Abstract
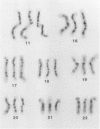
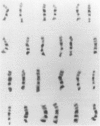
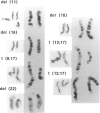
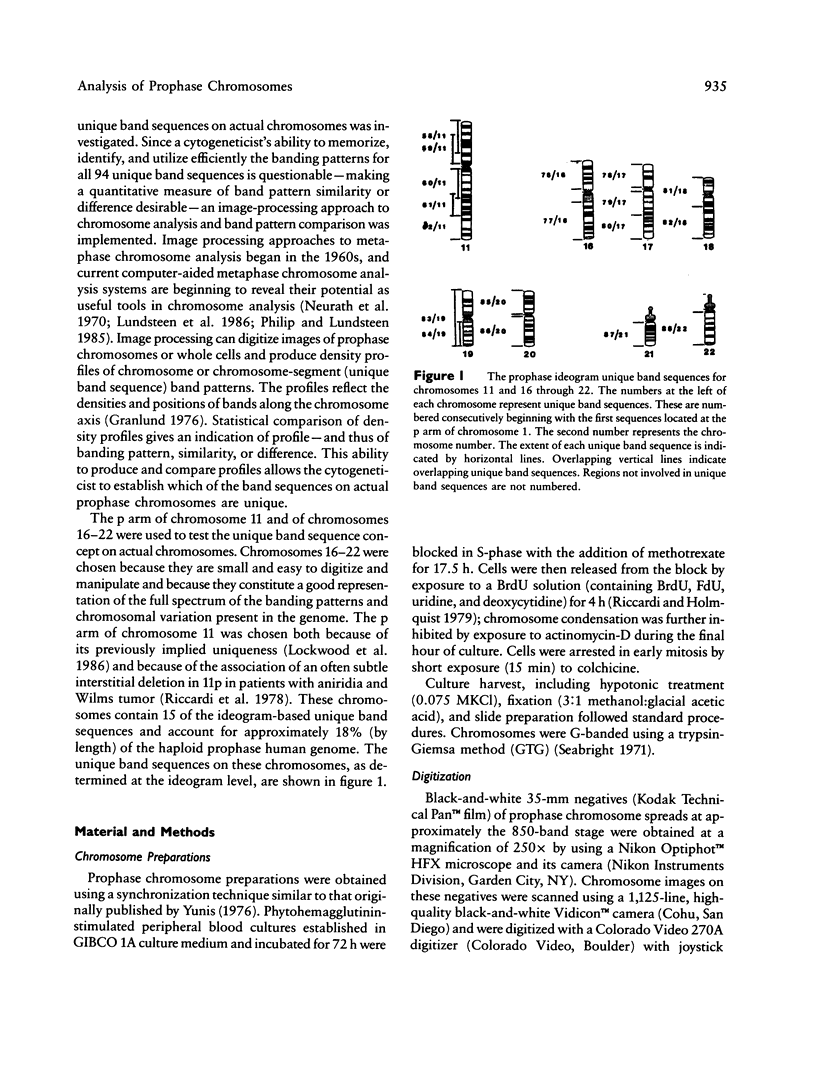
Using human prophase chromosome ideograms at the 850-band stage, we previously demonstrated that the 24 prophase ideograms can be divided into a set of 94 unique band sequences, each having a recognizable banding pattern distinct from other nonhomologous chromosome portions. Using actual prophase mitotic cells in this study, we analyzed the p arm of chromosome 11 and of chromosomes 16-22 and characterized a similar set of unique band sequences on actual chromosomes. This set of unique band sequences, a statistical comparison scheme, and image-processing techniques outlined in the present report can be used to identify and distinguish banding patterns of these chromosomes and to determine band pattern abnormalities.
Full text
PDF


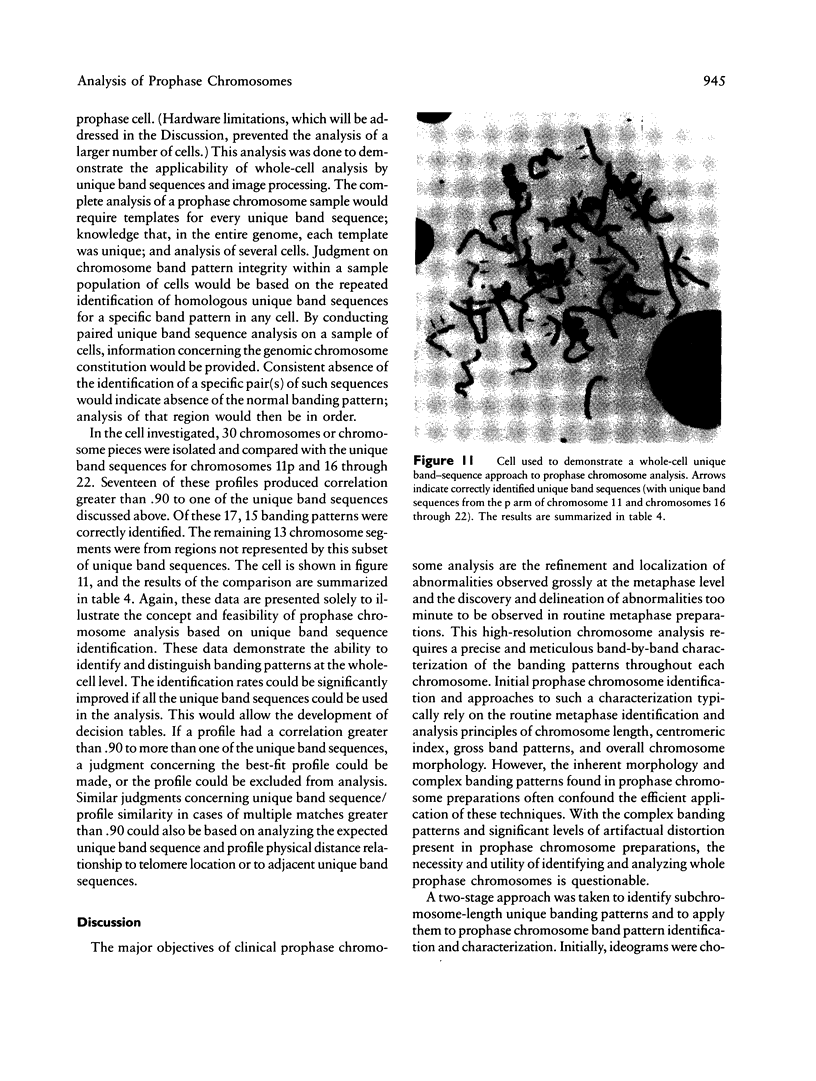
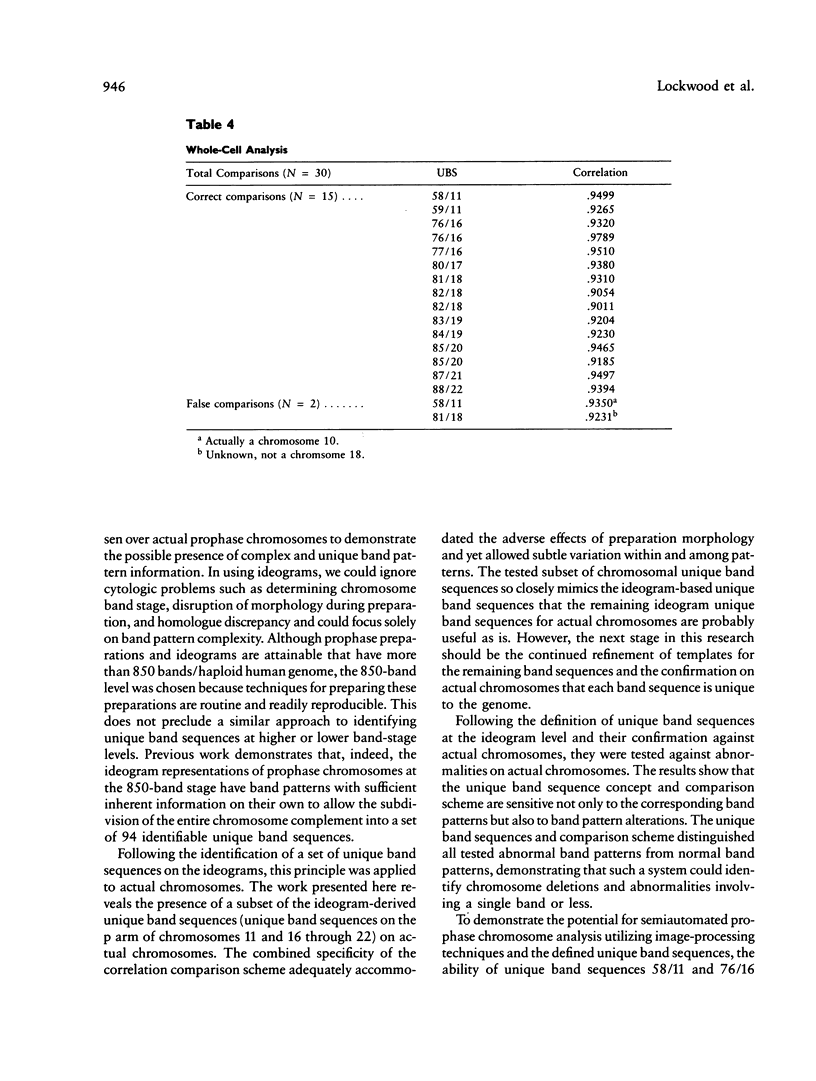

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Francke U. High-resolution ideograms of trypsin-Giemsa banded human chromosomes. Cytogenet Cell Genet. 1981;31(1):24–32. doi: 10.1159/000131622. [DOI] [PubMed] [Google Scholar]
- Granlund G. H. Identification of human chromosomes by using integrated density profiles. IEEE Trans Biomed Eng. 1976 May;23(3):182–192. doi: 10.1109/tbme.1976.324629. [DOI] [PubMed] [Google Scholar]
- Lockwood D. H., Riccardi V. M., Zimmerman S. O., Johnston D. A. Prophase chromosome unique band sequences: definition and utilization. Cytogenet Cell Genet. 1986;42(3):141–153. doi: 10.1159/000132267. [DOI] [PubMed] [Google Scholar]
- Lundsteen C., Gerdes T., Maahr J. Automatic classification of chromosomes as part of a routine system for clinical analysis. Cytometry. 1986 Jan;7(1):1–7. doi: 10.1002/cyto.990070102. [DOI] [PubMed] [Google Scholar]
- Neurath P. W., Ampola M. G., Low D. A., Selles W. D. Combined interactive computer measurement and automatic classification of human chromosomes. Cytogenetics. 1970;9(6):424–435. doi: 10.1159/000130112. [DOI] [PubMed] [Google Scholar]
- Philip J., Lundsteen C. Semiautomated chromosome analysis. A clinical test. Clin Genet. 1985 Feb;27(2):140–146. doi: 10.1111/j.1399-0004.1985.tb00201.x. [DOI] [PubMed] [Google Scholar]
- Riccardi V. M., Holmquist G. P. De novo 13q paracentric inversion in a boy with cleft palate and mental retardation. Hum Genet. 1979 Nov;52(2):211–215. [PubMed] [Google Scholar]
- Riccardi V. M., Sujansky E., Smith A. C., Francke U. Chromosomal imbalance in the Aniridia-Wilms' tumor association: 11p interstitial deletion. Pediatrics. 1978 Apr;61(4):604–610. [PubMed] [Google Scholar]
- Schwartz S., Palmer C. G. High-resolution chromosome analysis: I. Applications and limitations. Am J Med Genet. 1984 Oct;19(2):291–299. doi: 10.1002/ajmg.1320190211. [DOI] [PubMed] [Google Scholar]
- Seabright M. A rapid banding technique for human chromosomes. Lancet. 1971 Oct 30;2(7731):971–972. doi: 10.1016/s0140-6736(71)90287-x. [DOI] [PubMed] [Google Scholar]
- Yunis J. J. High resolution of human chromosomes. Science. 1976 Mar 26;191(4233):1268–1270. doi: 10.1126/science.1257746. [DOI] [PubMed] [Google Scholar]