Abstract
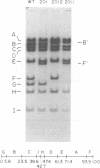
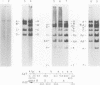
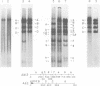
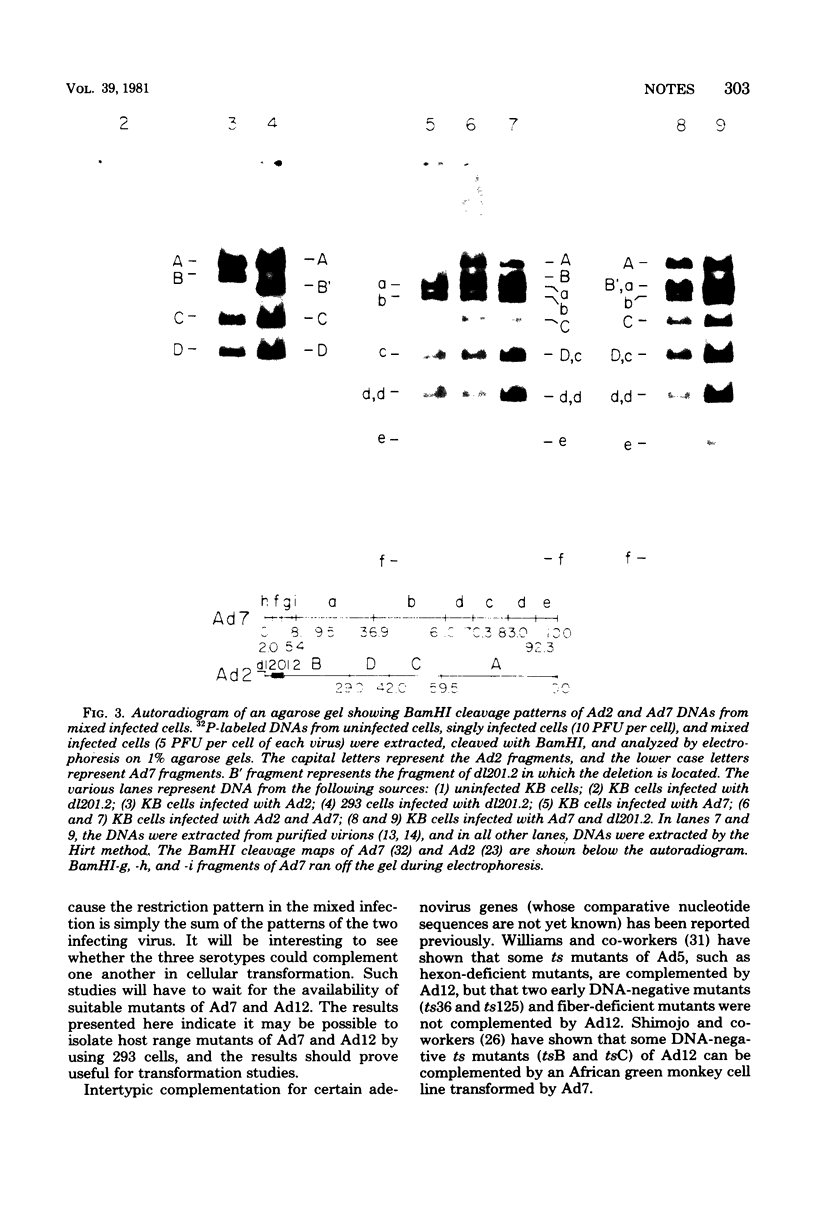
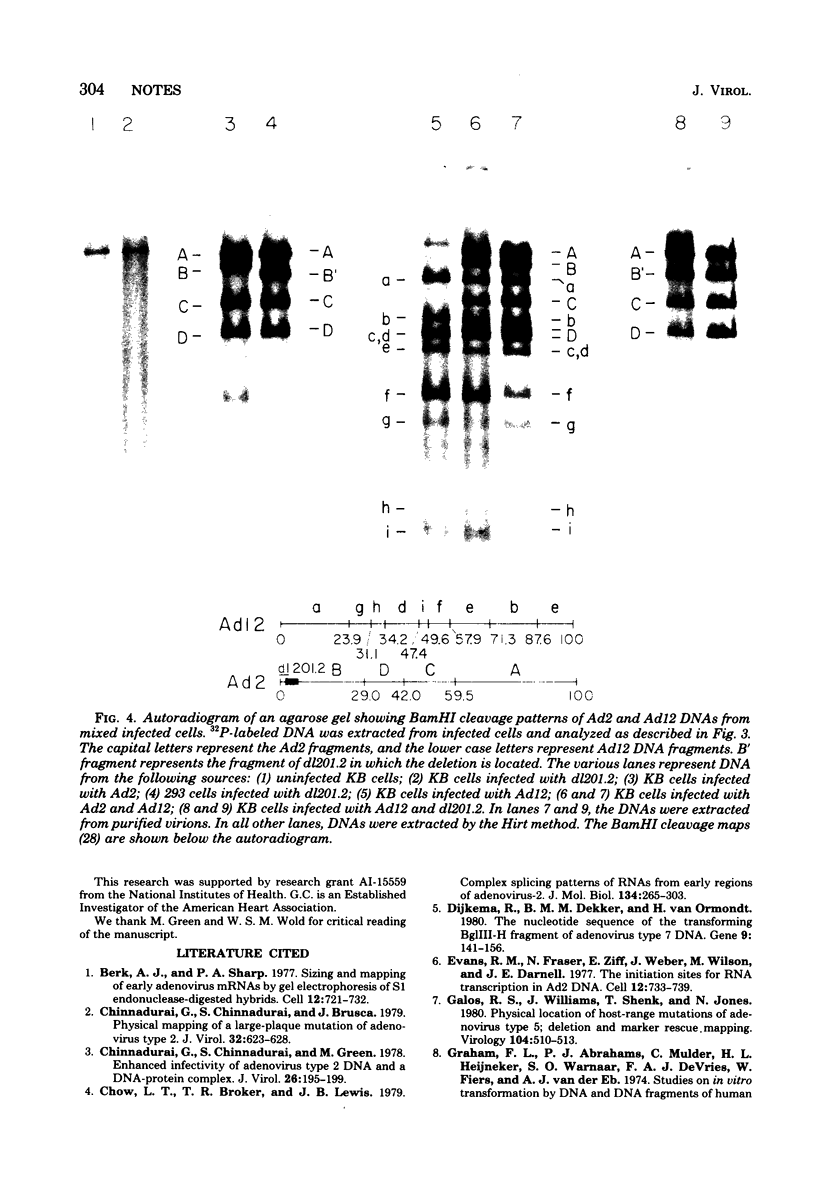
We have examined the functional similarity of the transforming genes for replicative functions among three different subgroups of human adenoviruses (A, B, and C), using mutant complementation as an assay. A host range deletion mutant (dl201.2) of Ad2 (nononcogenic subgroup C) lacking about 5% of the viral DNA covering two early gene blocks (E1a and E1b) involved in cellular transformation was isolated and tested for its ability to replicate in nonpermissive KB cells in the presence of Ad7 (weakly oncogenic group B) or ad12 (highly oncogenic group A). The complementation of the mutant defect was demonstrated by cleaving the viral DNA extracted from mixed infected cells or the DNA extracted from purified virions from mixed infected cells with restriction endonuclease BamHI, which produces a different cleavage pattern with the DNA of each serotype. It was found that the defects in E1a plus E1b of dl201.2 could be complemented by Ad7 and Ad12, indicating that these genes in Ad2, Ad7, and Ad12 have similar functions during productive infection.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Chinnadurai G., Chinnadurai S., Brusca J. Physical mapping of a large-plaque mutation of adenovirus type 2. J Virol. 1979 Nov;32(2):623–628. doi: 10.1128/jvi.32.2.623-628.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnadurai G., Chinnadurai S., Green M. Enhanced infectivity of adenovirus type 2 DNA and a DNA-protein complex. J Virol. 1978 Apr;26(1):195–199. doi: 10.1128/jvi.26.1.195-199.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow L. T., Broker T. R., Lewis J. B. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J Mol Biol. 1979 Oct 25;134(2):265–303. doi: 10.1016/0022-2836(79)90036-6. [DOI] [PubMed] [Google Scholar]
- Dijkema R., Dekker B. M., Van Ormondt H. The nucleotide sequence of the transforming BglII-H fragment of adenovirus type 7 DNA. Gene. 1980 Apr;9(1-2):141–156. doi: 10.1016/0378-1119(80)90172-9. [DOI] [PubMed] [Google Scholar]
- Evans R. M., Fraser N., Ziff E., Weber J., Wilson M., Darnell J. E. The initiation sites for RNA transcription in Ad2 DNA. Cell. 1977 Nov;12(3):733–739. doi: 10.1016/0092-8674(77)90273-2. [DOI] [PubMed] [Google Scholar]
- GREEN M., PINA M. BIOCHEMICAL STUDIES ON ADENOVIRUS MULTIPLICATION, VI. PROPERTIES OF HIGHLY PURIFIED TUMORIGENIC HUMAN ADENOVIRUSES AND THEIR DNA. Proc Natl Acad Sci U S A. 1964 Jun;51:1251–1259. doi: 10.1073/pnas.51.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GREEN M., PINA M. Biochemical studies on adenovirus multiplication. IV. Isolation, purification, and chemical analysis of adenovirus. Virology. 1963 May;20:199–207. doi: 10.1016/0042-6822(63)90157-0. [DOI] [PubMed] [Google Scholar]
- Galos R. S., Williams J., Shenk T., Jones N. Physical location of host-range mutations of adenovirus type 5; deletion and marker-rescue mapping. Virology. 1980 Jul 30;104(2):510–513. doi: 10.1016/0042-6822(80)90356-6. [DOI] [PubMed] [Google Scholar]
- Graham F. L., Harrison T., Williams J. Defective transforming capacity of adenovirus type 5 host-range mutants. Virology. 1978 May 1;86(1):10–21. doi: 10.1016/0042-6822(78)90003-x. [DOI] [PubMed] [Google Scholar]
- Graham F. L., Smiley J., Russell W. C., Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977 Jul;36(1):59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Green M., Mackey J. K., Wold W. S., Rigden P. Thirty-one human adenovirus serotypes (Ad1-Ad31) form five groups (A-E) based upon DNA genome homologies. Virology. 1979 Mar;93(2):481–492. doi: 10.1016/0042-6822(79)90251-4. [DOI] [PubMed] [Google Scholar]
- Green M. Oncogenic viruses. Annu Rev Biochem. 1970;39:701–756. doi: 10.1146/annurev.bi.39.070170.003413. [DOI] [PubMed] [Google Scholar]
- Harrison T., Graham F., Williams J. Host-range mutants of adenovirus type 5 defective for growth in HeLa cells. Virology. 1977 Mar;77(1):319–329. doi: 10.1016/0042-6822(77)90428-7. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Huebner R. J., Casey M. J., Chanock R. M., Schell K. Tumors induced in hamsters by a strain of adenovirus type 3: sharing of tumor antigens and "neoantigens" with those produced by adenovirus type 7 tumors. Proc Natl Acad Sci U S A. 1965 Aug;54(2):381–388. doi: 10.1073/pnas.54.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N., Shenk T. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell. 1979 Jul;17(3):683–689. doi: 10.1016/0092-8674(79)90275-7. [DOI] [PubMed] [Google Scholar]
- Maat J., Van Ormondt H. The nucleotide sequence of the transforming HindIII-G fragment of adenovirus type 5 DNA. The region between map positions 4.5 (HpaI site) and 8.0 (HindIII site). Gene. 1979 May;6(1):75–90. doi: 10.1016/0378-1119(79)90086-6. [DOI] [PubMed] [Google Scholar]
- Mackey J. K., Wold W. S., Rigden P., Green M. Transforming region of group A, B, and C adenoviruses: DNA homology studies with twenty-nine human adenovirus serotypes. J Virol. 1979 Mar;29(3):1056–1064. doi: 10.1128/jvi.29.3.1056-1064.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder C., Arrand J. R., Delius H., Keller W., Pettersson U., Roberts R. J., Sharp P. A. Cleavage maps of DNA from adenovirus types 2 and 5 by restriction endonucleases EcoRI and HpaI. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):397–400. doi: 10.1101/sqb.1974.039.01.051. [DOI] [PubMed] [Google Scholar]
- Sekikawa K., Shiroki K., Shimojo H., Ojima S., Fujinaga K. Transformation of a rat cell line by an adenovirus 7 DNA fragment. Virology. 1978 Jul 1;88(1):1–7. doi: 10.1016/0042-6822(78)90103-4. [DOI] [PubMed] [Google Scholar]
- Shiroki K., Handa H., Shimojo H., Yano S., Ojima S., Fujinaga K. Establishment and characterization of rat cell lines transformed by restriction endonuclease fragments of adenovirus 12 DNA. Virology. 1977 Oct 15;82(2):462–471. doi: 10.1016/0042-6822(77)90019-8. [DOI] [PubMed] [Google Scholar]
- Shiroki K., Shimojo H., Levine A. J., Sekikawa K., Rabek J. Suppression of the temperature-sensitive character of adenovirus 12 early mutants in monkey cells transformed by an adenovirus 7-simian virus 40 hybrid. Virology. 1976 Feb;69(2):431–437. doi: 10.1016/0042-6822(76)90474-8. [DOI] [PubMed] [Google Scholar]
- Sugisaki H., Sugimoto K., Takanami M., Shiroki K., Saito I., Shimojo H., Sawada Y., Uemizu Y., Uesugi S., Fujinaga K. Structure and gene organization in the transformed Hind III-G fragment of Ad12. Cell. 1980 Jul;20(3):777–786. doi: 10.1016/0092-8674(80)90324-4. [DOI] [PubMed] [Google Scholar]
- Van Ormondt H., Maat J., De Waard A., Van der Eb A. J. The nucleotide sequence of the transforming HpaI-E fragment of adenovirus type 5 DNA. Gene. 1978 Dec;4(4):309–328. doi: 10.1016/0378-1119(78)90048-3. [DOI] [PubMed] [Google Scholar]
- Willians J. F., Young C. S., Austin P. E. Genetic analysis of human adenovirus type 5 in permissive and nonpermissive cells. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):427–437. doi: 10.1101/sqb.1974.039.01.055. [DOI] [PubMed] [Google Scholar]
- van der Eb A. J., van Ormondt H., Schrier P. I., Lupker J. H., Jochemsen H., van den Elsen P. J., DeLeys R. J., Maat J., van Beveren C. P., Dijkema R. Structure and function of the transforming genes of human adenoviruses and SV40. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):383–399. doi: 10.1101/sqb.1980.044.01.043. [DOI] [PubMed] [Google Scholar]