Abstract
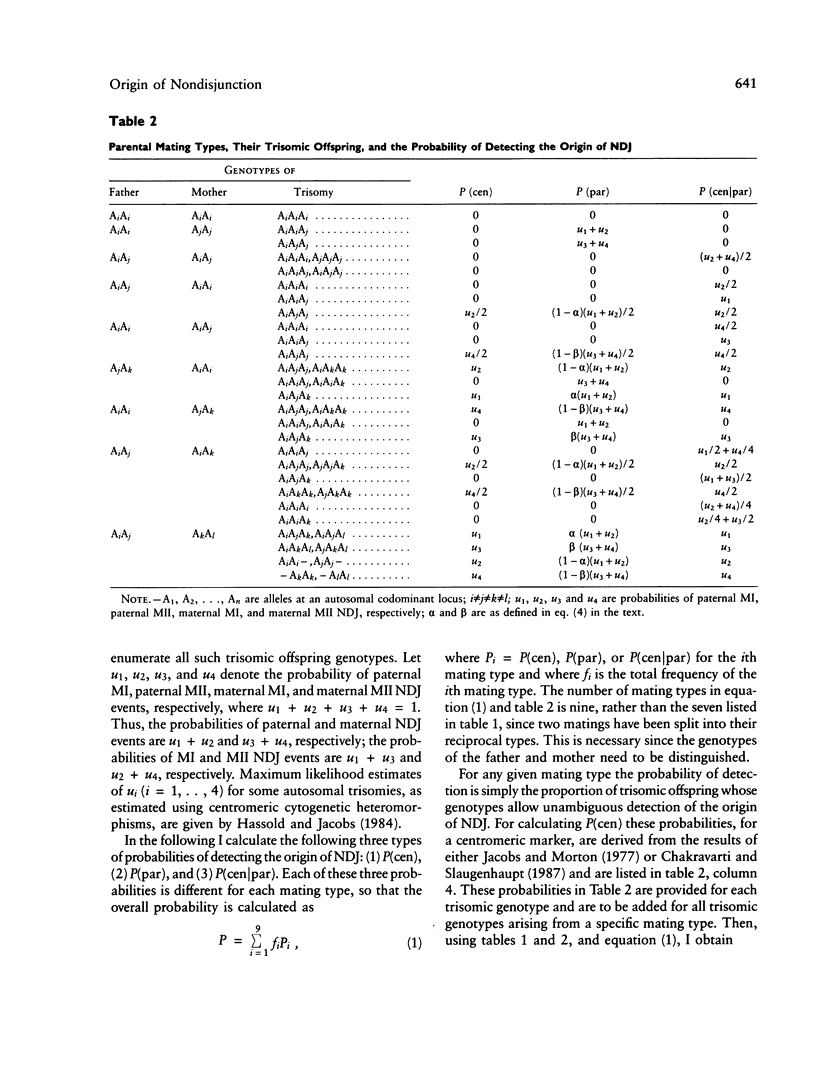
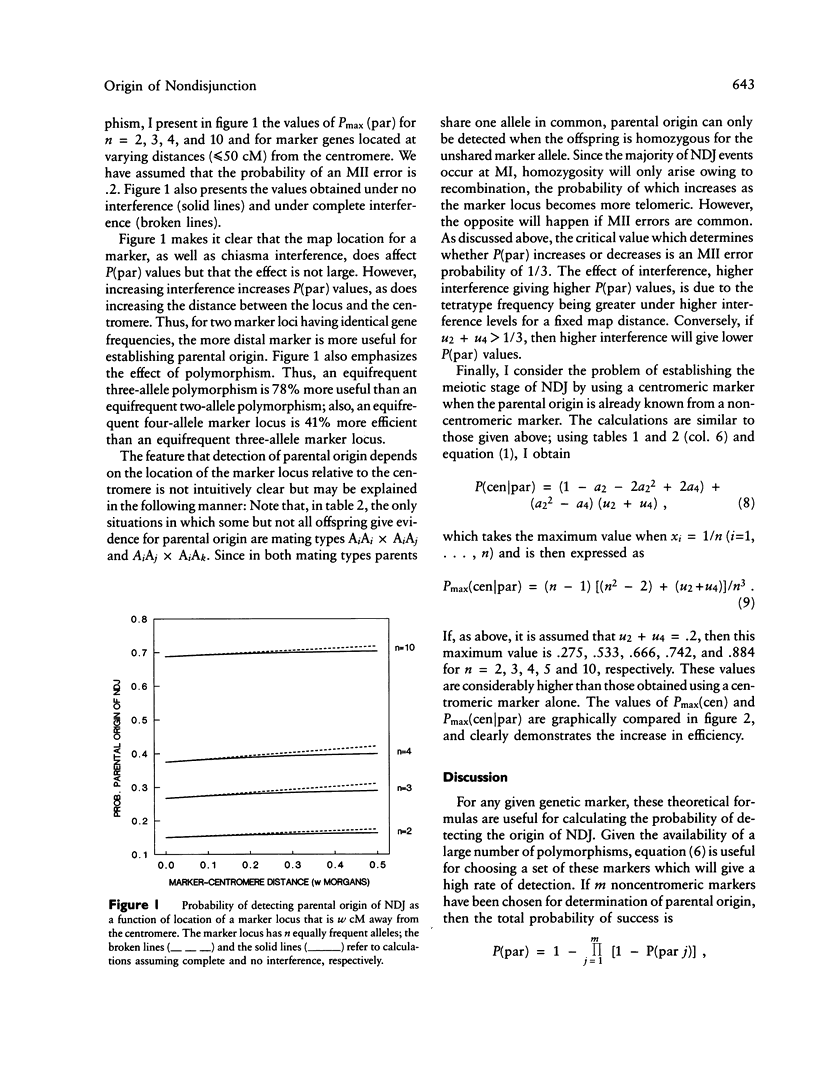
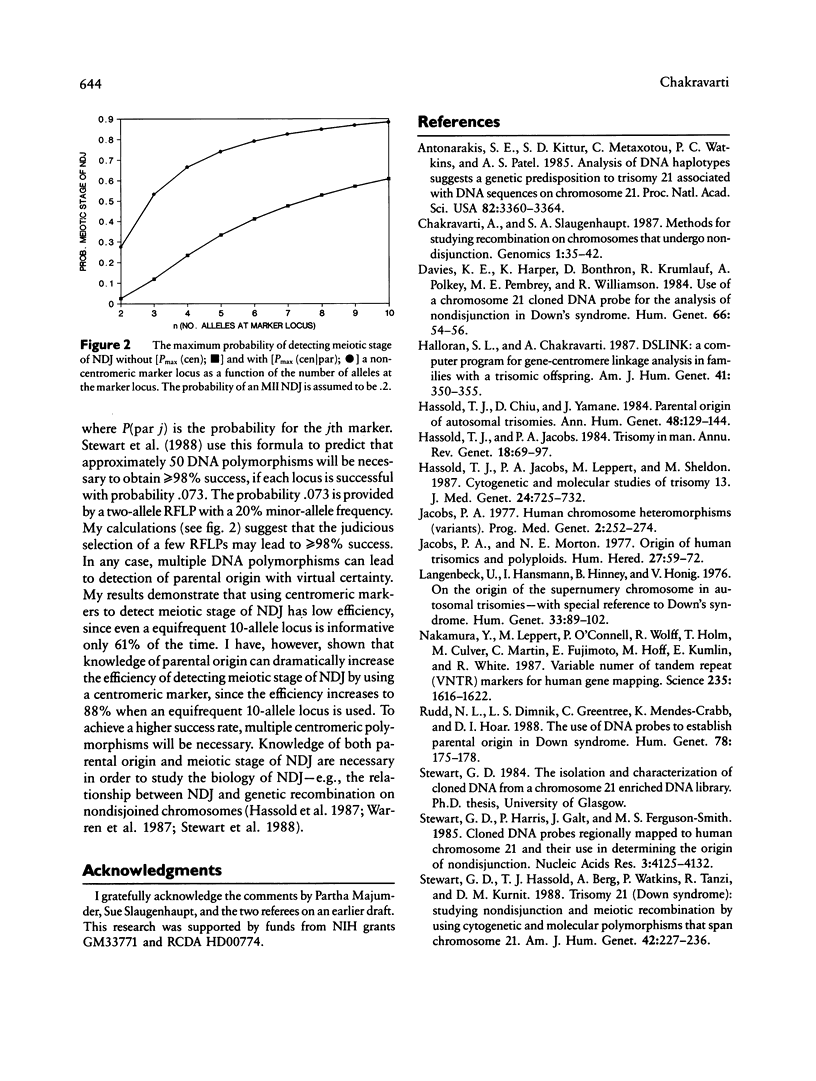
For studying the biology of autosomal trisomies it is necessary to establish the parental origin and meiotic stage of nondisjunction by using genetic markers. Theoretical formulas are obtained for calculating the probability of establishing (1) parental origin and meiotic stage of nondisjunction by using a centromeric marker, (2) parental origin of nondisjunction by using a noncentromeric marker, and (3) meiotic stage, given parental origin of nondisjunction. These theoretical calculations demonstrate that parental origin of nondisjunction can be identified with virtual certainty by utilizing multiple genetic markers along a chromosome arm. Centromeric markers are by themselves inefficient for determining meiotic stage of the error, but the efficiency can be considerably increased if parental origin is known with certainty. Even then, multiple centromeric markers may be necessary.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antonarakis S. E., Kittur S. D., Metaxotou C., Watkins P. C., Patel A. S. Analysis of DNA haplotypes suggests a genetic predisposition to trisomy 21 associated with DNA sequences on chromosome 21. Proc Natl Acad Sci U S A. 1985 May;82(10):3360–3364. doi: 10.1073/pnas.82.10.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti A., Slaugenhaupt S. A. Methods for studying recombination on chromosomes that undergo nondisjunction. Genomics. 1987 Sep;1(1):35–42. doi: 10.1016/0888-7543(87)90102-9. [DOI] [PubMed] [Google Scholar]
- Davies K. E., Harper K., Bonthron D., Krumlauf R., Polkey A., Pembrey M. E., Williamson R. Use of a chromosome 21 cloned DNA probe for the analysis of non-disjunction in Down syndrome. Hum Genet. 1984;66(1):54–56. doi: 10.1007/BF00275186. [DOI] [PubMed] [Google Scholar]
- Halloran S. L., Chakravarti A. DSLINK: a computer program for gene-centromere linkage analysis in families with a trisomic offspring. Am J Hum Genet. 1987 Sep;41(3):350–355. [PMC free article] [PubMed] [Google Scholar]
- Hassold T. J., Jacobs P. A. Trisomy in man. Annu Rev Genet. 1984;18:69–97. doi: 10.1146/annurev.ge.18.120184.000441. [DOI] [PubMed] [Google Scholar]
- Hassold T., Chiu D., Yamane J. A. Parental origin of autosomal trisomies. Ann Hum Genet. 1984 May;48(Pt 2):129–144. doi: 10.1111/j.1469-1809.1984.tb01008.x. [DOI] [PubMed] [Google Scholar]
- Hassold T., Jacobs P. A., Leppert M., Sheldon M. Cytogenetic and molecular studies of trisomy 13. J Med Genet. 1987 Dec;24(12):725–732. doi: 10.1136/jmg.24.12.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs P. A. Human chromosome heteromorphisms (variants). Prog Med Genet. 1977;2:251–274. doi: 10.1016/0378-1119(77)90004-x. [DOI] [PubMed] [Google Scholar]
- Jacobs P. A., Morton N. E. Origin of human trisomics and polyploids. Hum Hered. 1977;27(1):59–72. doi: 10.1159/000152852. [DOI] [PubMed] [Google Scholar]
- Langenbeck U., Hansmann I., Hinney B., Hönig V. On the origin of the supernumerary chromosome in autosomal trisomies--with special reference to Down's syndrome. A bias in tracing nondisjunction by chromosomal and biochemical polymorphisms. Hum Genet. 1976 Jul 27;33(2):89–102. doi: 10.1007/BF00281882. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Leppert M., O'Connell P., Wolff R., Holm T., Culver M., Martin C., Fujimoto E., Hoff M., Kumlin E. Variable number of tandem repeat (VNTR) markers for human gene mapping. Science. 1987 Mar 27;235(4796):1616–1622. doi: 10.1126/science.3029872. [DOI] [PubMed] [Google Scholar]
- Rudd N. L., Dimnik L. S., Greentree C., Mendes-Crabb K., Hoar D. I. The use of DNA probes to establish parental origin in Down syndrome. Hum Genet. 1988 Feb;78(2):175–178. doi: 10.1007/BF00278191. [DOI] [PubMed] [Google Scholar]
- Stewart G. D., Harris P., Galt J., Ferguson-Smith M. A. Cloned DNA probes regionally mapped to human chromosome 21 and their use in determining the origin of nondisjunction. Nucleic Acids Res. 1985 Jun 11;13(11):4125–4132. doi: 10.1093/nar/13.11.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G. D., Hassold T. J., Berg A., Watkins P., Tanzi R., Kurnit D. M. Trisomy 21 (Down syndrome): studying nondisjunction and meiotic recombination by using cytogenetic and molecular polymorphisms that span chromosome 21. Am J Hum Genet. 1988 Feb;42(2):227–236. [PMC free article] [PubMed] [Google Scholar]
- Warren A. C., Chakravarti A., Wong C., Slaugenhaupt S. A., Halloran S. L., Watkins P. C., Metaxotou C., Antonarakis S. E. Evidence for reduced recombination on the nondisjoined chromosomes 21 in Down syndrome. Science. 1987 Aug 7;237(4815):652–654. doi: 10.1126/science.2955519. [DOI] [PubMed] [Google Scholar]
- Willard H. F., Skolnick M. H., Pearson P. L., Mandel J. L. Report of the Committee on Human Gene Mapping by Recombinant DNA Techniques. Cytogenet Cell Genet. 1985;40(1-4):360–489. doi: 10.1159/000132180. [DOI] [PubMed] [Google Scholar]
- Willard H. F., Waye J. S., Skolnick M. H., Schwartz C. E., Powers V. E., England S. B. Detection of restriction fragment length polymorphisms at the centromeres of human chromosomes by using chromosome-specific alpha satellite DNA probes: implications for development of centromere-based genetic linkage maps. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5611–5615. doi: 10.1073/pnas.83.15.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]


