Abstract
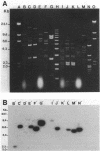
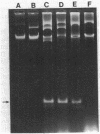
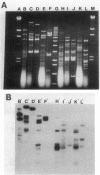
Plasmids from the first six reported beta-lactamase-producing (Bla+) enterococci were compared for genetic relatedness. Bla+ enterococcal plasmids from strains isolated in Houston, Tex.; Philadelphia, Pa.; Connecticut; and Pittsburgh, Pa., had heterogeneous HaeIII and MspI-ClaI restriction endonuclease digestion patterns. A staphylococcal beta-lactamase probe hybridized to all six Bla+ enterococcal plasmids, but hybridization was detected on different HaeIII and MspI-ClaI fragments of the six plasmids. An enterococcal gentamicin resistance (Gmr) probe hybridized to a common 3.9-kilobase HaeIII fragment from the five Gmr plasmids. The Houston plasmid was cross-hybridized to the other five strains, and moderate to extensive homology was demonstrated. Bla+ enterococcal plasmids from a broad geographic range are heterogeneous with respect to size and restriction endonuclease digestion patterns but contain homologous genetic material, including Bla+ and Gmr determinants.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Courvalin P., Carlier C., Collatz E. Plasmid-mediated resistance to aminocyclitol antibiotics in group D streptococci. J Bacteriol. 1980 Aug;143(2):541–551. doi: 10.1128/jb.143.2.541-551.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti J. J., Gilmore K. S., Courvalin P. Nucleotide sequence analysis of the gene specifying the bifunctional 6'-aminoglycoside acetyltransferase 2"-aminoglycoside phosphotransferase enzyme in Streptococcus faecalis and identification and cloning of gene regions specifying the two activities. J Bacteriol. 1986 Aug;167(2):631–638. doi: 10.1128/jb.167.2.631-638.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann S. A., Moellering R. C., Jr The enterococcus: "putting the bug in our ears". Ann Intern Med. 1987 May;106(5):757–761. doi: 10.7326/0003-4819-106-5-757. [DOI] [PubMed] [Google Scholar]
- Krogstad D. J., Korfhagen T. R., Moellering R. C., Jr, Wennersten C., Swartz M. N. Aminoglycoside-inactivating enzymes in clinical isolates of Streptococcus faecalis. An explanation for resistance to antibiotic synergism. J Clin Invest. 1978 Aug;62(2):480–486. doi: 10.1172/JCI109149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus beta-lactamase gene. J Biol Chem. 1981 Nov 10;256(21):11283–11291. [PubMed] [Google Scholar]
- Murray B. E., An F. Y., Clewell D. B. Plasmids and pheromone response of the beta-lactamase producer Streptococcus (Enterococcus) faecalis HH22. Antimicrob Agents Chemother. 1988 Apr;32(4):547–551. doi: 10.1128/aac.32.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray B. E., Church D. A., Wanger A., Zscheck K., Levison M. E., Ingerman M. J., Abrutyn E., Mederski-Samoraj B. Comparison of two beta-lactamase-producing strains of Streptococcus faecalis. Antimicrob Agents Chemother. 1986 Dec;30(6):861–864. doi: 10.1128/aac.30.6.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray B. E., Mederski-Samaroj B. Transferable beta-lactamase. A new mechanism for in vitro penicillin resistance in Streptococcus faecalis. J Clin Invest. 1983 Sep;72(3):1168–1171. doi: 10.1172/JCI111042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray B. E., Mederski-Samoraj B., Foster S. K., Brunton J. L., Harford P. In vitro studies of plasmid-mediated penicillinase from Streptococcus faecalis suggest a staphylococcal origin. J Clin Invest. 1986 Jan;77(1):289–293. doi: 10.1172/JCI112289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson J. E., Colodny S. M., Zervos M. J. Serious infection due to beta-lactamase-producing Streptococcus faecalis with high-level resistance to gentamicin. J Infect Dis. 1988 Nov;158(5):1144–1145. doi: 10.1093/infdis/158.5.1144. [DOI] [PubMed] [Google Scholar]
- Patterson J. E., Masecar B. L., Kauffman C. A., Schaberg D. R., Hierholzer W. J., Jr, Zervos M. J. Gentamicin resistance plasmids of enterococci from diverse geographic areas are heterogeneous. J Infect Dis. 1988 Jul;158(1):212–216. doi: 10.1093/infdis/158.1.212. [DOI] [PubMed] [Google Scholar]
- Patterson J. E., Masecar B. L., Zervos M. J. Characterization and comparison of two penicillinase-producing strains of Streptococcus (Enterococcus) faecalis. Antimicrob Agents Chemother. 1988 Jan;32(1):122–124. doi: 10.1128/aac.32.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson J. E., Zervos M. J. Susceptibility and bactericidal activity studies of four beta-lactamase-producing enterococci. Antimicrob Agents Chemother. 1989 Feb;33(2):251–253. doi: 10.1128/aac.33.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber D. A., Goering R. V. Tn4201, a beta-lactamase transposon in Staphylococcus aureus. Antimicrob Agents Chemother. 1988 Aug;32(8):1164–1169. doi: 10.1128/aac.32.8.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y., Kessler R. E., Shaw J. H., Lopatin D. E., An F., Clewell D. B. Plasmid content of Streptococcus faecalis strain 39-5 and identification of a pheromone (cPD1)-induced surface antigen. J Gen Microbiol. 1983 Apr;129(4):1207–1215. doi: 10.1099/00221287-129-4-1207. [DOI] [PubMed] [Google Scholar]
- Zervos M. J., Kauffman C. A., Therasse P. M., Bergman A. G., Mikesell T. S., Schaberg D. R. Nosocomial infection by gentamicin-resistant Streptococcus faecalis. An epidemiologic study. Ann Intern Med. 1987 May;106(5):687–691. doi: 10.7326/0003-4819-106-5-687. [DOI] [PubMed] [Google Scholar]