classification of “insufficiently close to any efficacy profile.” Three-dimensional graphical depiction of vehicle-treated control sample biomarker values placed them outside the minimum volume encompassing the respective drug classes (Figs. 1 and 2). Significantly, these control samples were omitted from the data sets used to construct the efficacy profiles. Thus, their effective partition supports the “insufficiently close to any efficacy profile” approach to novel outgroup identification. Control samples treated with compounds of other neuroactive classes (PCP and amphetamine) also fell distinctly outside the three drug class volumes in the RF depiction. In the CT depictions, although these neuroactive drugs fell technically outside the three classes, they were very close to the opioid treatments, perhaps because of shared aspects of cellular physiology. Any compound tested will necessarily have a position in n-dimensional biological descriptor space that falls closest to one of the centroids created by the different classes of drug treatment. In our present cheminformatic scheme, this closest centroid determines the class of the drug being tested. An optimized version of this assay may establish a minimum n-dimensional hypervolume surrounding all known members of a drug class, outside of which a given drug under evaluation may be classified as not belonging to any known therapeutic class. In practice, as new bioactive compounds are detected by the assay, the boundaries of each hypervolume may be empirically optimized by functionally assaying, in vivo, novel compounds that progressively define revised hypervolume borders.
Potentially of interest in analyses of this kind are trends in misclassification. For example, κ and μ opioid receptor agonist-induced expression profiles were predominantly correctly classified by both the RF and the first iteration of CT. However, δ opioid receptor agonist-induced profiles were predominantly classified as antidepressant profiles (Fig. 2). A question raised by this result is whether targeting the δ opioid receptor system would have utility in the treatment of depression. Studies of δ opioid agonist action in mouse models of depression, in fact, support the idea that this receptor system mediates depressive behavior (24). Thus, in vitro analyses of this kind may have utility in discovering novel therapeutic applications of drugs.
Another potential application of efficacy profiling is the validation of genes as therapeutic drug targets, an early step in the drug development process. Manipulation of a specific gene in culture, either by overexpression or gene silencing, may identify it as a drug target when a therapeutically relevant expression profile is induced. The derivation of biomarker profiles as signatures of specific pharmacological functions provides a foundation for in vitro assessment of the therapeutic relevance of presumptive disease-mediating biological molecules, and the efficacy of therapies devised to target them.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CT, classification tree; RF, random forest; PCP, phenylcyclidine; SSRI, selective serotonin reuptake inhibitor.
References
- 1.Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D. & Brown, P. O. (2000) Mol. Biol. Cell 11, 4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hughes, T. R., Marton, M. J., Jones, A. R., Roberts, C. J., Stoughton, R., Armour, C. D., Bennett, H. A., Coffey, E., Dai, H., He, Y. D., et al. (2000) Cell 102, 109-126. [DOI] [PubMed] [Google Scholar]
- 3.Sørlie, T., Perou, C. M., Tibshirani, R., Aas, T., Geisler, S., Johnsen, H., Hastie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 10869-10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pomeroy, S. L., Tamayo, P., Gaasenbeek, M., Sturla, L. M., Angelo, M., McLaughlin, M. E., Kim, J. Y., Goumnerova, L. C., Black, P. M., Lau, C., et al. (2002) Nature 415, 436-442. [DOI] [PubMed] [Google Scholar]
- 5.Perou, C. M., Jeffrey, S. S., van de Rijn, M., Rees, C. A., Eisen, M. B., Ross, D. T., Pergamenschikov, A., Williams, C. F., Zhu, S. X., Lee, J. C., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 9212-9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perou, C. M., Sorlie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., Rees, C. A., Pollack, J. R., Ross, D. T., Johnsen, H., Akslen, L. A., et al. (2000) Nature 406, 747-752. [DOI] [PubMed] [Google Scholar]
- 7.Scherf, U., Ross, D. T., Waltham, M., Smith, L. H., Lee, J. K., Tanabe, L., Kohn, K. W., Reinhold, W. C., Myers, T. G., Andrews, D. T., et al. (2000) Nat. Genet. 24, 236-244. [DOI] [PubMed] [Google Scholar]
- 8.Ross, D. T., Scherf, U., Eisen, M. B., Perou, C. M., Rees, C., Spellman, P., Iyer, V., Jeffrey, S. S., van de Rijn, M., Waltham, M., et al. (2000) Nat. Genet. 24, 227-235. [DOI] [PubMed] [Google Scholar]
- 9.Califano, A., Stolovitzky, G. & Tu, Y. (2000) Proc. Int. Conf. Intel. Syst. Mol. Biol. 8, 75-85. [PubMed] [Google Scholar]
- 10.Hamadeh, H. K., Bushel, P. R., Jayadev, S., DiSorbo, O., Bennett, L., Li, L., Tennant, R., Stoll, R., Barrett, J. C., Paules, R. S., et al. (2002) Toxicol. Sci. 67, 232-240. [DOI] [PubMed] [Google Scholar]
- 11.Hamadeh, H. K., Bushel, P. R., Jayadev, S., Martin, K., DiSorbo, O., Sieber, S., Bennett, L., Tennant, R., Stoll, R., Barrett, J. C., et al. (2002) Toxicol. Sci. 67, 219-231. [DOI] [PubMed] [Google Scholar]
- 12.Waring, J. F., Ciurlionis, R., Jolly, R. A., Heindel, M. & Ulrich, R. G. (2001) Toxicol. Lett. 120, 359-368. [DOI] [PubMed] [Google Scholar]
- 13.Waring, J. F. & Halbert, D. N. (2002) Curr. Opin. Mol. Ther. 4, 229-235. [PubMed] [Google Scholar]
- 14.Lakkis, M. M., DeCristofaro, M. F., Ahr, H. J. & Mansfield, T. A. (2002) Exp. Rev. Mol. Diagn. 2, 337-345. [DOI] [PubMed] [Google Scholar]
- 15.Mendez, M. A., Hodar, C., Vulpe, C., Gonzalez, M. & Cambiazo, V. (2002) FEBS Lett. 522, 24-28. [DOI] [PubMed] [Google Scholar]
- 16.Stephanopoulos, G., Hwang, D., Schmitt, W. A. & Misra, J. (2002) Bioinformatics 18, 1054-1063. [DOI] [PubMed] [Google Scholar]
- 17.Brown, M. P., Grundy, W. N., Lin, D., Cristianini, N., Sugnet, C. W., Furey, T. S., Ares, M., Jr., & Haussler, D. (2000) Proc. Natl. Acad. Sci. USA 97, 262-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu, Y., Selaru, F. M., Yin, J., Zou, T. T., Shustova, V., Mori, Y., Sato, F., Liu, T. C., Olaru, A., Wang, S., et al. (2002) Cancer Res. 62, 3493-3497. [PubMed] [Google Scholar]
- 19.Zhang, H. & Yu, C. Y. (2002) Front. Biosci. 7, c63-c67. [DOI] [PubMed] [Google Scholar]
- 20.Hastie, T. & Tibshirani, R. (2001) The Elements of Statistical Learning: Data Mining, Inference, and Prediction (Springer, Berlin).
- 21.Venables, W. N. & Ripley, B. D. (1999) Modern Applied Statistics with S-PLUS (Springer, Berlin).
- 22.Breiman, L. (2001) Machine Learning 45, 5-32. [Google Scholar]
- 23.Amit, Y. & Geman, D. (1997) Neural Comput. 9, 1545-1588. [Google Scholar]
- 24.Tejedor-Real, P., Mico, J. A., Smadja, C., Maldonado, R., Roques, B. P. & Gilbert-Rahola, J. (1998) Eur. J. Pharmacol. 354, 1-7. [DOI] [PubMed] [Google Scholar]