Abstract
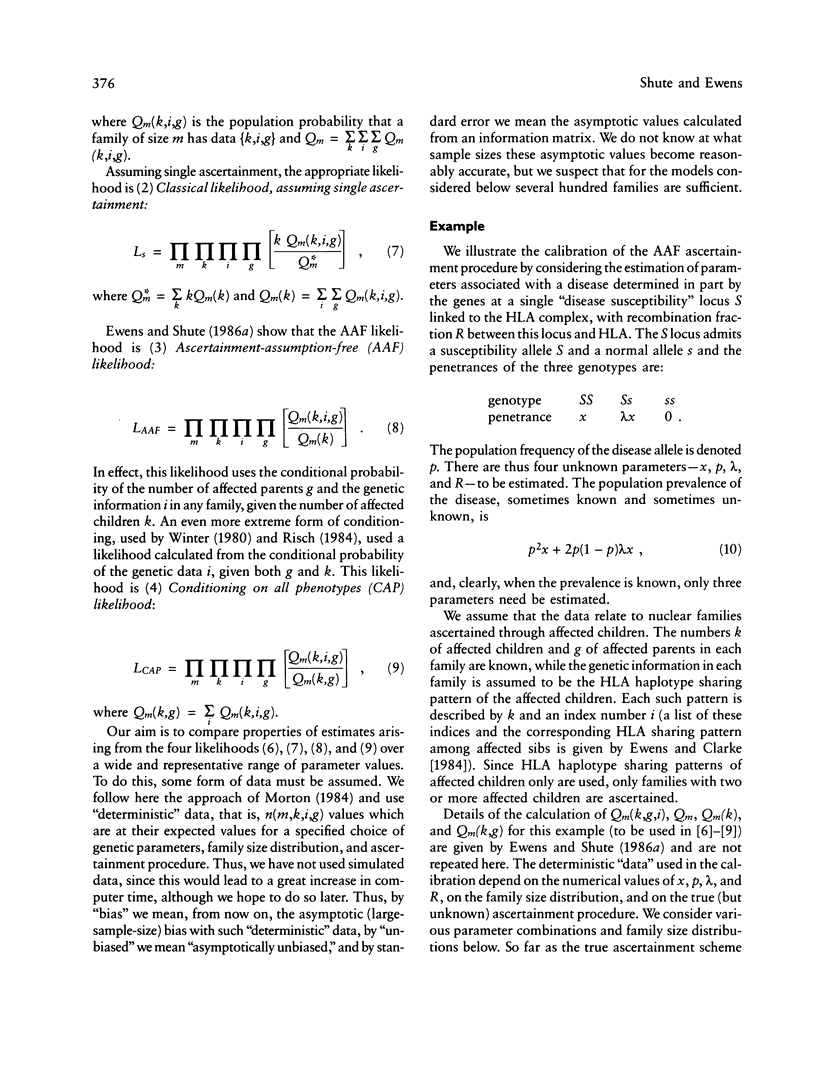
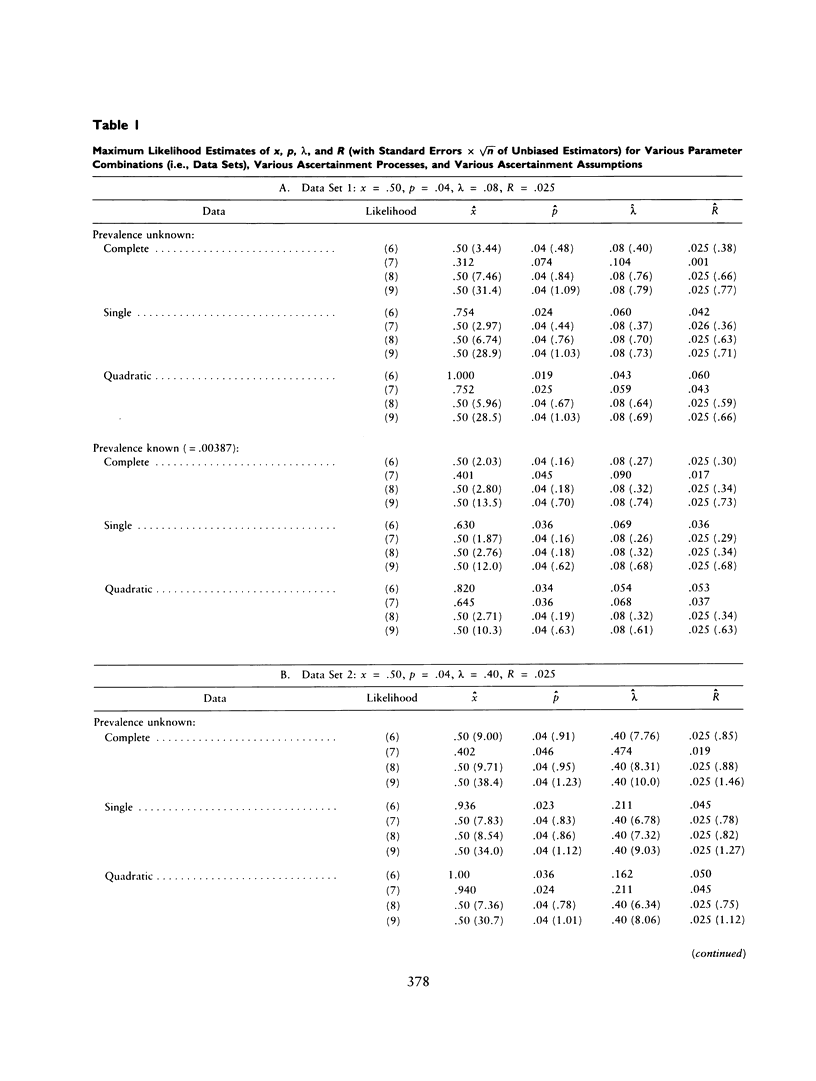
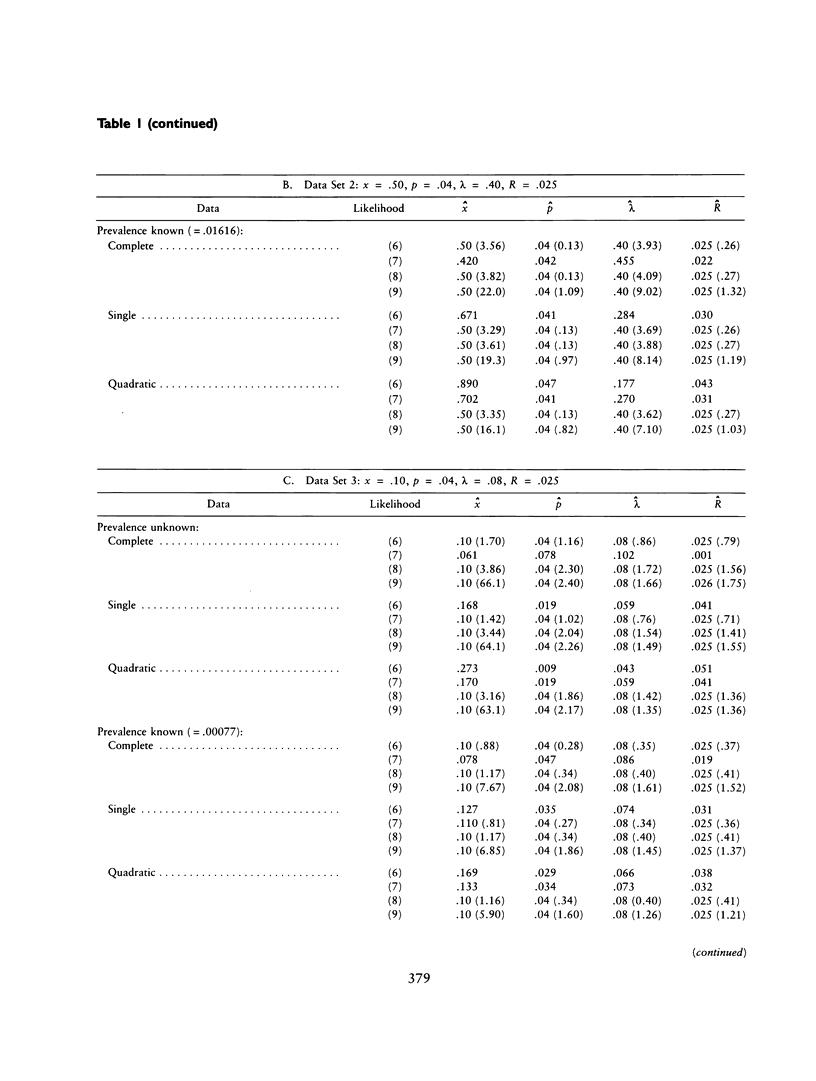
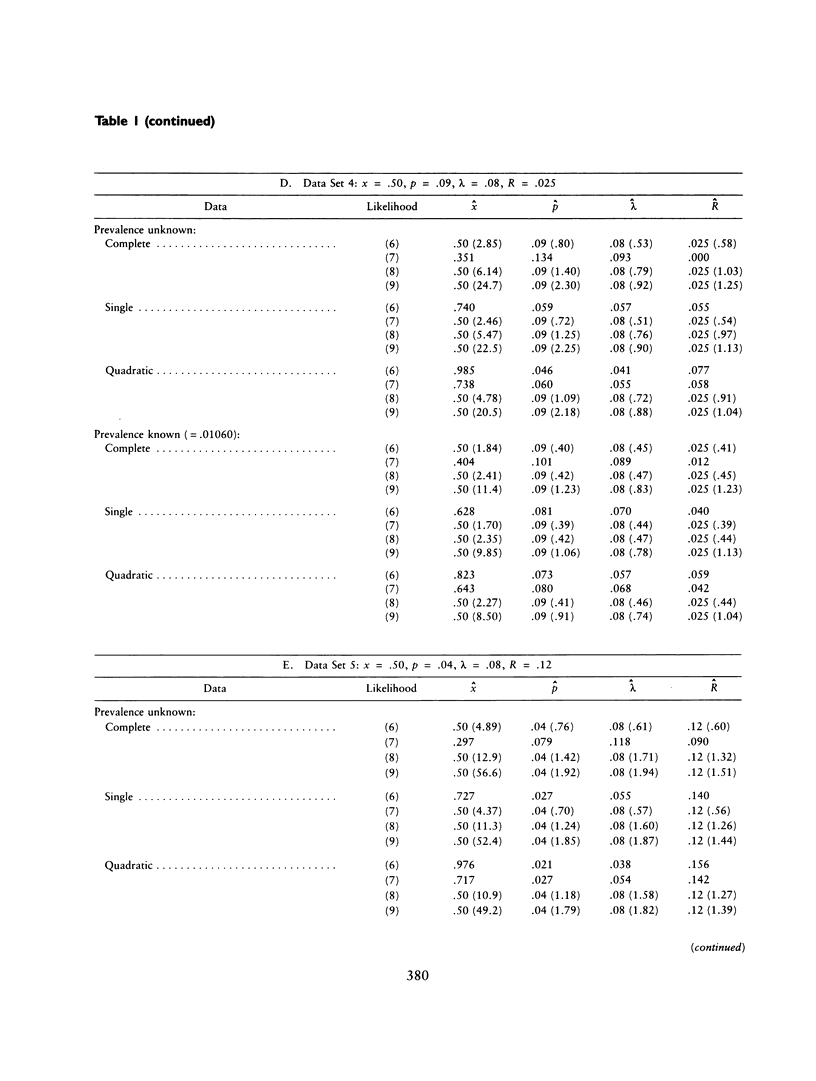
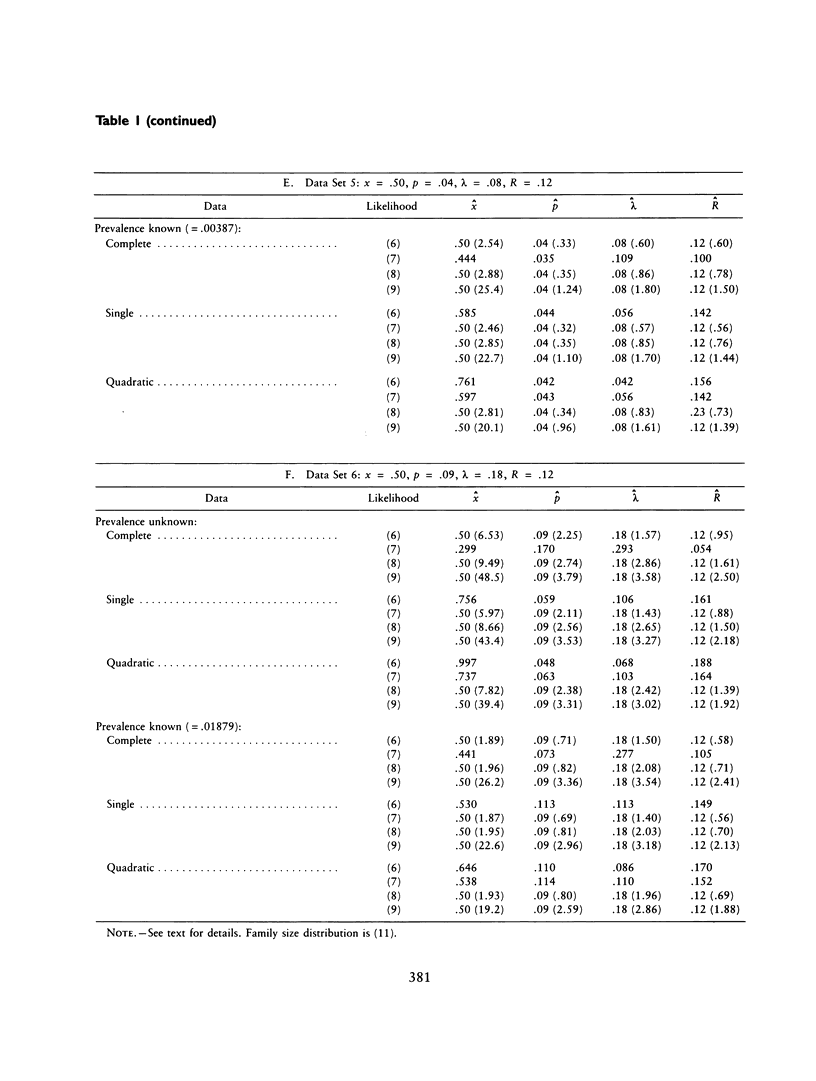
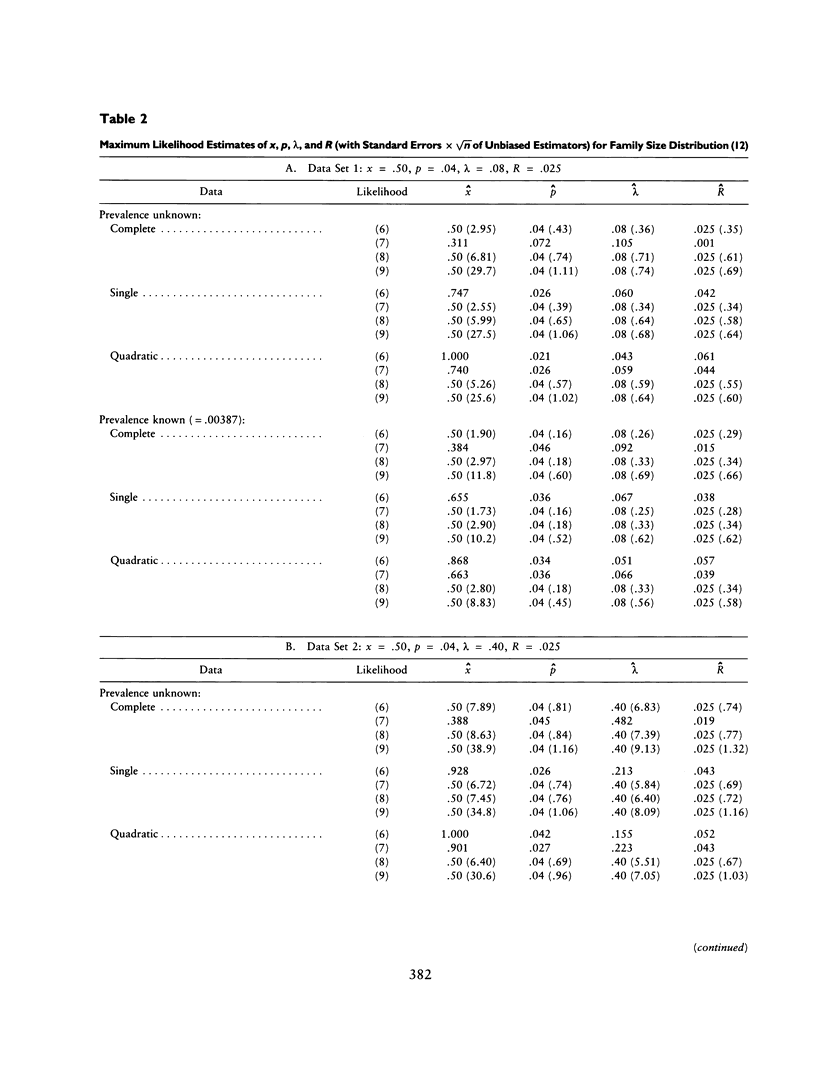
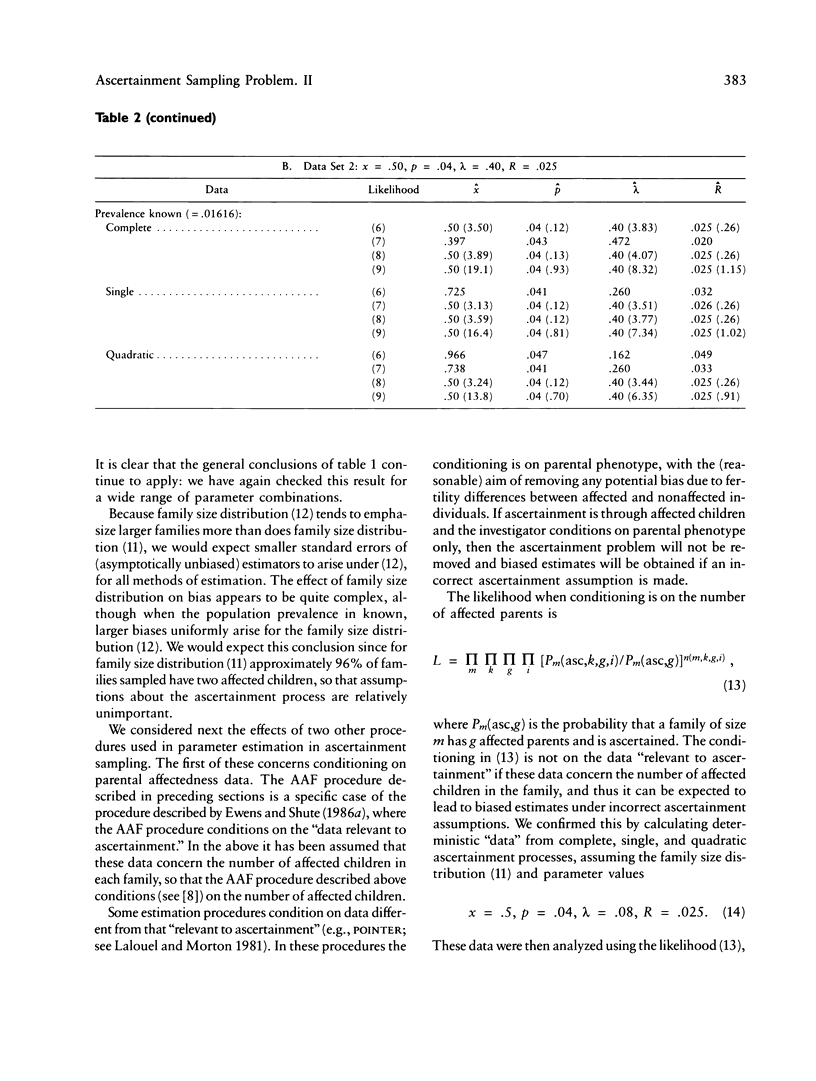
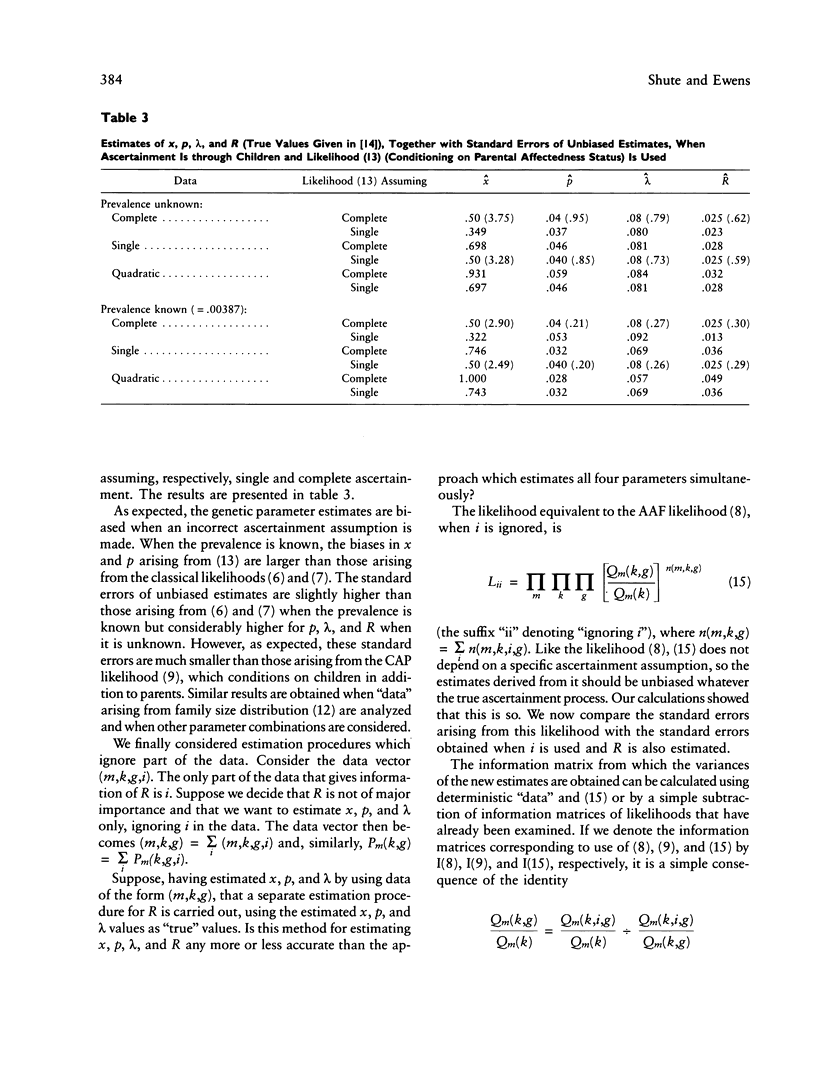
The ascertainment problem arises when families are sampled by a nonrandom process and some assumption about this sampling process must be made in order to estimate genetic parameters. Under classical ascertainment assumptions, estimation of genetic parameters cannot be separated from estimation of the parameters of the ascertainment process, so that any misspecification of the ascertainment process causes biases in estimation of the genetic parameters. Ewens and Shute proposed a resolution to this problem, involving conditioning the likelihood of the sample on the part of the data which is "relevant to ascertainment." The usefulness of this approach can only be assessed by examining the properties (in particular, bias and standard error) of the estimates which arise by using it for a wide range of parameter values and family size distributions and then comparing these biases and standard errors with those arising under classical ascertainment procedures. These comparisons are carried out in the present paper, and we also compare the proposed method with procedures which condition on, or ignore, parts of the data.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ewens W. J., Clarke C. P. Maximum likelihood estimation of genetic parameters of HLA-linked diseases using data from families of various sizes. Am J Hum Genet. 1984 Jul;36(4):858–872. [PMC free article] [PubMed] [Google Scholar]
- Ewens W. J., Shute N. C. A resolution of the ascertainment sampling problem. I. Theory. Theor Popul Biol. 1986 Dec;30(3):388–412. doi: 10.1016/0040-5809(86)90042-0. [DOI] [PubMed] [Google Scholar]
- Ewens W. J., Shute N. C., Cox N. J., Price R. A., Spielman R. S. Ascertainment considerations in the analysis of affected sib shared haplotype data. Genet Epidemiol Suppl. 1986;1:319–322. doi: 10.1002/gepi.1370030748. [DOI] [PubMed] [Google Scholar]
- Greenberg D. A. The effect of proband designation on segregation analysis. Am J Hum Genet. 1986 Sep;39(3):329–339. [PMC free article] [PubMed] [Google Scholar]
- Lalouel J. M., Morton N. E. Complex segregation analysis with pointers. Hum Hered. 1981;31(5):312–321. doi: 10.1159/000153231. [DOI] [PubMed] [Google Scholar]
- Risch N. Segregation analysis incorporating linkage markers. I. Single-locus models with an application to type I diabetes. Am J Hum Genet. 1984 Mar;36(2):363–386. [PMC free article] [PubMed] [Google Scholar]
- Winter R. M. The estimation of phenotype distributions from pedigree data. Am J Med Genet. 1980;7(4):537–542. doi: 10.1002/ajmg.1320070415. [DOI] [PubMed] [Google Scholar]


