Abstract
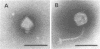
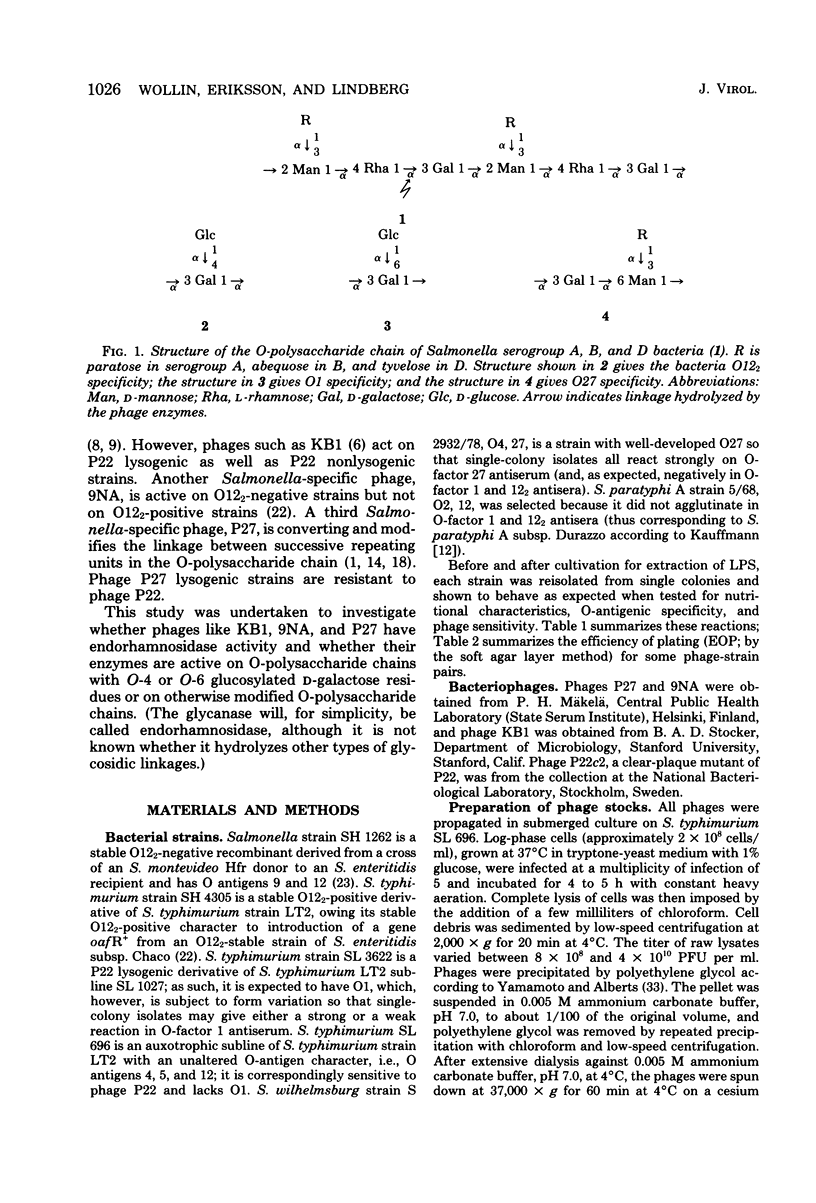
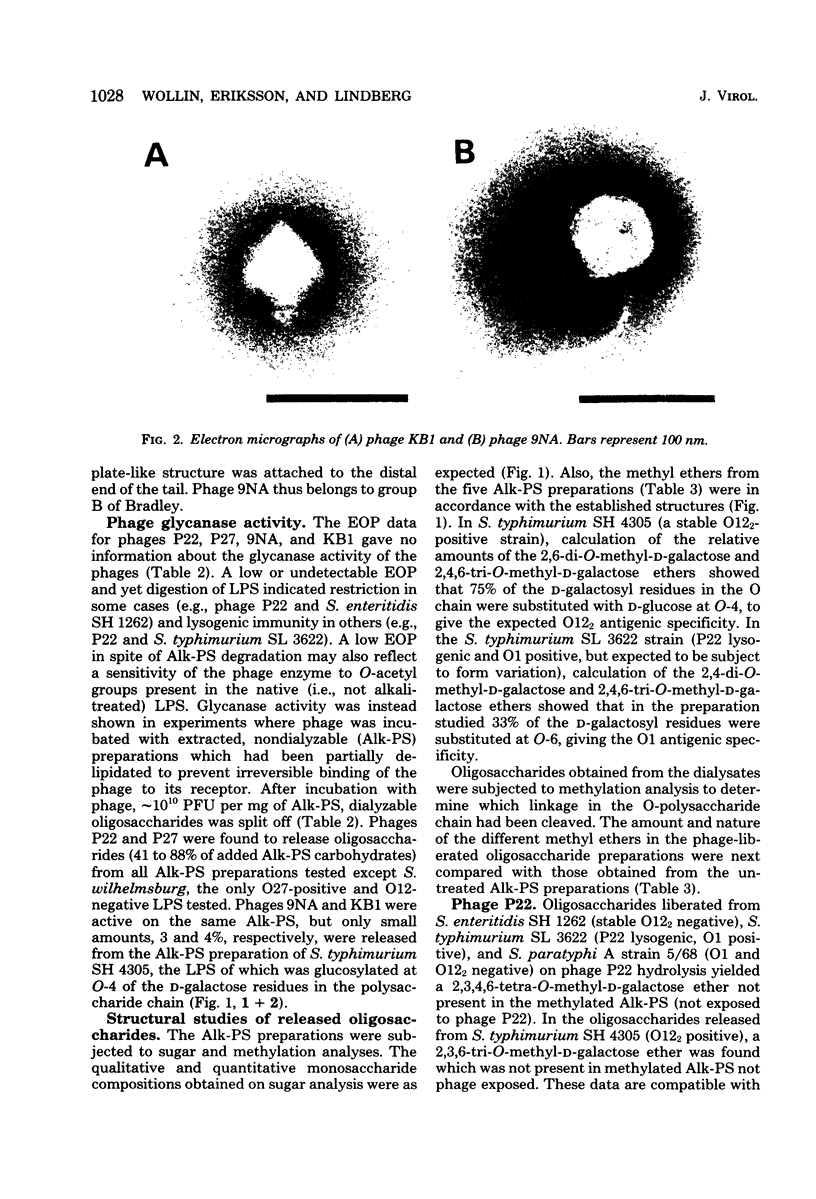
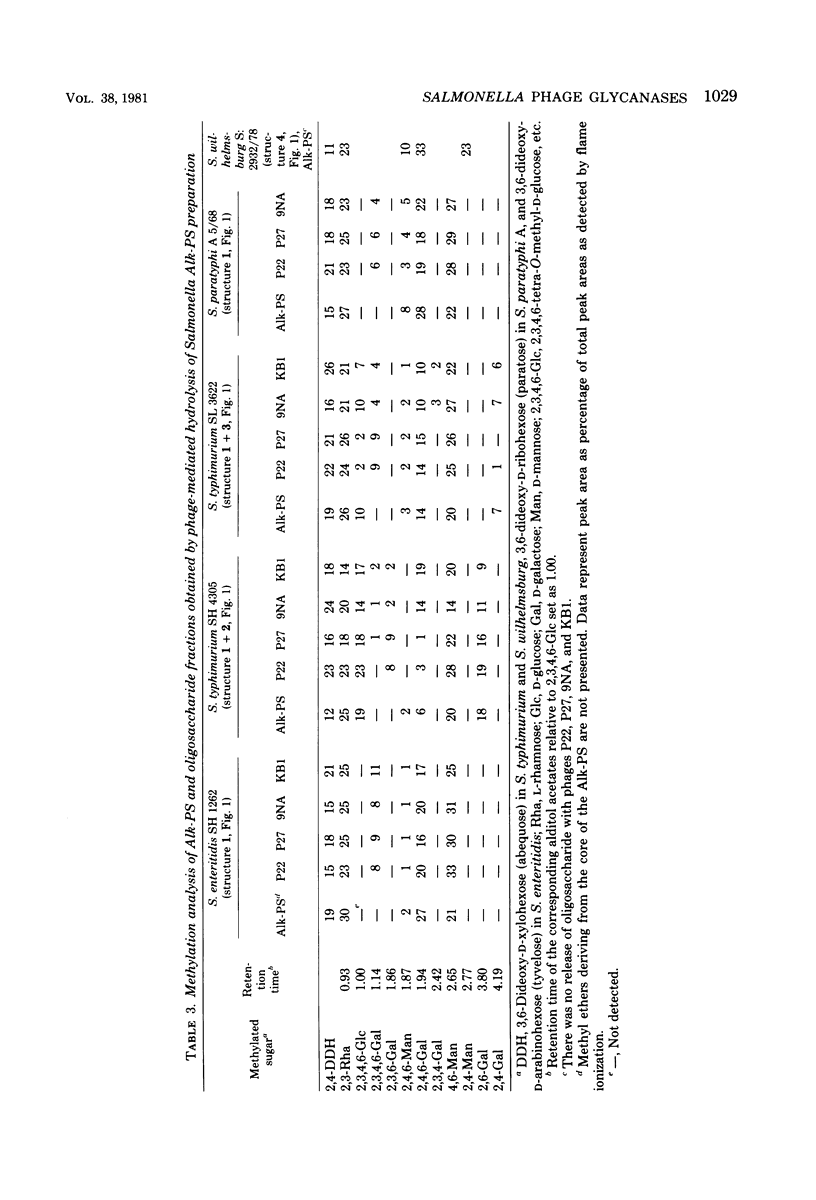
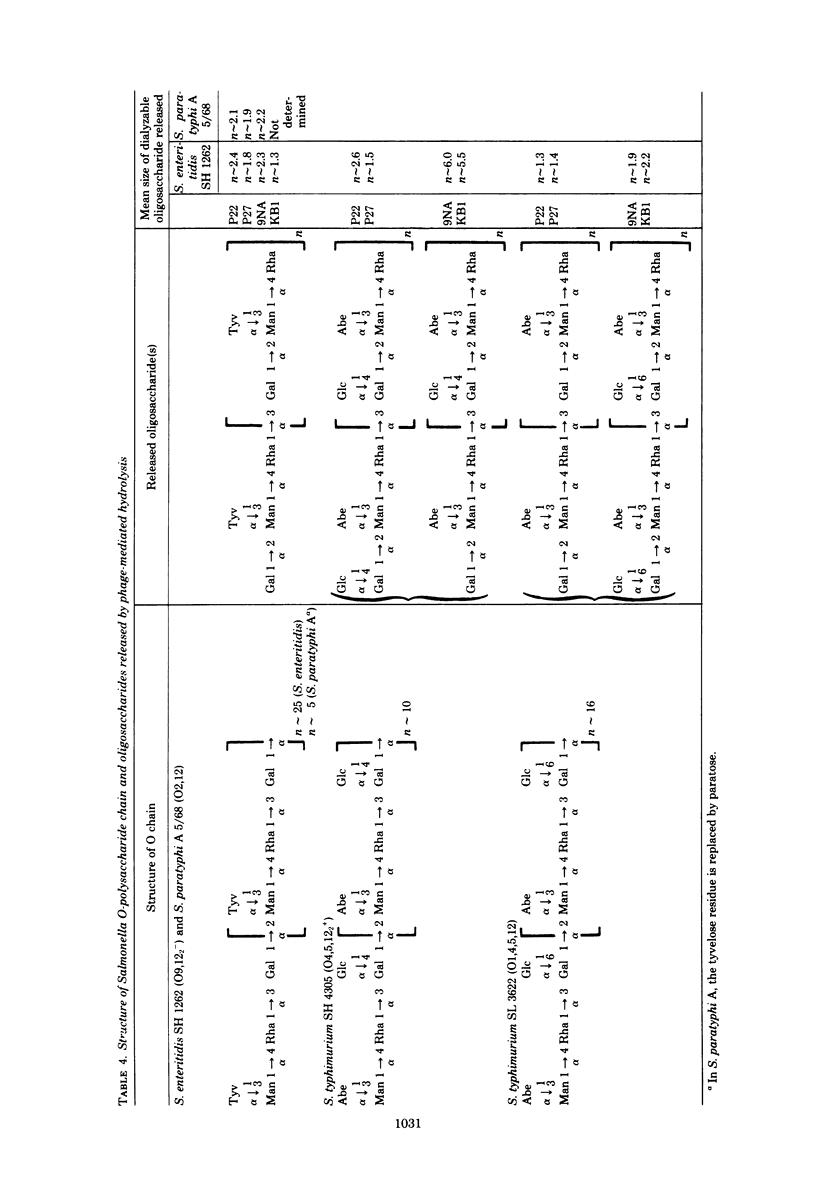
Four bacteriophages, P22, P27, 9NA, and KB1, active on smooth Salmonella strains belonging to serogroups A, B, and D1 were investigated for endoglycosidase activity and specificity in enzyme hydrolysis assays. Purified phage was incubated with phenol-water-extracted lipopolysaccharide preparations which had been partially delipidated. Dialyzable oligosaccharides, released by phage glycosidase activity, were analyzed by sugar and methylation analyses. Phages P27, 9NA, and KB1, as well as P22 assayed earlier (U. Eriksson et al., J. Gen. Virol. 43: 503-511, 1979; S. Iwashita and S. Kanegasaki, Biochem. Biophys. Res. Commun. 55:403-409, 1973), were all found to have phage-associated endorhamnosidase activity hydrolyzing the O-polysaccharide chain common to bacteria of serogroups A, B, and D1 [Formula: see text] between the l-rhamnose and d-galactose residues. The nature of the R monosaccharide, abequose, tyvelose, or paratose, had no effect on the activity or specificity of the endorhamnosidase, whereas a change of the d-galactose → d-mannose linkage from α1,2 to α1,6 made the O-polysaccharide chain resistant to the endorhamnosidases. Modification of the O chain by glucosylation of the d-galactose residue at O-4 or O-6 revealed two glycosidase specificities: the phage P22 and P27 enzymes hydrolyzed O chains glucosylated at O-4 but not O-6, whereas the phage 9NA and KB1 enzymes hydrolyzed chains glucosylated at O-6 but not O-4. Phage KB1, like P22 and P27, had a short, noncontractile tail containing a base plate with tail spikes (morphologically Bradley group C), whereas 9NA had a long, flexible tail ending with a base plate-like appendage (Bradley group B), which suggests that the endorhamnosidase activity can be associated with different tail structures.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdian G., Lüderitz O., Staub A. M. Immunochemical studies on Salmonella. XI. Chemical modification correlated with conversion of group B Salmonella by bacteriophage 27. Ann N Y Acad Sci. 1966 Jun 30;133(2):405–424. doi: 10.1111/j.1749-6632.1966.tb52380.x. [DOI] [PubMed] [Google Scholar]
- Bayer M. E., Takeda K., Uetake H. Effects of receptor destruction by Salmonella bacteriophages epsilon 15 and c341. Virology. 1980 Sep;105(2):328–337. doi: 10.1016/0042-6822(80)90034-3. [DOI] [PubMed] [Google Scholar]
- Bessler W., Fehmel F., Freund-Mölbert E., Knüfermann H., Stirm S. Escherichia coli capsule bacteriophages. IV. Free capsule depolymerase 29. J Virol. 1975 Apr;15(4):976–984. doi: 10.1128/jvi.15.4.976-984.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boro H., Brenchley J. E. A new generalized transducing phage for Salmonella typhimurium LT2. Virology. 1971 Sep;45(3):835–836. doi: 10.1016/0042-6822(71)90208-x. [DOI] [PubMed] [Google Scholar]
- Bradley D. E. Ultrastructure of bacteriophage and bacteriocins. Bacteriol Rev. 1967 Dec;31(4):230–314. doi: 10.1128/br.31.4.230-314.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson U., Lindberg A. A. Adsorption of phage P22 to Salmonella typhimurium. J Gen Virol. 1977 Feb;34(2):207–221. doi: 10.1099/0022-1317-34-2-207. [DOI] [PubMed] [Google Scholar]
- Eriksson U., Svenson S. B., Lönngren J., Lindberg A. A. Salmonella phage glycanases: substrate specificity of the phage P22 endo-rhamnosidase. J Gen Virol. 1979 Jun;43(3):503–511. doi: 10.1099/0022-1317-43-3-503. [DOI] [PubMed] [Google Scholar]
- Iwashita S., Kanegasaki S. Smooth specific phage adsorption: endorhamnosidase activity of tail parts of P22. Biochem Biophys Res Commun. 1973 Nov 16;55(2):403–409. doi: 10.1016/0006-291x(73)91101-7. [DOI] [PubMed] [Google Scholar]
- Kauffmann F. A Typhoid Variant and a New Serological Variation in the Salmonella Group. J Bacteriol. 1941 Feb;41(2):127–140. doi: 10.1128/jb.41.2.127-140.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski B., Beilharz H., Stirm S. Disruption of Vi bacteriophage III and localization of its deacetylase activity. J Gen Virol. 1975 Dec;29(3):267–280. doi: 10.1099/0022-1317-29-3-267. [DOI] [PubMed] [Google Scholar]
- Lindberg A. A. Bacteriophage receptors. Annu Rev Microbiol. 1973;27:205–241. doi: 10.1146/annurev.mi.27.100173.001225. [DOI] [PubMed] [Google Scholar]
- Lindberg A. A., Holme T. Evaluation of some extraction methods for the preparation of bacterial lipopolysaccharides for structural analysis. Acta Pathol Microbiol Scand B Microbiol Immunol. 1972;80(5):751–759. doi: 10.1111/j.1699-0463.1972.tb00203.x. [DOI] [PubMed] [Google Scholar]
- Lindberg A. A., Wollin R., Gemski P., Wohlhieter J. A. Interaction between bacteriophage Sf6 and Shigella flexner. J Virol. 1978 Jul;27(1):38–44. doi: 10.1128/jvi.27.1.38-44.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkelä P. H. Glucosylation of lipopolysaccharide in Salmonella: mutants negative for O antigen factor 1221. J Bacteriol. 1973 Nov;116(2):847–856. doi: 10.1128/jb.116.2.847-856.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger-Hug D., Choy Y. M., Schmidt G., Stirm S. Isolation of Enterobacteriaceae bacteriophage particles catalysing cell wall lipopolysaccharide degradation. J Gen Virol. 1977 Feb;34(2):381–385. doi: 10.1099/0022-1317-34-2-381. [DOI] [PubMed] [Google Scholar]
- STOCKER B. A., STAUB A. M., TENELLI R., KOPACKA B. [Immunochemical study on Salmonella. VI. Study of antigen 1 present on 2 Salmonella of the B and E4 groups]. Ann Inst Pasteur (Paris) 1960 Apr;98:505–523. [PubMed] [Google Scholar]
- Stirm S., Bessler W., Fehmel F., Freund-Mölbert E. Bacteriophage particles with endo-glycosidase activity. J Virol. 1971 Sep;8(3):343–346. doi: 10.1128/jvi.8.3.343-346.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenson S. B., Lönngren J., Carlin N., Lindberg A. A. Salmonella bacteriophage glycanases: endorhamnosidases of Salmonella typhimurium bacteriophages. J Virol. 1979 Nov;32(2):583–592. doi: 10.1128/jvi.32.2.583-592.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TINELLI R., STAUB A. M. [Immunochemical study on Salmonella. 7. Study of the products formed by the controlled acid hydrolysis of the polysaccharide extracted from S. typhi. Part 1. Analysis of antigen O: 12 in the Kauffmann-White scheme]. Bull Soc Chim Biol (Paris) 1960;42:583–599. [PubMed] [Google Scholar]
- Vieu J. F., Croissant O., Dauguet C. Structure des bactériophages responsables des phénomènes de conversion chez les Salmonella. Ann Inst Pasteur (Paris) 1965 Jul;109(1):160–166. [PubMed] [Google Scholar]
- Wilkinson R. G., Gemski P., Jr, Stocker B. A. Non-smooth mutants of Salmonella typhimurium: differentiation by phage sensitivity and genetic mapping. J Gen Microbiol. 1972 May;70(3):527–554. doi: 10.1099/00221287-70-3-527. [DOI] [PubMed] [Google Scholar]
- Wright A. Mechanism of conversion of the salmonella O antigen by bacteriophageepsilon 34. J Bacteriol. 1971 Mar;105(3):927–936. doi: 10.1128/jb.105.3.927-936.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]