Abstract
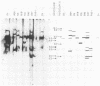
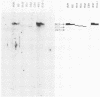
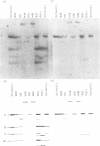
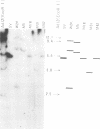
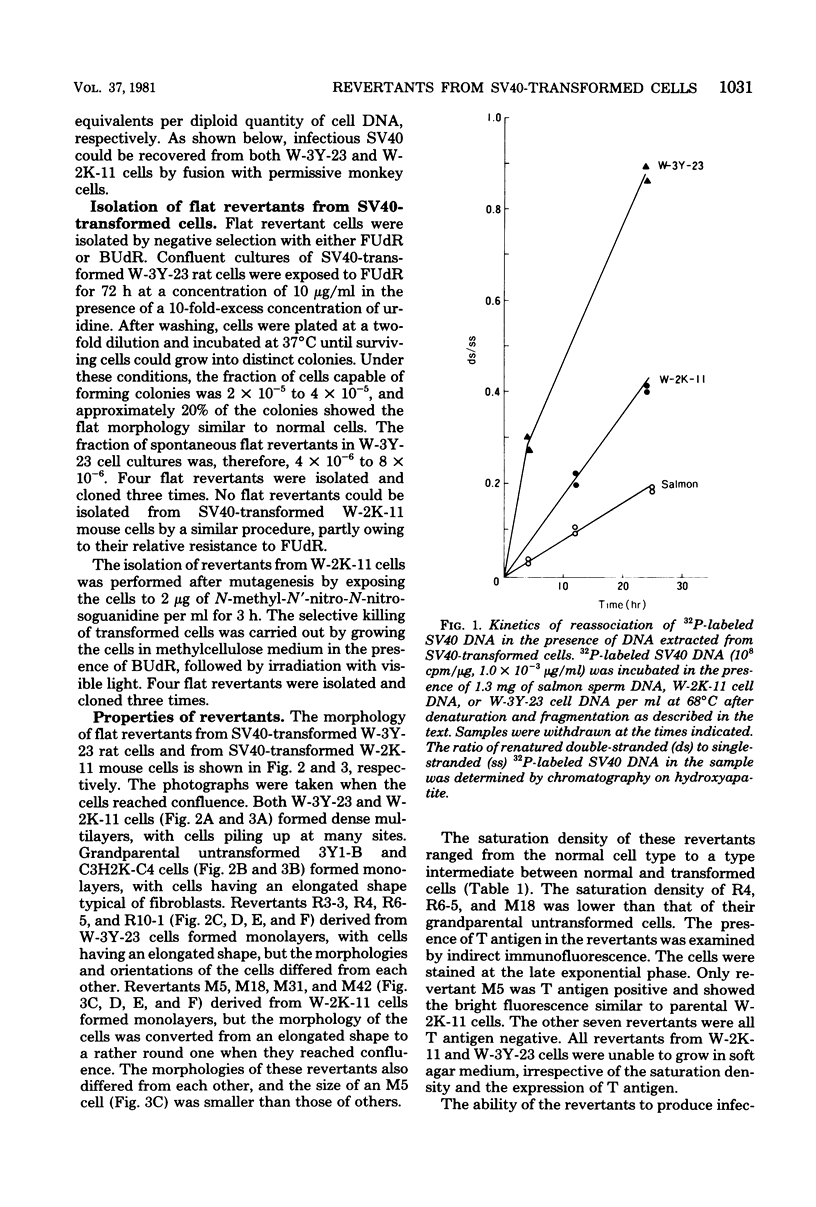
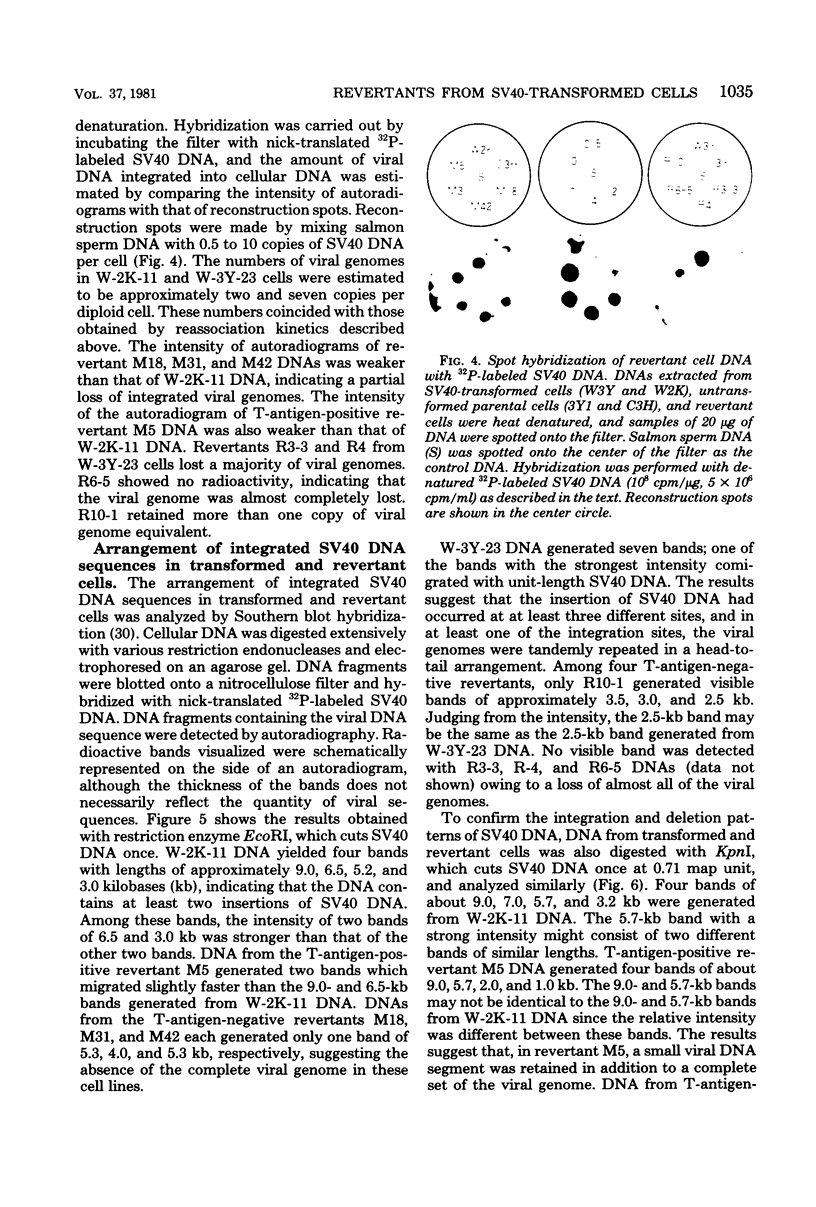
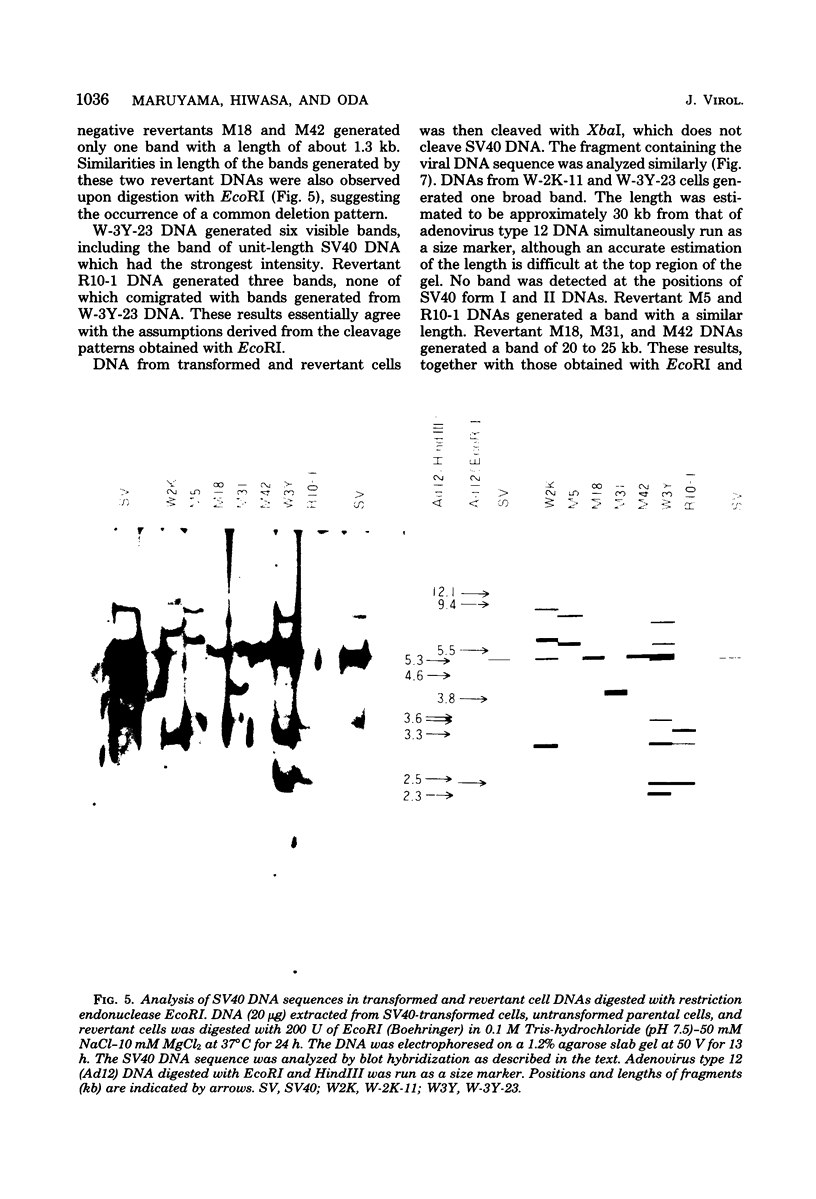
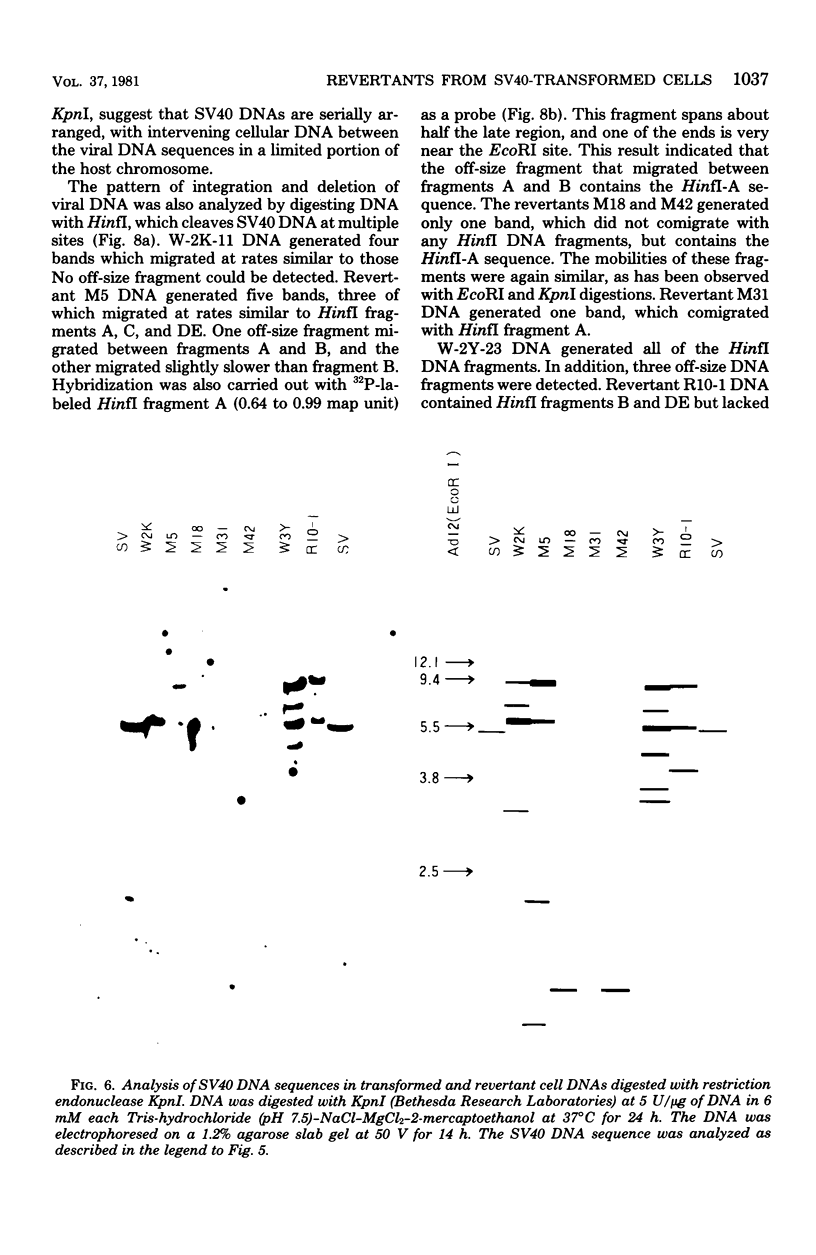
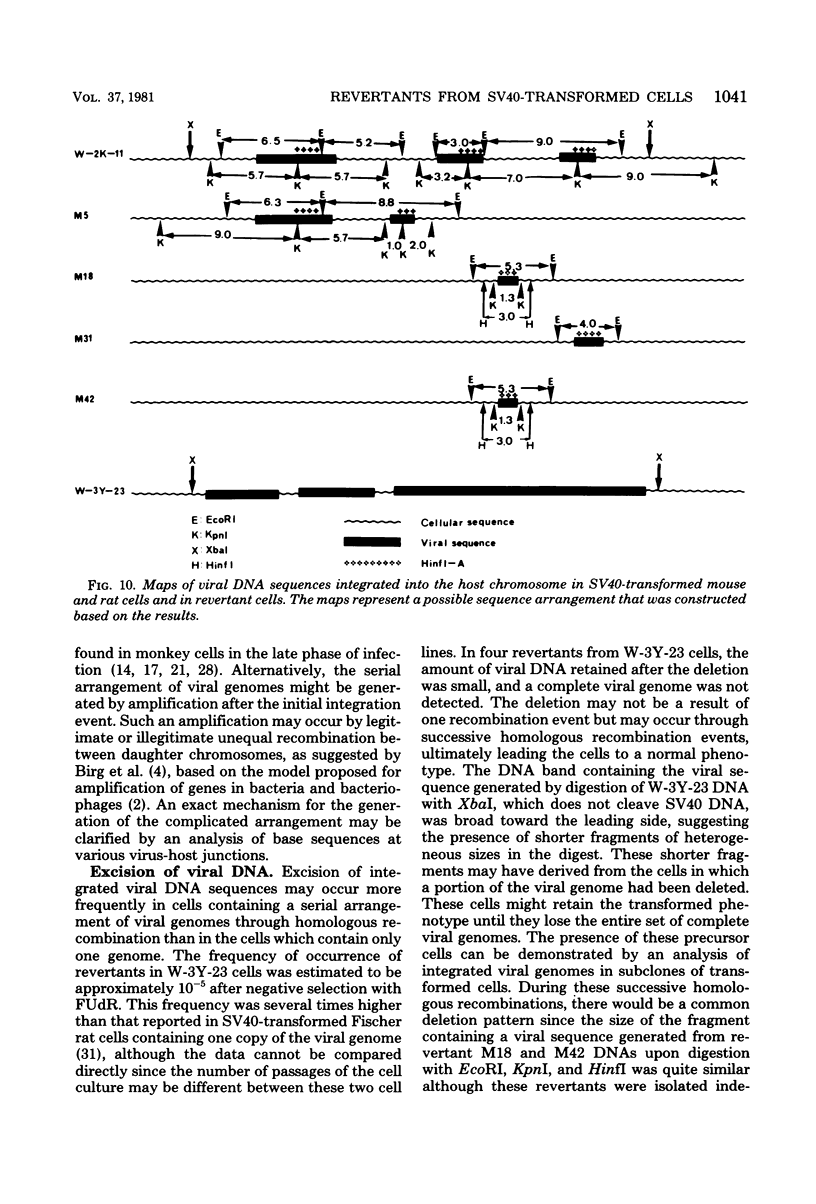
Eight clones of flat revertants were isolated by negative selection from simian virus 40 (SV40)-transformed mouse and rat cell lines in which two and six viral genome equivalents per cell were integrated, respectively. These revertants showed either a normal cell phenotype or a phenotype intermediate between normal and transformed cells as to cellular morphology and saturation density and were unable to grow in soft agar medium. One revertant derived from SV40-transformed mouse cells was T antigen positive, whereas the other seven revertants were T antigen negative. SV40 could be rescued only from the T-antigen-positive revertant by fusion with permissive monkey cells. The susceptibility of the revertants to retransformation by wild-type SV40 was variable among these revertants. T-antigen-negative revertants from SV40-transformed mouse cells were retransformed at a frequency of 3 to 10 times higher than their grandparental untransformed cells. In contrast, T-antigen-negative revertants from SV40-transformed rat cells could not be retransformed. The arrangement of viral genomes was analyzed by digestion of cellular DNA with restriction enzymes of different specificity, followed by detection of DNA fragments containing a viral sequence and rat cells were serially arranged within the length of about 30 kilobases, with at least two intervening cellular sequences. A head-to-tail tandem array of unit length viral genomes was present in at least one insertion site in the transformed rat cells. All of the revertants had undergone a deletion(s), and only a part of the viral genome was retained in T-antigen-negative revertants. A relatively high frequency of reversion in the transformed rat cells suggests that reversion occurs by homologous recombination between the integrated viral genomes.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alwine J. C., Reed S. I., Stark G. R. Characterization of the autoregulation of simian virus 40 gene A. J Virol. 1977 Oct;24(1):22–27. doi: 10.1128/jvi.24.1.22-27.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R. P., Roth J. R. Tandem genetic duplications in phage and bacteria. Annu Rev Microbiol. 1977;31:473–505. doi: 10.1146/annurev.mi.31.100177.002353. [DOI] [PubMed] [Google Scholar]
- Basilico C., Gattoni S., Zouzias D., Valle G. D. Loss of integrated viral DNA sequences in polyomatransformed cells is associated with an active viral A function. Cell. 1979 Jul;17(3):645–659. doi: 10.1016/0092-8674(79)90272-1. [DOI] [PubMed] [Google Scholar]
- Birg F., Dulbecco R., Fried M., Kamen R. State and organization of polyoma virus DNA sequences in transformed rat cell lines. J Virol. 1979 Feb;29(2):633–648. doi: 10.1128/jvi.29.2.633-648.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan M., Topp W., Sambrook J. The arrangement of simian virus 40 sequences in the DNA of transformed cells. Cell. 1976 Oct;9(2):269–287. doi: 10.1016/0092-8674(76)90118-5. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Brugge J. S., Butel J. S. Role of simian virus 40 gene A function in maintenance of transformation. J Virol. 1975 Mar;15(3):619–635. doi: 10.1128/jvi.15.3.619-635.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo M. S., Cameron I. R., Rogers M. E. Tandem integration of complete and defective SV40 genomes in mouse-human somatic cell hybrids. Cell. 1978 Dec;15(4):1411–1426. doi: 10.1016/0092-8674(78)90065-x. [DOI] [PubMed] [Google Scholar]
- Culp L. A., Black P. H. Contact-inhibited revertant cell lines isolated from simian virus 40-transformed cells. 3. Concanavalin A-selected revertant cells. J Virol. 1972 Apr;9(4):611–620. doi: 10.1128/jvi.9.4.611-620.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culp L. A., Grimes W. J., Black P. H. Contact-inhibited revertant cell lines isolated from SV40-transformed cells. I. Biologic, virologic, and chemical properties. J Cell Biol. 1971 Sep;50(3):682–690. doi: 10.1083/jcb.50.3.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga K., Sawada Y., Uemizu Y. A rapid screening for the specific DNA sequence: analysis of transforming DNA segments in adenovirus-transformed cells. Gan. 1979 Apr;70(2):239–243. [PubMed] [Google Scholar]
- Gelb L. D., Kohne D. E., Martin M. A. Quantitation of Simian virus 40 sequences in African green monkey, mouse and virus-transformed cell genomes. J Mol Biol. 1971 Apr 14;57(1):129–145. doi: 10.1016/0022-2836(71)90123-9. [DOI] [PubMed] [Google Scholar]
- Goff S. P., Berg P. Structure and formation of circular dimers of simian virus 40 DNA. J Virol. 1977 Oct;24(1):295–302. doi: 10.1128/jvi.24.1.295-302.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney E. G., Gurney T., Jr Density dependent inhibition of both growth and T-antigen expression in revertants isolated from simian virus 40-transformed mouse SVT2 cells. J Virol. 1979 Nov;32(2):667–671. doi: 10.1128/jvi.32.2.667-671.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitotsumachi S., Rabinowitz Z., Sachs L. Ciromosomal control of reversion in transformed cells. Nature. 1971 Jun 25;231(5304):511–514. doi: 10.1038/231511a0. [DOI] [PubMed] [Google Scholar]
- Jaenisch R., Levine A. J. DNA replication of SV40-infected cells. VII. Formation of SV40 catenated and circular dimers. J Mol Biol. 1973 Jan 10;73(2):199–212. doi: 10.1016/0022-2836(73)90323-9. [DOI] [PubMed] [Google Scholar]
- Kelly T. J., Jr, Lewis A. M., Jr, Levine A. S., Siegel S. Structure of two adenovirus-simian virus 40 hybrids which contain the entire SV40 genome. J Mol Biol. 1974 Oct 15;89(1):113–126. doi: 10.1016/0022-2836(74)90165-x. [DOI] [PubMed] [Google Scholar]
- Khoury G., May E. Regulation of early and late simian virus 40 transcription: overproduction of early viral RNA in the absence of a functional T-antigen. J Virol. 1977 Jul;23(1):167–176. doi: 10.1128/jvi.23.1.167-176.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura G., Itagaki A., Summers J. Rat cell line 3y1 and its virogenic polyoma- and sv40- transformed derivatives. Int J Cancer. 1975 Apr 15;15(4):694–706. doi: 10.1002/ijc.2910150419. [DOI] [PubMed] [Google Scholar]
- Martin M. A., Howley P. M., Byrne J. C., Garon C. F. Characterization of supercoiled oligomeric SV40 DNA molecules in productively infected cells. Virology. 1976 May;71(1):28–40. doi: 10.1016/0042-6822(76)90091-x. [DOI] [PubMed] [Google Scholar]
- Martin R. G., Chou J. Y. Simian virus 40 functions required for the establishment and maintenance of malignant transformation. J Virol. 1975 Mar;15(3):599–612. doi: 10.1128/jvi.15.3.599-612.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn M., Weber K. Simian virus 40 gene A function and maintenance of transformation. J Virol. 1975 Mar;15(3):636–644. doi: 10.1128/jvi.15.3.636-644.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozanne B. Variants of simian virus 40-transformed 3T3 cells that are resistant to concanavalin A. J Virol. 1973 Jul;12(1):79–89. doi: 10.1128/jvi.12.1.79-89.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack R. E., Green H., Todaro G. J. Growth control in cultured cells: selection of sublines with increased sensitivity to contact inhibition and decreased tumor-producing ability. Proc Natl Acad Sci U S A. 1968 May;60(1):126–133. doi: 10.1073/pnas.60.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack R., Wolman S., Vogel A. Reversion of virus-transformed cell lines: hyperploidy accompanies retention of viral genes. Nature. 1970 Dec 5;228(5275):938–passim. doi: 10.1038/228938a0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Berg P. Does simian virus 40 DNA integrate into cellular DNA during productive infection? J Virol. 1978 Nov;28(2):475–489. doi: 10.1128/jvi.28.2.475-489.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rigby P. The transforming genes of SV40 and polyoma. Nature. 1979 Dec 20;282(5741):781–784. doi: 10.1038/282781a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steinberg B., Pollack R., Topp W., Botchan M. Isolation and characterization of T antigen-negative revertants from a line of transformed rat cells containing one copy of the SV40 genome. Cell. 1978 Jan;13(1):19–32. doi: 10.1016/0092-8674(78)90134-4. [DOI] [PubMed] [Google Scholar]
- Stoker M., O'Neill C., Berryman S., Waxman V. Anchorage and growth regulation in normal and virus-transformed cells. Int J Cancer. 1968 Sep 15;3(5):683–693. doi: 10.1002/ijc.2910030517. [DOI] [PubMed] [Google Scholar]
- Tegtmeyer P. Function of simian virus 40 gene A in transforming infection. J Virol. 1975 Mar;15(3):613–618. doi: 10.1128/jvi.15.3.613-618.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer P. Simian virus 40 deoxyribonucleic acid synthesis: the viral replicon. J Virol. 1972 Oct;10(4):591–598. doi: 10.1128/jvi.10.4.591-598.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A., Pollack R. Isolation and characterization of revertant cell lines. IV. Direct selection of serum-revertant sublines of SV40-transformed 3T3 mouse cells. J Cell Physiol. 1973 Oct;82(2):189–198. doi: 10.1002/jcp.1040820207. [DOI] [PubMed] [Google Scholar]
- Vogel A., Pollack R. Isolation and characterization of revertant cell lines. VI. Susceptibility of revertants to retransformation by simian virus 40 and murine sarcoma virus. J Virol. 1974 Dec;14(6):1404–1410. doi: 10.1128/jvi.14.6.1404-1410.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A., Risser R., Pollack R. Isolation and characterization of revertant cell lines. 3. Isolation of density-revertants of SV40-transformed 3T3 cells using colchicine. J Cell Physiol. 1973 Oct;82(2):181–188. doi: 10.1002/jcp.1040820206. [DOI] [PubMed] [Google Scholar]
- Wyke J. A method of isolating cells incapable of multiplication in suspension culture. Exp Cell Res. 1971 May;66(1):203–208. doi: 10.1016/s0014-4827(71)80030-7. [DOI] [PubMed] [Google Scholar]
- Yoshikura H., Hirokawa Y. Induction of cell replication. Exp Cell Res. 1968 Oct;52(2):439–444. doi: 10.1016/0014-4827(68)90485-0. [DOI] [PubMed] [Google Scholar]