Abstract
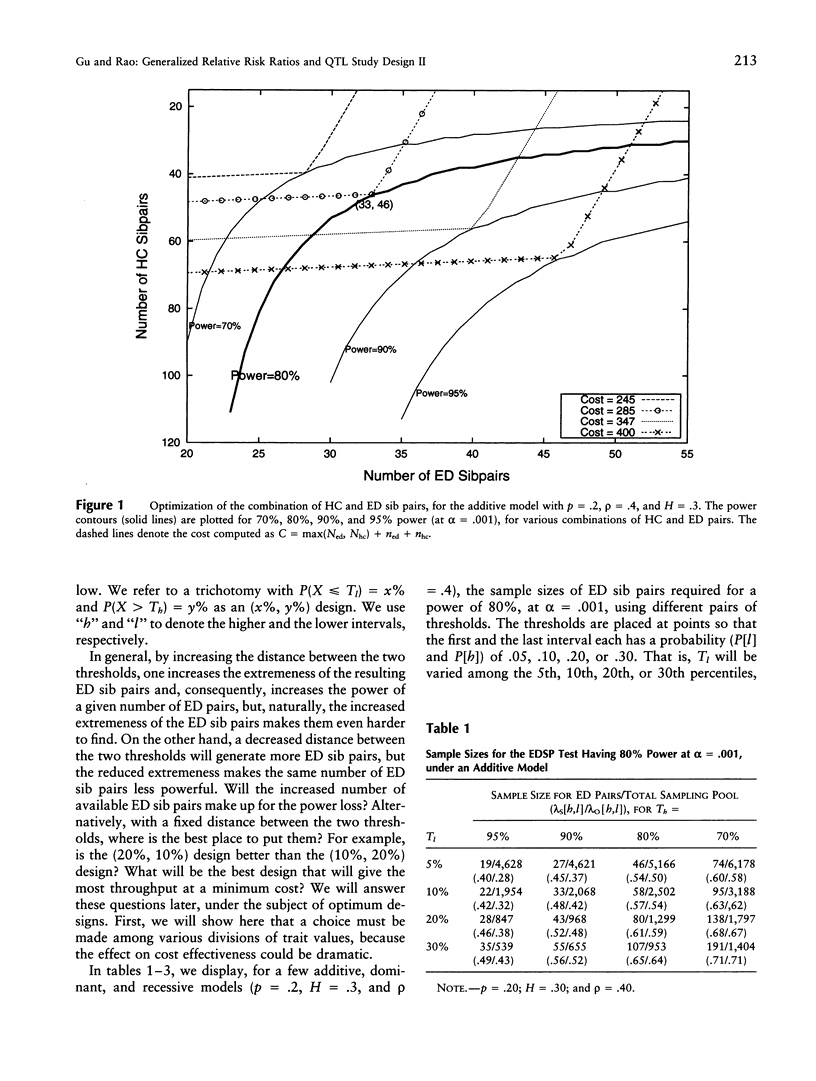
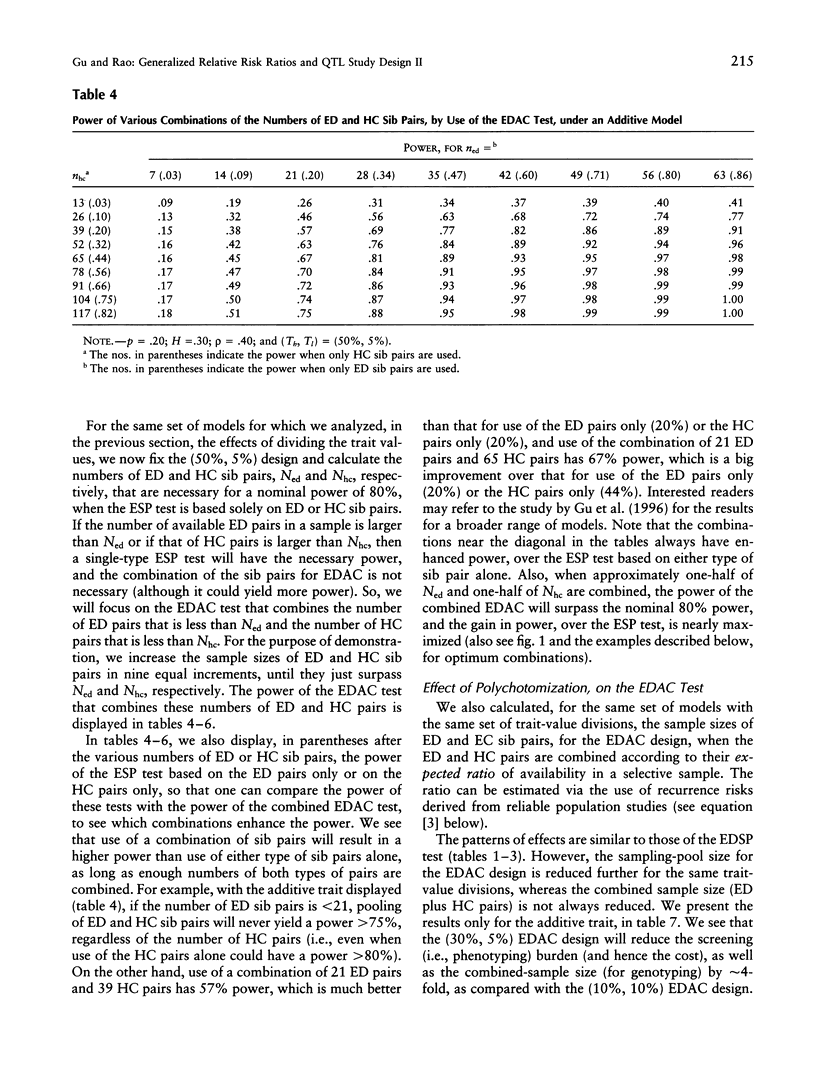
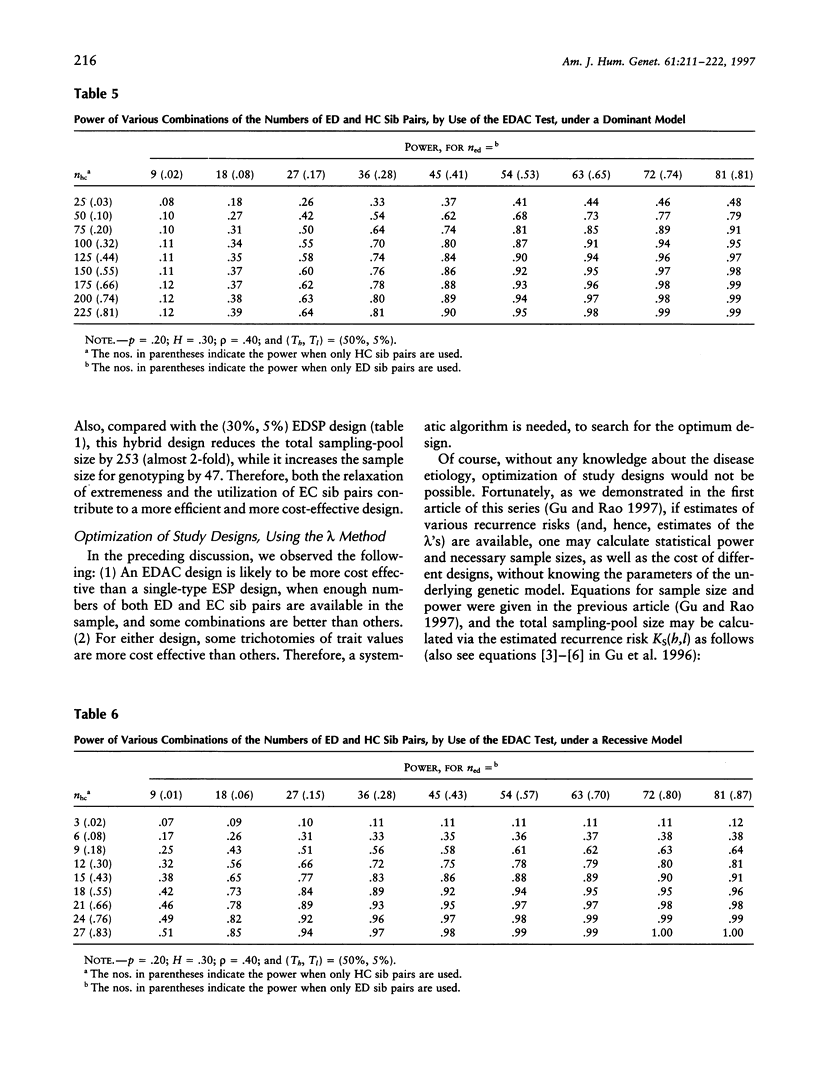
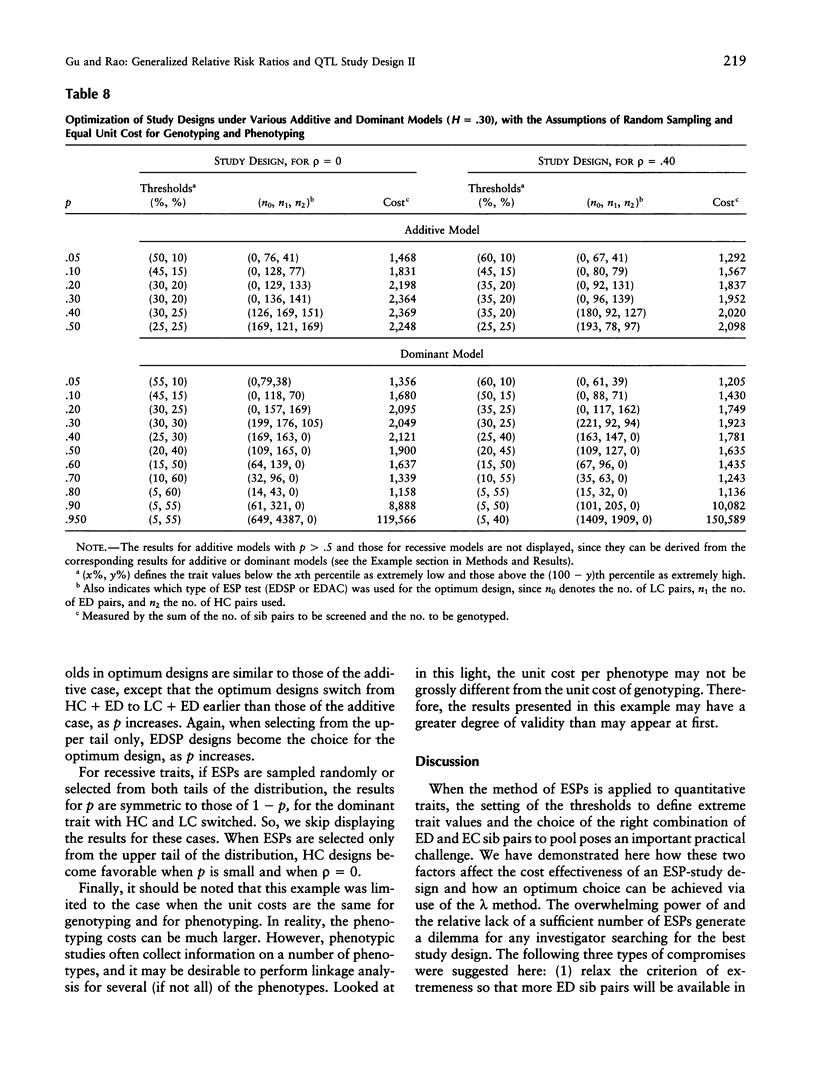
We are concerned here with practical issues in the application of extreme sib-pair (ESP) methods to quantitative traits. Two important factors-namely, the way extreme trait values are defined and the proportions in which different types of ESPs are pooled, in the analysis-are shown to determine the power and the cost effectiveness of a study design. We found that, in general, combining reasonable numbers of both extremely discordant and extremely concordant sib pairs that were available in the sample is more powerful and more cost effective than pursuing only a single type of ESP. We also found that dividing trait values with a less extreme threshold at one end or at both ends of the trait distribution leads to more cost-effective designs. The notion of generalized relative risk ratios (the lambda methods, as described in the first part of this series of two articles) is used to calculate the power and sample size for various choices of polychotomization of trait values and for the combination of different types of ESPs. A balance then can be struck among these choices, to attain an optimum design.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blackwelder W. C., Elston R. C. A comparison of sib-pair linkage tests for disease susceptibility loci. Genet Epidemiol. 1985;2(1):85–97. doi: 10.1002/gepi.1370020109. [DOI] [PubMed] [Google Scholar]
- Carey G., Williamson J. Linkage analysis of quantitative traits: increased power by using selected samples. Am J Hum Genet. 1991 Oct;49(4):786–796. [PMC free article] [PubMed] [Google Scholar]
- Eaves L., Meyer J. Locating human quantitative trait loci: guidelines for the selection of sibling pairs for genotyping. Behav Genet. 1994 Sep;24(5):443–455. doi: 10.1007/BF01076180. [DOI] [PubMed] [Google Scholar]
- Gu C., Rao D. C. A linkage strategy for detection of human quantitative-trait loci. I. Generalized relative risk ratios and power of sib pairs with extreme trait values. Am J Hum Genet. 1997 Jul;61(1):200–210. doi: 10.1086/513908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C., Todorov A., Rao D. C. Combining extremely concordant sibpairs with extremely discordant sibpairs provides a cost effective way to linkage analysis of quantitative trait loci. Genet Epidemiol. 1996;13(6):513–533. doi: 10.1002/(SICI)1098-2272(1996)13:6<513::AID-GEPI1>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Risch N., Zhang H. Extreme discordant sib pairs for mapping quantitative trait loci in humans. Science. 1995 Jun 16;268(5217):1584–1589. doi: 10.1126/science.7777857. [DOI] [PubMed] [Google Scholar]
- Zhang H., Risch N. Mapping quantitative-trait loci in humans by use of extreme concordant sib pairs: selected sampling by parental phenotypes. Am J Hum Genet. 1996 Oct;59(4):951–957. [PMC free article] [PubMed] [Google Scholar]


