Abstract
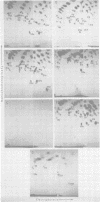
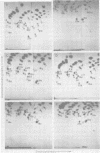
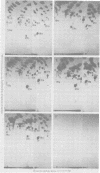
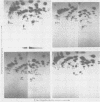
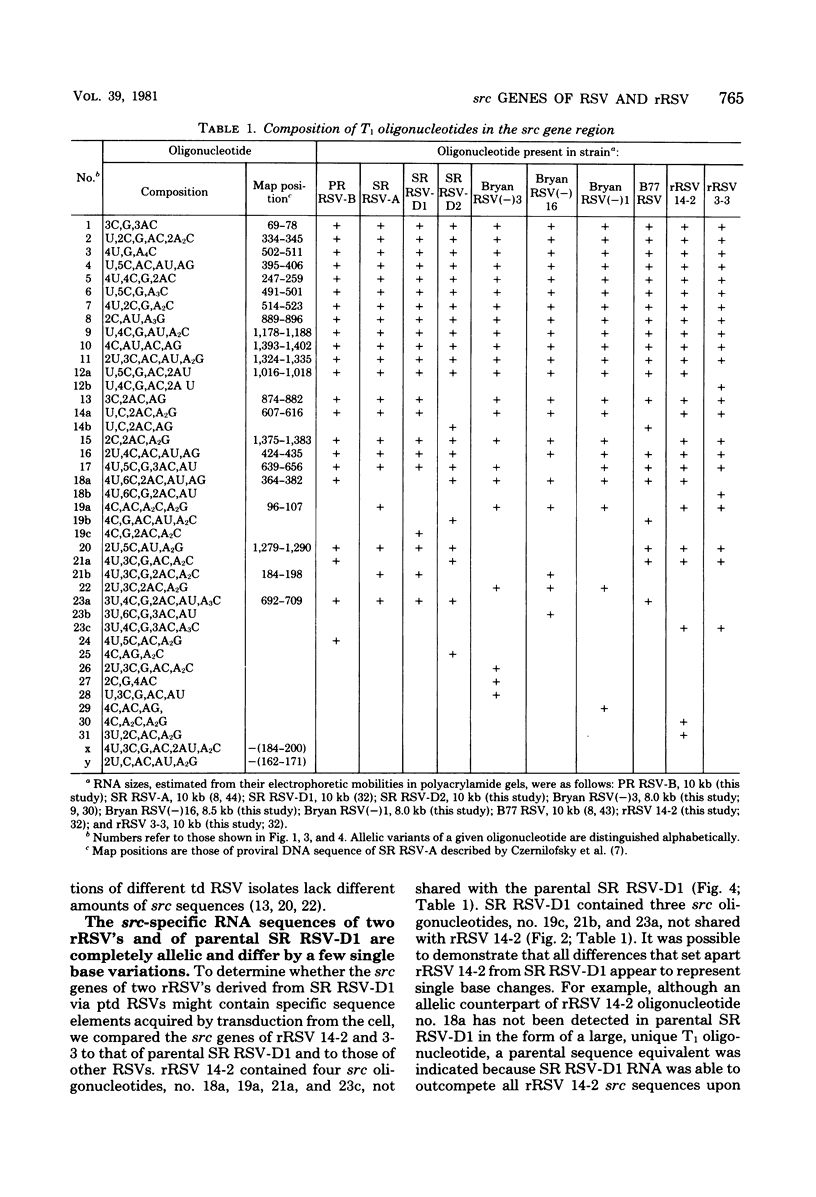
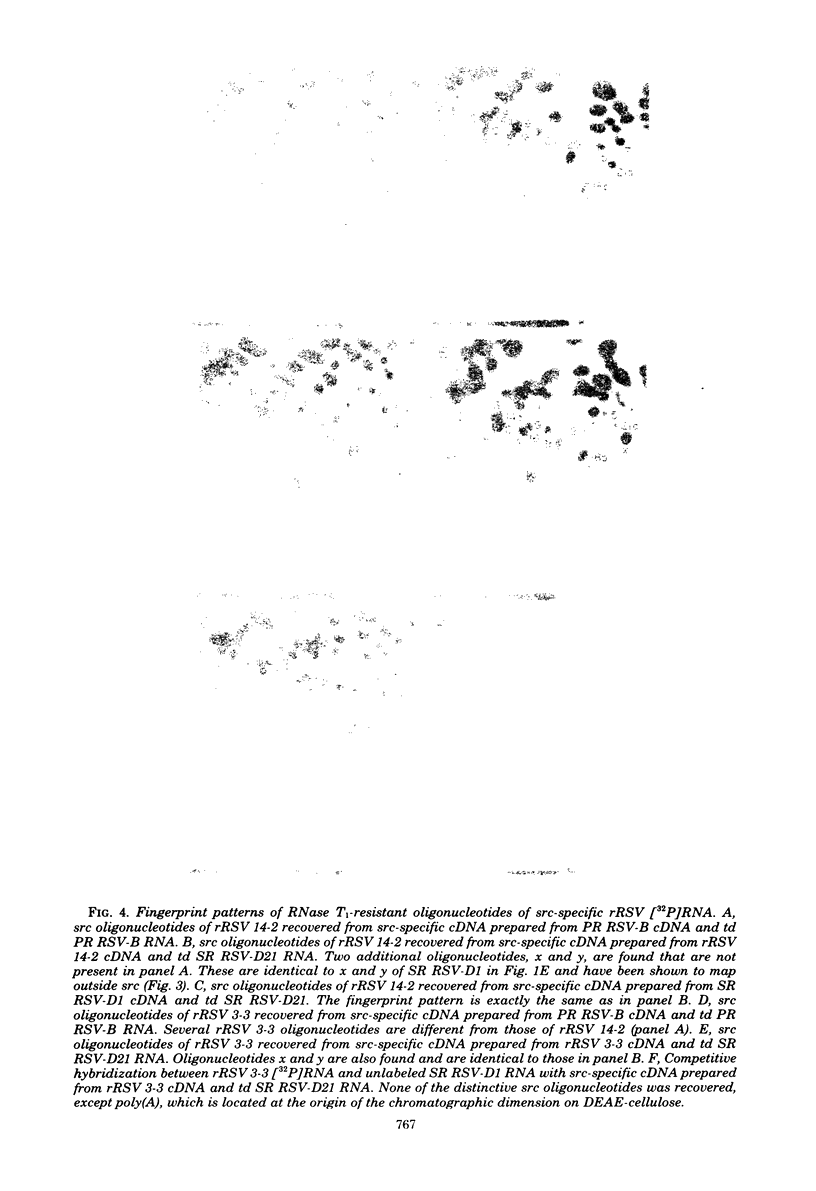
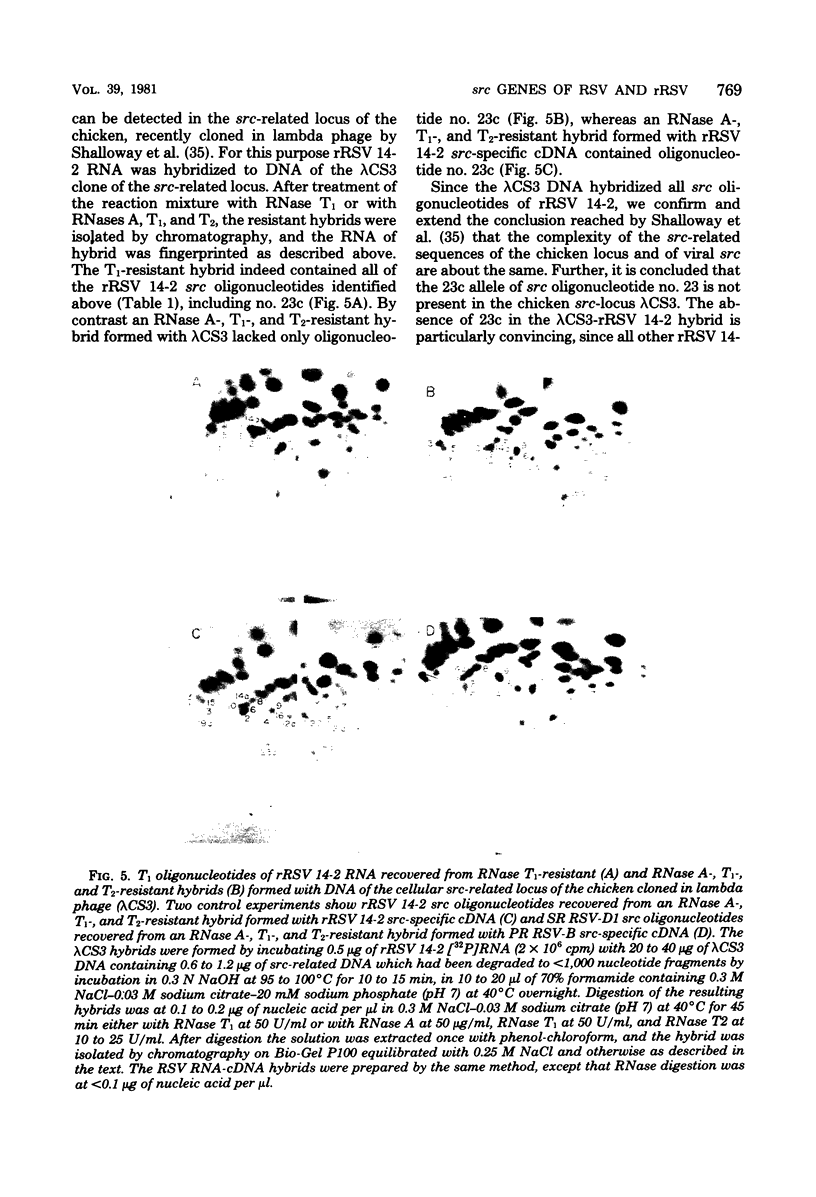
The src genes of different Rous sarcoma virus (RSV) strains have been reported to be highly conserved by some investigators using RNA-cDNA hybridization, whereas others using oligonucleotide, peptide, and serological analyses have judged src genes to be variable in 30 to 50% of the respective markers. Moreover, distinctive src oligonucleotides and peptides of so-called recovered RSVs (rRSV's) whose src genes were reported to be experimentally transduced from the cell are thought to represent specific markers of host-derived src sequences. By contrast, we have pointed out previously that these markers may represent point mutations of parental equivalents. Here we have compared the src-specific sequences of eight RSV strains and of two rRSV's to each other and to a molecular clone of the src-related chicken locus. Our comparisons are based on RNase T1-resistant oligonucleotides of RNA hybridized to src-specific cDNA, which was prepared by hybridizing RSV cDNA with RNA of isogenic src deletion mutants, or to a cloned cellular src-related DNA. All of the approximately 20 src-oligonucleotides of a given RSV strain were recovered by src-specific cDNA's of all other RSV strains or by cellular src-related DNA. The number of oligonucleotides varied slightly with the length of the src deletion used to prepare src-specific cDNA, thus providing a measure for src deletion mutants. Our data indicate that the src genes of all RSV strains tested, including the two reportedly transduced from the cell, are about 98% conserved and completely allelic with only scattered single nucleotide differences in certain variable regions which are subject to point mutations. Hence, based on the src oligonucleotide markers analyzed by us and others, we cannot distinguish between a cellular and viral origin of rRSV's. However, the following are not compatible with a cellular origin of rRSV's. (i) The only putative oligonucleotide marker which is exclusively shared by the two rRSV's studied and which differs from a parental counterpart in a single base was not detectable in cellular src-related DNA. (ii) The number of different allelic src markers observed by us and others in rRSV's was too large to derive from one or two known cellular src-related loci. (iii) The known absence of linkage of the cellular src-related locus with other virion sequences was extended to all non-src oligonucleotides, including some mapping directly adjacent to src. This is difficult to reconcile with the claim that transformation-defective, partial src deletion mutants of RSV which contain both, one, or, as we show here, possibly no src termini nevertheless transduce at the same frequencies, even though homologous, single or double illegitimate recombinations would be involved. Given (i) our evidence that src genes are subject to point mutation under selective conditions similar to those prevailing when rRSV's were generated and (ii) the lack of absolute evidence for the clonal purity of the transformation-defective, partial src deletion mutants of RSV used to generate rRSV's, we submit that the src genes of rRSV's could have been generated by cross-reactivation of nonoverlapping src deletions or mutation of src variants possibly present in transformation-defective, partial src deletion mutants of RSV. To prove experimental transduction, unambiguous markers need to be identified, or it would be necessary to generate rRSV's with molecularly cloned transformation-defective, partial src deletion mutants of RSV. Although our evidence casts doubt on the idea that specific src sequences of rRSV's originated by transduction, the close relationship between viral src and cellular src-related sequences argues that src genes originated at one time in evolution from the cell by events that involved illegitimate recombination and deletion of non-src sequences that interrupt the cellular src locus.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beemon K., Hunter T., Sefton B. M. Polymorphism of avian sarcoma virus src proteins. J Virol. 1979 Apr;30(1):190–200. doi: 10.1128/jvi.30.1.190-200.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand C., Palese P. Sequential passage of influenza virus in embryonated eggs or tissue culture: emergence of mutants. Virology. 1980 Dec;107(2):424–433. doi: 10.1016/0042-6822(80)90309-8. [DOI] [PubMed] [Google Scholar]
- Brugge J. S., Erikson R. L. Identification of a transformation-specific antigen induced by an avian sarcoma virus. Nature. 1977 Sep 22;269(5626):346–348. doi: 10.1038/269346a0. [DOI] [PubMed] [Google Scholar]
- Coffin J. M., Billeter M. A. A physical map of the Rous sarcoma virus genome. J Mol Biol. 1976 Jan 25;100(3):293–318. doi: 10.1016/s0022-2836(76)80065-4. [DOI] [PubMed] [Google Scholar]
- Coffin J. M., Champion M., Chabot F. Nucleotide sequence relationships between the genomes of an endogenous and an exogenous avian tumor virus. J Virol. 1978 Dec;28(3):972–991. doi: 10.1128/jvi.28.3.972-991.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin J. M., Tsichlis P. N., Barker C. S., Voynow S., Robinson H. L. Variation in avian retrovirus genomes. Ann N Y Acad Sci. 1980;354:410–425. doi: 10.1111/j.1749-6632.1980.tb27982.x. [DOI] [PubMed] [Google Scholar]
- Czernilofsky A. P., Levinson A. D., Varmus H. E., Bishop J. M., Tischer E., Goodman H. M. Nucleotide sequence of an avian sarcoma virus oncogene (src) and proposed amino acid sequence for gene product. Nature. 1980 Sep 18;287(5779):198–203. doi: 10.1038/287198a0. [DOI] [PubMed] [Google Scholar]
- Dierks P. M., Highfield P. E., Parsons J. T. Deletion mutant of the Bratislava-77 strain of Rous sarcoma virus containing a fusion of the group-specific antigen and envelope genes. J Virol. 1979 Nov;32(2):567–582. doi: 10.1128/jvi.32.2.567-582.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesberg P. H., Bister K., Moscovici C. Avian acute leukemia virus MC29: conserved and variable RNA sequences and recombination with helper virus. Virology. 1979 Nov;99(1):121–134. doi: 10.1016/0042-6822(79)90043-6. [DOI] [PubMed] [Google Scholar]
- Duesberg P. H., Kawai S., Wang L. H., Vogt P. K., Murphy H. M., Hanafusa H. RNA of replication-defective strains of Rous sarcoma virus. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1569–1573. doi: 10.1073/pnas.72.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesberg P. H. Transforming genes of retroviruses. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):13–29. doi: 10.1101/sqb.1980.044.01.005. [DOI] [PubMed] [Google Scholar]
- Evans L. H., Duesberg P. H., Troxler D. H., Scolnick E. M. Spleen focus-forming Friend virus: identification of genomic RNA and its relationship to helper virus RNA. J Virol. 1979 Jul;31(1):133–146. doi: 10.1128/jvi.31.1.133-146.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincham V. J., Neiman P. E., Wyke J. A. Novel nonconditional mutants in the src gene of Rous sarcoma virus: isolation and preliminary characterization. Virology. 1980 May;103(1):99–111. doi: 10.1016/0042-6822(80)90129-4. [DOI] [PubMed] [Google Scholar]
- Friis R. R. Abortive infection of Japanese quail cells with avian sarcoma viruses. Virology. 1972 Dec;50(3):701–712. doi: 10.1016/0042-6822(72)90424-2. [DOI] [PubMed] [Google Scholar]
- Hanafusa H., Halpern C. C., Buchhagen D. L., Kawai S. Recovery of avian sarcoma virus from tumors induced by transformation-defective mutants. J Exp Med. 1977 Dec 1;146(6):1735–1747. doi: 10.1084/jem.146.6.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseltine W. A., Kleid D. G., Panet A., Rothenberg E., Baltimore D. Ordered transcription of RNA tumor virus genomes. J Mol Biol. 1976 Sep 5;106(1):109–131. doi: 10.1016/0022-2836(76)90303-x. [DOI] [PubMed] [Google Scholar]
- Hughes S. H., Payvar F., Spector D., Schimke R. T., Robinson H. L., Payne G. S., Bishop J. M., Varmus H. E. Heterogeneity of genetic loci in chickens: analysis of endogenous viral and nonviral genes by cleavage of DNA with restriction endonucleases. Cell. 1979 Oct;18(2):347–359. doi: 10.1016/0092-8674(79)90054-0. [DOI] [PubMed] [Google Scholar]
- Hughes S. H., Stubblefield E., Payvar F., Engel J. D., Dodgson J. B., Spector D., Cordell B., Schimke R. T., Varmus H. E. Gene localization by chromosome fractionation: globin genes are on at least two chromosomes and three estrogen-inducible genes are on three chromosomes. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1348–1352. doi: 10.1073/pnas.76.3.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess R. E., Hanafusa H. Viral and cellular src genes contribute to the structure of recovered avian sarcoma virus transforming protein. Cell. 1981 Apr;24(1):155–164. doi: 10.1016/0092-8674(81)90511-0. [DOI] [PubMed] [Google Scholar]
- Kawai S., Duesberg P. H., Hanafusa H. Transformation-defective mutants of Rous sarcoma virus with src gene deletions of varying length. J Virol. 1977 Dec;24(3):910–914. doi: 10.1128/jvi.24.3.910-914.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M., Duesberg P. H., Horst J., Vogt P. K. Avian tumor virus RNA: a comparison of three sarcoma viruses and their transformation-defective derivatives by oligonucleotide fingerprinting and DNA-RNA hybridization. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2266–2270. doi: 10.1073/pnas.70.8.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M., Hu S. S., Vogt P. K. Occurrence of partial deletion and substitution of the src gene in the RNA genome of avian sarcoma virus. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4781–4785. doi: 10.1073/pnas.74.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. H., Bister K., Pawson A., Robins T., Moscovici C., Duesberg P. H. Fujinami sarcoma virus: an avian RNA tumor virus with a unique transforming gene. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2018–2022. doi: 10.1073/pnas.77.4.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson A. D., Oppermann H., Levintow L., Varmus H. E., Bishop J. M. Evidence that the transforming gene of avian sarcoma virus encodes a protein kinase associated with a phosphoprotein. Cell. 1978 Oct;15(2):561–572. doi: 10.1016/0092-8674(78)90024-7. [DOI] [PubMed] [Google Scholar]
- Maisel J., Dina D., Duesberg P. Murine sarcoma viruses: the helper-independence reported for a Moloney variant is unconfirmed; distinct strains differ in the size of their RNAs. Virology. 1977 Jan;76(1):295–312. doi: 10.1016/0042-6822(77)90304-x. [DOI] [PubMed] [Google Scholar]
- Martin G. S., Lee W. H., Duesberg P. H. Generation of nondefective Rous sarcoma virus by asymmetric recombination between deletion mutants. J Virol. 1980 Nov;36(2):591–594. doi: 10.1128/jvi.36.2.591-594.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter J. A. Genetic studies of the ploidy of Moloney murine leukemia virus. J Virol. 1977 Apr;22(1):9–15. doi: 10.1128/jvi.22.1.9-15.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon P., Pawson A., Bister K., Martin G. S., Duesberg P. H. Specific RNA sequences and gene products of MC29 avian acute leukemia virus. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5874–5878. doi: 10.1073/pnas.75.12.5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E. C., Jr, Arlinghaus R. B. Tryptic peptide analyses of polypeptides generated by premature termination of cell-free protein synthesis allow a determination of the Rauscher leukemia virus gag gene order. J Virol. 1978 Dec;28(3):929–935. doi: 10.1128/jvi.28.3.929-935.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy H. M. A new replication-defective variant of the Bryan high-titer strain Rous sarcoma virus. Virology. 1977 Apr;77(2):705–721. doi: 10.1016/0042-6822(77)90493-7. [DOI] [PubMed] [Google Scholar]
- Purchio A. F., Erikson E., Brugge J. S., Erikson R. L. Identification of a polypeptide encoded by the avian sarcoma virus src gene. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1567–1571. doi: 10.1073/pnas.75.3.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins T., Duesberg P. Specific RNA sequences of Rous sarcoma virus (RSV) recovered from tumors induced by transformation-defective RSV deletion mutants. Virology. 1979 Mar;93(2):427–434. doi: 10.1016/0042-6822(79)90246-0. [DOI] [PubMed] [Google Scholar]
- Sefton B. M., Beemon K., Hunter T. Comparison of the expression of the src gene of Rous sarcoma virus in vitro and in vivo. J Virol. 1978 Dec;28(3):957–971. doi: 10.1128/jvi.28.3.957-971.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefton B. M., Hunter T., Beemon K. Relationship of polypeptide products of the transforming gene of Rous sarcoma virus and the homologous gene of vertebrates. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2059–2063. doi: 10.1073/pnas.77.4.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalloway D., Zelenetz A. D., Cooper G. M. Molecular cloning and characterization of the chicken gene homologous to the transforming gene of Rous sarcoma virus. Cell. 1981 May;24(2):531–541. doi: 10.1016/0092-8674(81)90344-5. [DOI] [PubMed] [Google Scholar]
- Shine J., Czernilofsky A. P., Friedrich R., Bishop J. M., Goodman H. M. Nucleotide sequence at the 5' terminus of the avian sarcoma virus genome. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1473–1477. doi: 10.1073/pnas.74.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D. H., Varmus H. E., Bishop J. M. Nucleotide sequences related to the transforming gene of avian sarcoma virus are present in DNA of uninfected vertebrates. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4102–4106. doi: 10.1073/pnas.75.9.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehelin D., Guntaka R. V., Varmus H. E., Bishop J. M. Purification of DNA complementary to nucleotide sequences required for neoplastic transformation of fibroblasts by avian sarcoma viruses. J Mol Biol. 1976 Mar 5;101(3):349–365. doi: 10.1016/0022-2836(76)90152-2. [DOI] [PubMed] [Google Scholar]
- Stoll E., Billeter M. A., Palmenberg A., Weissmann C. Avian myeloblastosis virus RNA is terminally redundant: implications for the mechanism of retrovirus replication. Cell. 1977 Sep;12(1):57–72. doi: 10.1016/0092-8674(77)90185-4. [DOI] [PubMed] [Google Scholar]
- Tsichlis P. N., Conklin K. F., Coffin J. M. Mutant and recombinant avian retroviruses with extended host range. Proc Natl Acad Sci U S A. 1980 Jan;77(1):536–540. doi: 10.1073/pnas.77.1.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigne R., Breitman M. L., Moscovici C., Vogt P. K. Restitution of fibroblast-transforming ability in src deletion mutants of avian sarcoma virus during animal passage. Virology. 1979 Mar;93(2):413–426. doi: 10.1016/0042-6822(79)90245-9. [DOI] [PubMed] [Google Scholar]
- Vigne R., Neil J. C., Breitman M. L., Vogt P. K. Recovered src genes are polymorphic and contain host markers. Virology. 1980 Aug;105(1):71–85. doi: 10.1016/0042-6822(80)90157-9. [DOI] [PubMed] [Google Scholar]
- Wang L. H., Duesberg P. H., Kawai S., Hanafusa H. Location of envelope-specific and sarcoma-specific oligonucleotides on RNA of Schmidt-Ruppin Rous sarcoma virus. Proc Natl Acad Sci U S A. 1976 Feb;73(2):447–451. doi: 10.1073/pnas.73.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. H., Duesberg P. H., Robins T., Yokota H., Vogt P. K. The terminal oligonucleotides of avian tumor virus RNAs are genetically linked. Virology. 1977 Oct 15;82(2):472–492. doi: 10.1016/0042-6822(77)90020-4. [DOI] [PubMed] [Google Scholar]
- Wang L. H., Duesberg P., Beemon K., Vogt P. K. Mapping RNase T1-resistant oligonucleotides of avian tumor virus RNAs: sarcoma-specific oligonucleotides are near the poly(A) end and oligonucleotides common to sarcoma and transformation-defective viruses are at the poly(A) end. J Virol. 1975 Oct;16(4):1051–1070. doi: 10.1128/jvi.16.4.1051-1070.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. H., Duesberg P., Mellon P., Vogt P. K. Distribution of envelope-specific and sarcoma-specific nucleotide sequences from different parents in the RNAs of avian tumor virus recombinants. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1073–1077. doi: 10.1073/pnas.73.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. H., Halpern C. C., Nadel M., Hanafusa H. Recombination between viral and cellular sequences generates transforming sarcoma virus. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5812–5816. doi: 10.1073/pnas.75.12.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. H., Moscovici C., Karess R. E., Hanafusa H. Analysis of the src gene of sarcoma viruses generated by recombination between transformation-defective mutants and quail cellular sequences. J Virol. 1979 Nov;32(2):546–556. doi: 10.1128/jvi.32.2.546-556.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. H., Snyder P., Hanafusa T., Hanafusa H. Evidence for the common origin of viral and cellular sequences involved in sarcomagenic transformation. J Virol. 1980 Jul;35(1):52–64. doi: 10.1128/jvi.35.1.52-64.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. H., Snyder P., Hanafusa T., Moscovici C., Hanafusa H. Comparative analysis of cellular and viral sequences related to sarcomagenic cell transformation. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 2):755–764. doi: 10.1101/sqb.1980.044.01.081. [DOI] [PubMed] [Google Scholar]
- Wang S. Y., Hayward W. S., Hanafusa H. Genetic variation in the RNA transcripts of endogenous virus genes in uninfected chicken cells. J Virol. 1977 Oct;24(1):64–73. doi: 10.1128/jvi.24.1.64-73.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]