Abstract
Dynamic regulation of ion channels is critical for maintaining fluid balance in epithelial tissues. Cystic fibrosis, a genetic disease characterized by impaired fluid transport in epithelial tissues, is caused by dysfunctional cystic fibrosis transmembrane conductance regulator (CFTR) Cl- channel activity. Recent studies have shown that binding of PSD-95/Dlg/ZO-1 (PDZ) domain proteins to CFTR is important for retaining it at the apical membrane and for regulating its channel activity. Here, we describe a phosphorylation mechanism that regulates CFTR channel activity, which is mediated by PDZ domains. The Na+/H+ exchanger regulatory factor (NHERF) binds to CFTR and increases its open probability (Po). Protein kinase C disrupts the stimulatory effect of NHERF on CFTR channel Po. Phosphorylation by PKC of Ser-162 in the PDZ2 domain of NHERF is critical for this functional effect. Furthermore, a mutation in PDZ2 that mimics phosphorylation decreases CFTR binding and disrupts the ability of NHERF PDZ1–2 to stimulate CFTR channel Po. Our results identify a role for PKC and suggest that phosphorylation of NHERF PDZ2 domain may be an important mechanism for regulating CFTR channel activity.
The cystic fibrosis transmembrane conductance regulator (CFTR) is a cAMP-stimulated chloride ion channel that is expressed in the apical membrane of epithelial cells, where its activity is critical for transepithelial salt and fluid transport. The structure of CFTR resembles that of other ATP-binding cassette transporters in having two homologous motifs, each consisting of six transmembrane helices followed by a cytoplasmic ATP-binding domain. The two motifs are connected by a central regulatory (R) domain, containing numerous consensus sites for phosphorylation by protein kinases A (PKA) and C (PKC). Gating of CFTR channels is tightly regulated by phosphorylation, and by binding and hydrolysis of ATP (1). Whereas PKA activates the channel by directly phosphorylating sites within the R-domain, PKC does not activate CFTR substantially but seems to increase its responsiveness to PKA (2). However, the molecular mechanisms that account for PKC regulation of CFTR are poorly understood.
CFTR seems to exist within a multiprotein complex, in which its activity is regulated by interactions with other proteins. CFTR has been demonstrated to bind to several PSD-95/Dlg/ZO-1 (PDZ) domain proteins via its C-terminal tail (3–7). PDZ domain interactions may anchor CFTR to the cytoskeleton to help localize and retain CFTR specifically at the apical membrane of epithelial cells (8, 9). In addition, binding of PDZ domains of Na+/H+ exchanger regulatory factor (NHERF, also known as EBP50) and CAP70 directly regulates CFTR channel activity (6, 10). The binding and cross-linking of two C-terminal tails of CFTR by two PDZ domains in a bivalent molecule allosterically enhances CFTR gating, whereas PDZ domain binding in the absence of cross-linking does not. Based on the observed biphasic concentration dependence of the regulation of CFTR open probability (Po) by NHERF, we had proposed that CFTR channel activity can be regulated by the stoichiometry of CFTR–NHERF interactions (11). Nevertheless, it is unknown whether modulation of CFTR–NHERF interactions is exploited as a physiological mechanism for regulating CFTR channel activity. Multiprotein complexes are expected to be dynamic, in which protein interactions may be transient and stoichiometries variable. However, little is known regarding physiological modulation of protein interactions in multiprotein complexes in general, or of the CFTR–NHERF interaction specifically. In principle, these stoichiometries could be varied by changes in either relative concentrations or binding affinities of proteins within the complex.
Recent studies have revealed that phosphorylation of residues near the C terminus of target proteins is a mechanism for regulating PDZ domain interactions (11). The -2 residue of PDZ-binding sites is frequently a serine or threonine. The C-terminal PDZ-binding site of the inward rectifier K+ channel Kir2.3 contains a Ser at the -2 position within a consensus motif for PKA phosphorylation. Phosphorylation of this residue abolishes Kir2.3 interaction with the PSD-95 PDZ3 domain (12). Phosphorylation of the β2 adrenergic receptor C terminus on Ser at the -2 position disrupts its PDZ interactions with NHERF, which affects its plasma membrane recycling (13). Similar phosphorylation-dependent interactions between α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor subunits and cytosolic PDZ domain-containing proteins influence synaptic plasticity, including long-term depression (14) and long-term potentiation (15). Because bivalent NHERF PDZ interactions profoundly modulate CFTR channel gating, we have considered that phosphorylation-dependent regulation of NHERF–CFTR interactions could provide an important mechanism to regulate CFTR activity.
Here, we identify PKC as a modulator of the interaction between NHERF and CFTR. Whereas PKC is without effect on PKA-activated CFTR channel gating, it strongly inhibits the gating induced by NHERF. The effects of PKC are mediated by a mechanism involving phosphorylation of an NHERF PDZ domain. PKC phosphorylates Ser-162 within the PDZ2 domain of NHERF, which lowers its affinity for CFTR C terminus and disrupts the bivalent PDZ domain interaction of NHERF. Consequently, phosphorylated NHERF is unable to cross-link the channel and stimulate the activity of CFTR. Our findings suggest a mechanism for regulation of CFTR activity, mediated by modulation of NHERF PDZ domain interactions.
Materials and Methods
Creation of NHERF Mutants. NHERF mutations (T156A, S162A, S162D, S170A, and S290A) were constructed by using the QuikChange (Stratagene) mutagenesis strategy. All cDNAs were cloned into either pcDNA 3.0 (Invitrogen) or pGEX6P-1 (Amersham Biosciences) and verified by sequencing. Fusion proteins of GST fused to PDZ1 (1–139), PDZ2 (132–299), and PDZ1–2 (1–299) were expressed in Escherichia coli and purified as described (10).
Electrophysiology and Kinetic Analysis. Patch-clamp recordings of Calu-3 cells in excised inside-out patches were performed as described (10). The bath solution contained 140 mM NaCl, 2 mM MgCl2, 0.1 mM CaCl2, 1 mM EGTA, 2 mM MgATP, and 10 mM Hepes (pH 7.5) with 200 units/ml PKA catalytic subunit (Promega). The pipette solution lacked ATP and PKA. In experiments involving PKC, 2 μl of PKC (25 μg/ml; 2,200 units/mg) (Promega) along with 5 μM DiC8 (Sigma) were added to the bath solution. All experiments were performed at room temperature at ±60 mV transmembrane potential. Single-channel currents were filtered at 100 Hz, digitized at 2 kHz, and recorded by using PULSE+PULSEFIT software (HEKA Electronics, Lambrecht, Germany). Current records of at least a 5-min duration were analyzed for each experimental condition by TAC software (Bruxton, Seattle) for Po evaluation, as described (10). Data are presented as mean ± SEM. Student's paired t test was used to determine significance (P < 0.01).
In Vitro Phosphorylation Assays. Purified WT and mutant NHERF PDZ domains were examined for the ability to be phosphorylated by PKC. Typically, 1 μg of purified substrate was mixed with 25 milliunits of PKC, and kinase assays were carried out at 30°C in a volume of 20 μl, with 20 mM Hepes (pH 7.4), 1 mM DTT, 10 mM MgCl2, 1.7 mM CaCl2, and 0.05 mM ATP (5 μCi of [γ-32P]ATP per reaction; 1 Ci = 37 GBq) in the presence and absence of 600 μg/ml phosphatidylserine for 30 min before termination by addition of sample buffer. The samples were run on 10% Nu/PAGE gels, fixed, dried, and exposed to film. The extent of phosphorylation was quantified by densitometric analysis either by using a PhosphorImager system or scanning the film.
Metabolic Labeling and Immunoprecipitation of Epitope-Tagged NHERF. Chinese hamster ovary cells, plated onto 35-mm dishes and grown to ≈50% confluence, were transfected with 2 μg of hemagglutinin-NHERF cDNA (FuGENE 6, Roche Molecular Biochemicals). After 36–48 h, the medium was replaced with 2 ml of phosphate-free DMEM supplemented with 0.1 mCi of [32P]orthophosphate. After 90 min, the cells were incubated either with 100 nM phorbol 12-myristate 13-acetate (PMA) (Sigma) or DMSO for 20 min. Cells were washed three times with 2 ml of ice-cold PBS and lysed by adding 0.25 ml of ice-cold lysis buffer containing 150 mM NaCl, 50 mM Tris (pH 8.0), 1% Triton X-100, 0.1% SDS, 2 mM EDTA, 10 mM NaF, 10 mM sodium pyrophosphate, and 0.1 mM PMSF. The insoluble material was removed by centrifugation (14,000 × g for 10 min), and to the supernatants corresponding to equal amounts of total protein, α-hemagglutinin (Covance, Princeton) or α-EBP50 (Affinity Bioreagents, Neshanic Station, NJ) antibody was added (1:150). After rocking (4°C, 1 h), 40 μl of protein G-agarose (50%) was added, and the samples were rocked at 4°C for 1 h. The beads were washed three times with lysis buffer. SDS-sample buffer was added and heated at 65°C for 10 min. Proteins were separated on a 10% Nu/PAGE gel. After drying, phosphorylated proteins were detected by autoradiography and quantified by densitometric analysis using a PhosphorImager system or by scanning films.
Chemical Cross-Linking Experiments. PBS (50 μl, pH 7.5) containing 1 μg of purified PDZ domains or purified GST-PDZ fusion proteins was treated with 1 mM (final concentration) of the thiol-cleavable cross-linker 3,3′-dithiobis(sulfosuccinimidyl propionate) (Pierce) for 1 h on ice. Cross-linked products were split into two aliquots (3:2) and were denatured and analyzed in a 4–12% gradient gel (Nu/PAGE) under nonreducing conditions (without DTT) or after cleaving the cross-linker by treatment with DTT (100 mM) for 10 min; they were then visualized by Simply Blue safe stain (Invitrogen).
In Vitro Binding Assays. Purified WT and mutant (S162D) GST-PDZ2 proteins at various concentrations in 0.5 ml of PBS were incubated with a biotinylated peptide (1 μM) corresponding to the terminal 18 aa of CFTR for 1 h at 37°C. After incubation, 40 μl of 50% UltraLink streptavidin beads (Pierce) was added and gently mixed for 30 min. The beads were washed once (15 min) with PBS and twice (15 min each) with PBS containing 500 mM NaCl and eluted with SDS sample buffer. Eluted proteins were resolved on 10% Nu/PAGE gel, stained with Simply Blue safe stain, and quantified by densitometric analysis.
Results
PKC Modulates NHERF-Mediated Stimulation of CFTR Channel Po. Endogenous CFTR channels were recorded in excised inside-out membrane patches from Calu-3 cells, a lung submucosal gland cell line. We previously demonstrated that addition of a recombinant NHERF PDZ1–2 peptide to the cytoplasmic face of a patch containing a PKA-activated CFTR channel results in a marked (80–100%) increase in channel Po (10). Both PDZ domains present in a bivalent construct were required for this functional effect. The bivalent PDZ domain-mediated enhancement of channel activity occurred regardless of the initial Po or the number of channels in a patch, and it was not associated with changes in either the single-channel current amplitude or the number of channels in the patch (Fig. 1A; ref. 10). Thus, binding and cross-linking of two CFTR C termini by a bivalent NHERF molecule allosterically stimulates gating of the channel. The biphasic concentration dependence of the regulation of CFTR Po by NHERF suggested that CFTR channel activity could be regulated by the stoichiometry of CFTR–NHERF interactions. Therefore, we examined the effects of PKC on the modulation of CFTR channel activity by NHERF.
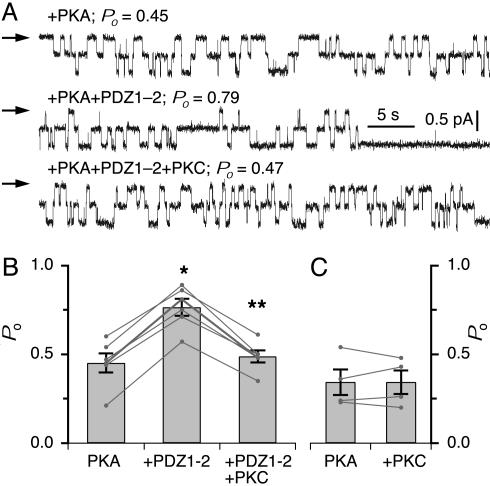
Fig. 1.
PKC inhibits NHERF PDZ domain-mediated stimulation of CFTR channel activity. (A) Representative current traces of CFTR in the presence of PKA, and after addition of 60 nM PDZ1–2, and PKC. Channel closed level indicated by arrows. (B) Mean Po of CFTR in the presence of PKA, after addition of PDZ1–2, and PKC. (C) Mean Po of CFTR before and after addition of PKC. * and ** indicate significant difference of Po after addition of PDZ1–2 peptide and after addition of PKC, respectively.
Application of PKC to the cytoplasmic face of a patch containing PKA-activated CFTR channel(s) that had been stimulated by NHERF PDZ1–2 markedly reduced channel Po (percent inhibition = 36 ± 2; n = 7; P < 0.01). Channel Po was decreased to a level that was comparable to the initial Po before the stimulation by NHERF PDZ1–2 (Fig. 1 A and B), indicating that PKC completely reversed the NHERF stimulation. In contrast, PKC was without effect on PKA-activated CFTR channel activities that had not been stimulated with NHERF (Fig. 1C), consistent with earlier studies (16). Furthermore, addition of NHERF PDZ1–2 peptide that has been either phosphorylated with PKC or in the presence of PKC was without effect on channel Po (data not shown, n = 3). These results suggest that PKC specifically inhibits the stimulatory effect of NHERF on CFTR channel Po.
Inhibition of NHERF Stimulation of CFTR Channel Activity by PKC Is Mediated via PDZ2 of NHERF. We hypothesized that PKC inhibited PDZ-mediated stimulation of CFTR channel gating by phosphorylating either CFTR or NHERF (or both) and disrupting the bivalent PDZ-domain interaction of NHERF with the channel. Phosphorylation of Ser/Thr at position -2 of consensus PDZ-binding sites disrupts binding of several proteins to PDZ domains (see Introduction). Thus, we speculated that Thr-1478 at the -2 position in the C terminus of CFTR could be a substrate for PKC and that its phosphorylation would disrupt binding of the tail of CFTR to NHERF PDZ domains. Alternatively, the NHERF PDZ domains could be substrates for PKC, and their phosphorylation may disrupt their binding to CFTR. However, such a mechanism for regulation of PDZ domain interactions has not been previously demonstrated.
To identify the functionally relevant phosphorylation substrate, we adopted the following strategy. We previously showed that a bivalent PDZ domain-mediated interaction is required to stimulate CFTR channel Po (10), whereas individual (monovalent) PDZ domains fail to stimulate CFTR gating, even though they are capable of binding to CFTR. GST exists as a stable dimer (for review, see refs. 17 and 18). It has been used as a dimerizing agent that can confer bivalency to proteins fused to it (19). Therefore, we predicted that individual (monovalent) PDZ domains fused to GST should effectively function as bivalent PDZ domain proteins because of the dimer formation of GST and thus become competent to stimulate CFTR gating. We reasoned that examination of the effects of PKC on the Po of CFTR channels that were stimulated by GST-PDZ fusion proteins would enable us to distinguish among the putative targets of PKC. Specifically, if the CFTR C terminus is the substrate, then PKC should decrease the Po of CFTR that has been stimulated by either GST-PDZ1 or GST-PDZ2. Alternatively, if PKC inhibits the stimulatory effects of only one of the GST-PDZ fusion proteins, then specific phosphorylation of that PDZ domain is likely to be responsible for the observed PKC inhibition of NHERF-stimulated CFTR activity.
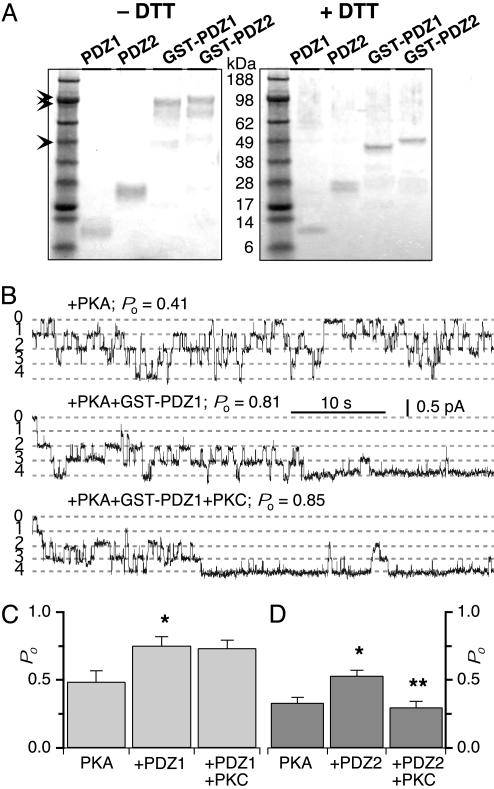
The individual GST-PDZ fusion proteins formed stable dimers, as expected (Fig. 2A). Both GST-PDZ1 and GST-PDZ2 significantly enhanced CFTR channel activity (Fig. 2B). In contrast, addition of either GST or the individual PDZ domains (monomer; Fig. 2 A) alone were without effects on channel Po (10). These results supported our prediction that a bivalent PDZ-domain complex, formed by dimerization of GST, would mimic the functional effects of the bivalent NHERF PDZ1–2 peptide. When PKC was subsequently added to the cytoplasmic bath solution, the Po of CFTR channel(s) that had been stimulated by GST-PDZ1 was not affected (Fig. 2 A and B). This result suggests, first, that CFTR is not the target of PKC and, second, that PDZ1 was also not the site of action. In contrast, PKC decreased the Po of CFTR that had been stimulated by GST-PDZ2 (Fig. 2C). These results therefore suggest that PKC inhibition of bivalent PDZ-domain stimulation of CFTR channel gating is mediated specifically via the NHERF PDZ2 domain.
Fig. 2.
Oligomeric status and the regulation of CFTR channel activity by GST-PDZ1 and GST-PDZ2. (A) Analysis of chemically cross-linked proteins under nonreducing (-DTT) and reducing (+DTT) conditions by SDS/PAGE. In the absence of DTT (Left), the top bands (double arrowheads) represent cross-linked dimers of GST-PDZ1 and GST-PDZ2, and the (faint) bottom band (single arrowhead) represents uncross-linked monomer. In contrast, no bands corresponding to dimers of individual PDZ domains are seen, suggesting that they exist as monomers. The dimers migrate as doublets due to the presence of intramolecular cross-links. In the presence of DTT (Right), only monomeric species are seen. (B) Functional effects of GST-PDZ1 and GST-PDZ2 on CFTR channel activity. Representative experiment containing four CFTR channels and their stimulation by addition of GST-PDZ1 and effect of subsequent addition of PKC on CFTR Po. Numbers indicate number of open channel levels. Summary of CFTR Po under various experimental conditions using GST-PDZ1 (C) and GST-PDZ2 (D). * and ** indicate significant difference of Po after addition of PDZ1–2 peptide and after addition of PKC, respectively.
PKC Phosphorylates NHERF PDZ2 Domain. The previous experiment strongly suggested that PKC inhibition of bivalent PDZ domain-mediated CFTR stimulation might involve phosphorylation of the PDZ2 domain of NHERF. To test this hypothesis, in vitro phosphorylation experiments were performed to determine whether the PDZ domains were substrates for PKC. Both PDZ2 and PDZ1–2 peptides were efficiently phosphorylated by PKC, whereas PDZ1 peptide phosphorylation was not detectable (Fig. 3A). This result indicates that the PDZ2 domain of NHERF is a specific substrate for PKC and suggests that phosphorylation of PDZ2 is responsible for disrupting the stimulatory effect of PDZ1–2 on CFTR activity. Although NHERF was phosphorylated under basal conditions in Calu-3 cells, its phosphorylation was increased by treatment of the cells with PMA (142 ± 17% of control; P < 0.05) (Fig. 3C). These results suggest that PKC phosphorylates NHERF in epithelial cells.
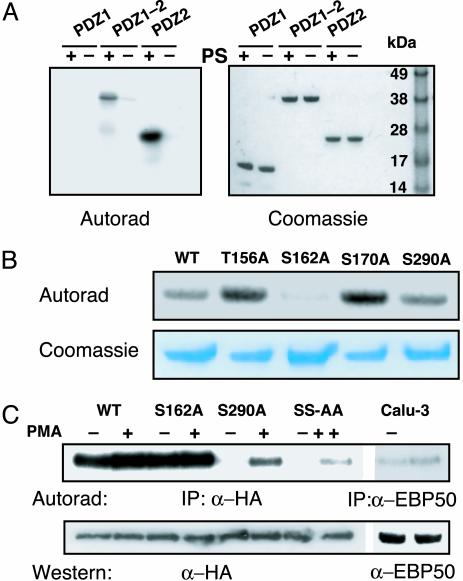
Fig. 3.
PKC phosphorylates NHERF. (A) PKC phosphorylates PDZ2 but not PDZ1 of NHERF. Shown is an autoradiogram of in vitro phosphorylation reaction of purified PDZ1, PDZ1–2, and PDZ2 peptides with PKC in the presence (+) and absence (-) of phosphatidylserine. Results are representative of four independent experiments. (B) Mutation of Ser-162 reduces phosphorylation of NHERF PDZ1–2 by PKC. The 32P content of WT PDZ1–2 is compared in this autoradiogram to the 32P contents of four different mutants. Results are representative of three experiments. (C) Metabolic labeling of cells with [32P]orthophosphate. Cells were treated with either DMSO (-) or PMA (100 nM) for 20 min (SS-AA, S162A, S290A). Results are representative of three independent experiments.
PKC Phosphorylates Ser-162 of NHERF. Potential phosphorylation sites within the NHERF PDZ2 domain were identified by using NETPHOS 2.0 (20). The sequences surrounding Thr-156 and Ser-170 are consensus PKC sites (S/T-x-K/R). Ser-290 was also of interest since phosphorylation of this residue has been postulated to promote oligomerization of NHERF (21). WT and various NHERF PDZ1–2 domains containing individual alanine substitutions (T156A, S162A, S170A, S290A) were examined for their ability to be phosphorylated by PKC in vitro. Mutations T156A, S170A, and S290A did not affect the ability of PKC to phosphorylate NHERF (Fig. 3B). For reasons that are not clear, we observed a small (10–20%) but reproducible increase in the phosphorylation of T156A and S170A by PKC. In contrast, the Ser-162 to Ala substitution significantly reduced (to 17 ± 4% of WT) the amount of phosphorylation, suggesting that Ser-162 is the primary site of PKC phosphorylation.
To determine whether PKC can phosphorylate NHERF on Ser-162 in vivo, hemagglutinin-NHERF-transfected Chinese hamster ovar y cells were metabolically labeled with [32P]orthophosphate and then stimulated to activate PKC. To quantify the relative degree of NHERF phosphorylation, the intensity of NHERF radioactivity was normalized to the intensity of labeling with α-hemagglutinin antibody. NHERF is constitutively phosphorylated in vivo (Fig. 3C), but treatment of the cells with phorbol esters caused a small increase in its phosphorylation (128 ± 15% of control; P < 0.05). In contrast, no increase was observed in the S162A mutant (104 ± 9% of control). Because the high level of constitutive phosphorylation could mask phosphorylation by PKC, we repeated the experiments with a NHERF construct that minimized the constitutive phosphorylation. It was shown previously that mouse NHERF is constitutively phosphorylated on Ser-289 (22), which corresponds to Ser-290 in human NHERF. Constitutive phosphorylation of NHERF was greatly reduced in a S290A mutant (12 ± 4% of WT; Fig. 3C), consistent with the results from mouse. PMA stimulation increased the amount of phosphorylated S290A NHERF by 434 ± 27% (Fig. 3C). Mutation of Ser-162 to alanine in the S290A background (SS-AA) drastically reduced the amount of PMA-stimulated phosphorylation ≈70% (27 ± 2% of S290A). Taken together, these results indicate that the primary site of PKC phosphorylation in NHERF is Ser-162 within the second PDZ domain.
Phosphorylation of Ser-162 Is Critical for PKC-Mediated Inhibition of NHERF-Stimulated CFTR Channel Po. If phosphorylation of Ser-162 is responsible for the inhibition by PKC, then the phospho-deficient (S162A) mutant should eliminate the inhibitory effect of PKC on NHERF-stimulated CFTR channel activity. Addition of PDZ1–2 S162A to excised patches stimulated CFTR channel Po to an extent that was comparable to that elicited by the WT PDZ1–2 (Fig. 4 A and B). However, subsequent addition of PKC failed to inhibit the stimulatory effect (Fig. 4 A and B). These results suggest that PKC inhibits the NHERF-mediated stimulation of CFTR channel activity by specific phosphorylation of Ser-162 within the PDZ2 domain of NHERF.
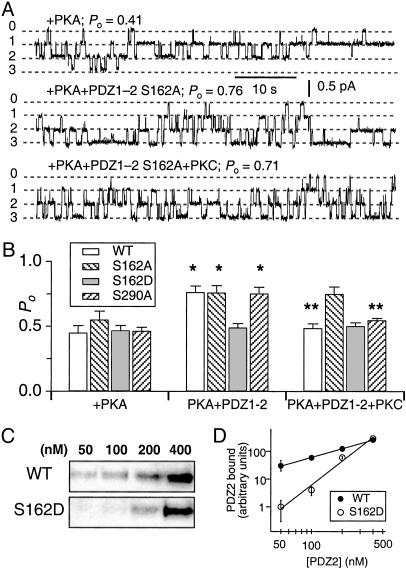
Fig. 4.
Phosphorylation of Ser-162 is critical for the inhibitory effect of PKC. (A) Representative current traces of CFTR channels in the presence of PKA and after addition of PDZ1–2–S162A and PKC. (B) Summary of CFTR Po in the presence of PKA and after addition of the indicated PDZ1–2 peptide (60 nM) and subsequent addition of PKC. * and ** indicate significant difference of Po after addition of PDZ1–2 peptide and after addition of PKC, respectively. (C) Binding of PDZ2 (WT and S162D) domain to CFTR C terminus at the indicated concentrations and (D) quantified by densitometry of gels (n = 3) with data normalized to binding observed in 50 nM (S162D).
The effects of specific amino acid phosphorylation can often be mimicked by mutation of that residue to an acidic one, which imparts an appropriate negative charge. We hypothesized that mutating Ser-162 to Asp (D) would cause the mutant peptide to mimic the behavior of the PKC-phosphorylated WT peptide. In agreement, the phospho-mimic (S162D) PDZ1–2 peptide failed to stimulate channel Po, and subsequent addition of PKC was without effect (Fig. 4B). These results suggest that Ser-162 is a critical residue for the stimulatory effect of NHERF on CFTR channel activity and that PKC-mediated phosphorylation abrogates this stimulating activity by imparting a negative charge on this residue.
It has been reported that NHERF can self-associate (23–25) and that the association is promoted by phosphorylation of Ser-290 (21). We therefore considered the possibility that oligomerization of NHERF might play a role in the observed regulation of CFTR channel activity. To address this issue, we tested the effects of PDZ1–2 S290A on CFTR channel activity. This mutant version of NHERF (S290A), which cannot be phosphorylated on Ser-290 but can be phosphorylated by PKC on Ser-162 (Fig. 3 B and C), stimulated CFTR Po to a level that was similar to that of WT NHERF. Furthermore, the stimulation could be abolished by PKC (Fig. 4B). These results suggest that NHERF oligomerization or its modulation is not relevant for its regulation of CFTR channel activity.
Replacement of Ser-162 with Aspartate Disrupts NHERF PDZ2 Interaction with CFTR. To understand the molecular consequences of Ser-162 phosphorylation, we examined the interaction between a PDZ2 (S162D) domain and CFTR, by performing pull-down assays using purified recombinant GST-fusion peptides. The WT PDZ2 interacted strongly with CFTR, whereas binding of PDZ2 (S162D) was impaired (Fig. 4C). More than 10-fold reduction in binding was observed at the concentrations (50–100 nM) that produced functional effects on CFTR gating (Fig. 4D). This result suggests that phosphorylation at Ser-162 interferes with the binding of PDZ2 to CFTR.
Discussion
PDZ domains are small protein interaction modules that have emerged as critical regulators of transport, localization, and assembly of supramolecular signaling complexes (26). Proteins that contain more than one PDZ domain can act as “hubs” in protein complexes by recruiting and organizing different proteins, facilitating their interactions to promote new and efficient functionality. Because the composition of such complexes will be determined by the concentrations of all of the interacting proteins and their binding affinities, the functionality of proteins within them will be regulated by mechanisms that influence these parameters.
The binding of the bivalent PDZ domain-containing protein NHERF to CFTR has profound influences on its channel gating that depend on the stoichiometry of the interaction. Thus, simultaneous binding of both PDZ domains of a single NHERF molecule stimulates gating, whereas binding of either individual domain does not. Because NHERF PDZ domains have been demonstrated to interact with a number of different proteins (22, 27–31), the effects of NHERF binding on CFTR channel activity will depend on the ability of CFTR to compete with other proteins for these interactions. We have considered that post-translational modification of either CFTR or NHERF could represent a mechanism to regulate such interactions. Here, we have investigated the role of PKC-mediated phosphorylation on the functional consequences of the interaction of NHERF and CFTR. PKC regulates CFTR, although the mechanisms have remained obscure. Of note, NHERF has been shown to interact with receptor for activated C-kinase (RACK1), a PKC-interacting protein (32). A protein complex of activated PKC and RACK1 bound to NHERF suggests a regulatory complex in which NHERF might act as a scaffold to facilitate phosphorylation of substrates, including CFTR and itself.
Regulation of apical membrane CFTR channels controls, in part, the rate of transepithelial Cl- secretion. PKC activity influences the rate of epithelial Cl- secretion. For example, phorbol esters stimulate a submaximal level of Cl- secretion and inhibit cAMP-dependent secretion (33, 34). The responses to PKC activation may be complex, as it can regulate CFTR by several mechanisms (2, 35). Our results show that PKC modulates the physical interaction between NHERF and CFTR, with functional consequences. The PDZ2 domain of NHERF is a substrate for PKC phosphorylation, and phosphorylated PDZ2 is impaired in its ability to bind CFTR. We therefore propose a regulatory role for PKC in which it negatively regulates CFTR channel activity by disrupting the stimulatory interactions between CFTR and NHERF.
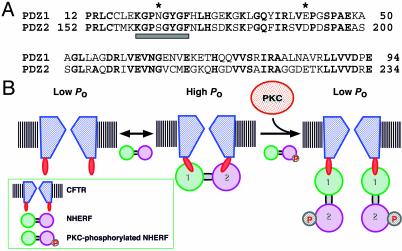
How can phosphorylation of Ser-162 disrupt the interaction between PDZ2 domain of NHERF and CFTR? The crystal structure of the PDZ1 domain of NHERF in complex with CFTR C terminus provides a clue (36). By mapping the PDZ2 sequence onto this structure, the role of specific residues within PDZ2 in its interaction with CFTR can be deduced. Ser-162 corresponds to Asn-22 in PDZ1, which is located in the conserved carboxylate-binding loop of the PDZ domain (Fig. 5A). In the PDZ1 structure, Asn-22 and Glu-43 interact with the guanido group of arginine (-1) in the C terminus (DTRL) of CFTR, contributing to the high affinity of this interaction. Indeed, mutagenesis of arginine (-1) to leucine or glutamic acid decreased the affinity of PDZ1-ligand interaction by 2- to 4-fold (5). Superposition of the PDZ1 domain bound to β2AR, platelet-derived growth factor receptor, and CFTR peptides reveals that the side chains of Asn-22 and Glu-43 exhibit large conformational changes upon binding (37). It is likely that Ser-162, as the homologous residue in PDZ2, participates in a similar role in mediating the interaction of PDZ2 with the tail of CFTR. Accordingly, phosphorylation of Ser-162 would be expected to disrupt interactions between the Arg (-1) guanido group and residues Ser-162 and Asp-183 in PDZ2 and account for the observed decrease in binding affinity of NHERF PDZ2 for CFTR.
Fig. 5.
(A) Sequence alignment of hNHERF PDZ1 and PDZ2. The carboxylate-binding loop (boxed below) and the amino acids (*) proposed to interact with Arg (-1) of the CFTR C terminus are denoted. Identical residues are in bold. (B) An equilibrium model illustrating the regulation of CFTR. A CFTR channel with two PDZ-binding sites and the two PDZ domains of NHERF are represented. See text for details.
We previously proposed a simple equilibrium model to account for the biphasic regulation of CFTR channel Po by NHERF (10). In this model (Fig. 5B), a single functional CFTR channel contains two PDZ-binding sites. The PDZ1 domain of NHERF initially binds to one of the sites, whereas the second PDZ-binding site on the channel could be bound either by the PDZ2 domain of the bound NHERF or the PDZ1 domain of a second molecule of NHERF. The activity of the channel depends on whether it is bound and cross-linked by single molecule of NHERF (1:1 CFTR channel/NHERF stoichiometry) or is bound by two NHERF molecules (1:2). At optimal NHERF concentrations, favoring the 1:1 stoichiometry, the channel is maximally activated. Because PKC phosphorylation of the PDZ2 domain reduces its binding affinity, PKC shifts the equilibrium from a 1:1 stoichiometry to a 1:2 stoichiometry, in which a single CFTR channel is bound to two NHERF molecules. In this mode, the CFTR channel is not cross-linked and therefore has an unstimulated Po. Thus, this phosphorylation regulation is mediated by a change in the stoichiometry of the PDZ-domain protein interaction with its ligand, which is distinct from previously described PDZ-binding site regulation, in which phosphorylation disrupts the interactions between the two binding partners. In agreement, stimulation of PKC activity did not significantly alter the amount of NHERF associated with CFTR in vivo (data not shown).
Our data provide evidence for a mechanism in which phosphorylation of a PDZ domain alters its interaction with a target protein. In the presence of multiple binding targets that differ only in their penultimate (-1) amino acid, the phosphorylation status of a PDZ domain could possibly provide a mechanism for selecting specific sets of binding partners. A database search for PDZ domains with serine or threonine in the carboxylate binding loop revealed several proteins that belong to the Discs-large (Dlg) superfamily, including SAP97, Chapsyn-110, SAP102, and PSD95. Of note, the Ser/Thr in the carboxylate-binding loops of these domains are contained within consensus Ca2+/calmodulin-dependent kinase II (CaMKII) phosphorylation motif (R/KXXS/T). Both PSD95 and SAP90 are CaMKII substrates in the postsynaptic density (38). Because CaMKII has been implicated in synaptic signaling and plasticity, it is worth considering the possibility that CaMKII mediates some of its effects by phosphorylation of PSD95 PDZ domains.
NHERF family proteins bind via their PDZ2 domains to a number of membrane-associated proteins, including the PTH 1 receptor (31) and NHE3 (27), as well as soluble proteins including PLC-β3 (29) and Yes-associated protein 65 (28). Therefore, we speculate that phosphorylation of NHERF PDZ2 domain will likely modify NHERF interactions with these proteins, as shown here for CFTR, and thus participate in regulating the activities and/or localization of these proteins as well. Thus, it will be of interest to analyze the role of PKC and Ser-162 phosphorylation in protein complexes containing NHERF. In summary, we have shown that CFTR channel activity that is modulated by PDZ domain-mediated interactions with NHERF family proteins depends on the stoichiometry of the interactions and that this stoichiometry is a target for regulation by PKC-mediated phosphorylation. The phosphorylation regulation of PDZ domain interactions identified here may be important in physiological regulation of signal transduction complexes.
Acknowledgments
This work was supported by Cystic Fibrosis Foundation Grants RAGHUR02I0 (to V.R.) and Comp III RDP and a grant from the National Institutes of Health (to J.K.F.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CFTR, cystic fibrosis transmembrane conductance regulator; NHERF, Na+/H+ exchanger regulatory factor; PMA, phorbol 12-myristate 13-acetate; CaMKII, Ca2+/calmodulin-dependent kinase II; PDZ, PSD-95/Dlg/ZO-1; PKA, protein kinase A; Po, open probability.
References
- 1.Sheppard, D. N. & Welsh, M. J. (1999) Physiol. Rev. 79, S23-S45. [DOI] [PubMed] [Google Scholar]
- 2.Jia, Y., Mathews, C. J. & Hanrahan, J. W. (1997) J. Biol. Chem. 272, 4978-4984. [DOI] [PubMed] [Google Scholar]
- 3.Wang, S., Raab, R. W., Schatz, P. J., Guggino, W. B. & Li, M. (1998) FEBS Lett. 427, 103-108. [DOI] [PubMed] [Google Scholar]
- 4.Short, D. B., Trotter, K. W., Reczek, D., Kreda, S. M., Bretscher, A., Boucher, R. C., Stutts, M. J. & Milgram, S. L. (1998) J. Biol. Chem. 273, 19797-19801. [DOI] [PubMed] [Google Scholar]
- 5.Hall, R. A., Ostedgaard, L. S., Premont, R. T., Blitzer, J. T., Rahman, N., Welsh, M. J. & Lefkowitz, R. J. (1998) Proc. Natl. Acad. Sci. USA 95, 8496-8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang, S., Yue, H., Derin, R. B., Guggino, W. B. & Li, M. (2000) Cell 103, 169-179. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, J., Moyer, B. D., Milewski, M., Loffing, J., Ikeda, M., Mickle, J. E., Cutting, G. R., Li, M., Stanton, B. A. & Guggino, W. B. (2002) J. Biol. Chem. 277, 3520-3529. [DOI] [PubMed] [Google Scholar]
- 8.Moyer, B. D., Duhaime, M., Shaw, C., Denton, J., Reynolds, D., Karlson, K. H., Pfeiffer, J., Wang, S., Mickle, J. E., Milewski, M., et al. (2000) J. Biol. Chem. 275, 27069-27074. [DOI] [PubMed] [Google Scholar]
- 9.Swiatecka-Urban, A., Duhaime, M., Coutermarsh, B., Karlson, K. H., Collawn, J., Milewski, M., Cutting, G. R., Guggino, W. B., Langford, G. & Stanton, B. A. (2002) J. Biol. Chem. 277, 40099-40105. [DOI] [PubMed] [Google Scholar]
- 10.Raghuram, V., Mak, D. D. & Foskett, J. K. (2001) Proc. Natl. Acad. Sci. USA 98, 1300-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung, H. J., Xia, J., Scannevin, R. H., Zhang, X. & Huganir, R. L. (2000) J. Neurosci. 20, 7258-7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen, N. A., Brenman, J. E., Snyder, S. H. & Bredt, D. S. (1996) Neuron 17, 759-767. [DOI] [PubMed] [Google Scholar]
- 13.Cao, T. T., Deacon, H. W., Reczek, D., Bretscher, A. & von Zastrow, M. (1999) Nature 401, 286-290. [DOI] [PubMed] [Google Scholar]
- 14.Kim, C. H., Chung, H. J., Lee, H. K. & Huganir, R. L. (2001) Proc. Natl. Acad. Sci. USA 98, 11725-11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi, Y., Shi, S. H., Esteban, J. A., Piccini, A., Poncer, J. C. & Malinow, R. (2000) Science 287, 2262-2267. [DOI] [PubMed] [Google Scholar]
- 16.Lansdell, K. A., Kidd, J. F., Delaney, S. J., Wainwright, B. J. & Sheppard, D. N. (1998) J. Physiol. (London) 512, 751-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dirr, H., Reinemer, P. & Huber, R. (1994) Eur. J. Biochem. 230, 614-620. [DOI] [PubMed] [Google Scholar]
- 18.Armstrong, R. N. (1997) Chem. Res. Toxicol. 10, 2-18. [DOI] [PubMed] [Google Scholar]
- 19.Tudyka, T. & Skerra, A. (1997) Protein Sci. 6, 2180-2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blom, N., Gammeltoft, S. & Brunak, S. (1999) J. Mol. Biol. 294, 1351-1362. [DOI] [PubMed] [Google Scholar]
- 21.Lau, A. G. & Hall, R. A. (2001) Biochemistry 40, 8572-8580. [DOI] [PubMed] [Google Scholar]
- 22.Hall, R. A., Spurney, R. F., Premont, R. T., Rahman, N., Blitzer, J. T., Pitcher, J. A. & Lefkowitz, R. J. (1999) J. Biol. Chem. 274, 24328-24334. [DOI] [PubMed] [Google Scholar]
- 23.Fouassier, L., Yun, C. C., Fitz, J. G. & Doctor, R. B. (2000) J. Biol. Chem. 275, 25039-25045. [DOI] [PubMed] [Google Scholar]
- 24.Maudsley, S., Zamah, A. M., Rahman, N., Blitzer, J. T., Luttrell, L. M., Lefkowitz, R. J. & Hall, R. A. (2000) Mol. Cell. Biol. 20, 8352-8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shenolikar, S., Minkoff, C. M., Steplock, D. A., Evangelista, C., Liu, M. & Weinman, E. J. (2001) FEBS Lett. 489, 233-236. [DOI] [PubMed] [Google Scholar]
- 26.Sheng, M. & Sala, C. (2001) Annu. Rev. Neurosci. 24, 1-29. [DOI] [PubMed] [Google Scholar]
- 27.Yun, C. H., Oh, S., Zizak, M., Steplock, D., Tsao, S., Tse, C. M., Weinman, E. J. & Donowitz, M. (1997) Proc. Natl. Acad. Sci. USA 94, 3010-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohler, P. J., Kreda, S. M., Boucher, R. C., Sudol, M., Stutts, M. J. & Milgram, S. L. (1999) J. Cell Biol. 147, 879-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang, J. I., Heo, K., Shin, K. J., Kim, E., Yun, C., Ryu, S. H., Shin, H. S. & Suh, P. G. (2000) J. Biol. Chem. 275, 16632-16637. [DOI] [PubMed] [Google Scholar]
- 30.Gisler, S. M., Stagljar, I., Traebert, M., Bacic, D., Biber, J. & Murer, H. (2001) J. Biol. Chem. 276, 9206-9213. [DOI] [PubMed] [Google Scholar]
- 31.Mahon, M. J., Donowitz, M., Yun, C. C. & Segre, G. V. (2002) Nature 417, 858-861. [DOI] [PubMed] [Google Scholar]
- 32.Liedtke, C. M., Yun, C. H., Kyle, N. & Wang, D. (2002) J. Biol. Chem. 277, 22925-22933. [DOI] [PubMed] [Google Scholar]
- 33.Welsh, M. J. (1987) Am. J. Physiol. 253, C828-C834. [DOI] [PubMed] [Google Scholar]
- 34.Barthelson, R. A., Jacoby, D. B. & Widdicombe, J. H. (1987) Am. J. Physiol. 253, C802-C808. [DOI] [PubMed] [Google Scholar]
- 35.Breuer, W., Glickstein, H., Kartner, N., Riordan, J. R., Ausiello, D. A. & Cabantchik, I. Z. (1993) J. Biol. Chem. 268, 13935-13939. [PubMed] [Google Scholar]
- 36.Karthikeyan, S., Leung, T. & Ladias, J. A. (2001) J. Biol. Chem. 276, 19683-19686. [DOI] [PubMed] [Google Scholar]
- 37.Karthikeyan, S., Leung, T. & Ladias, J. A. (2002) J. Biol. Chem. 277, 18973-18978. [DOI] [PubMed] [Google Scholar]
- 38.Yoshimura, Y., Aoi, C. & Yamauchi, T. (2000) Brain Res. 81, 118-128. [DOI] [PubMed] [Google Scholar]