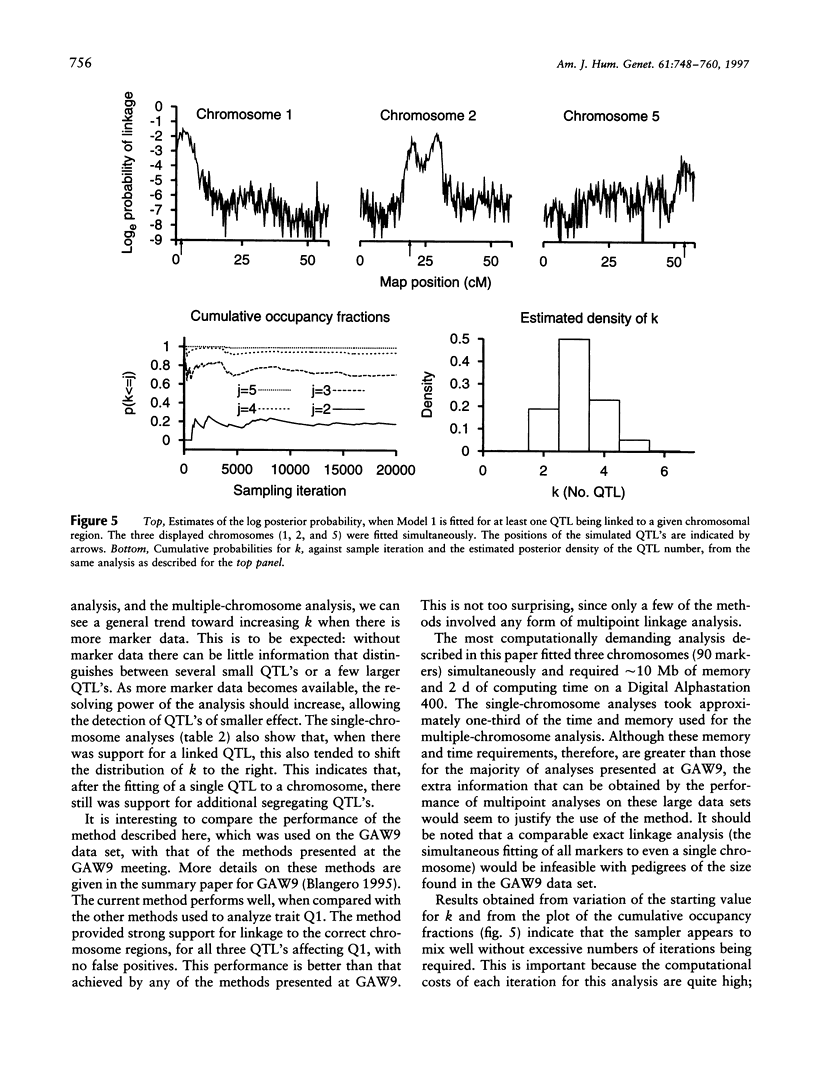
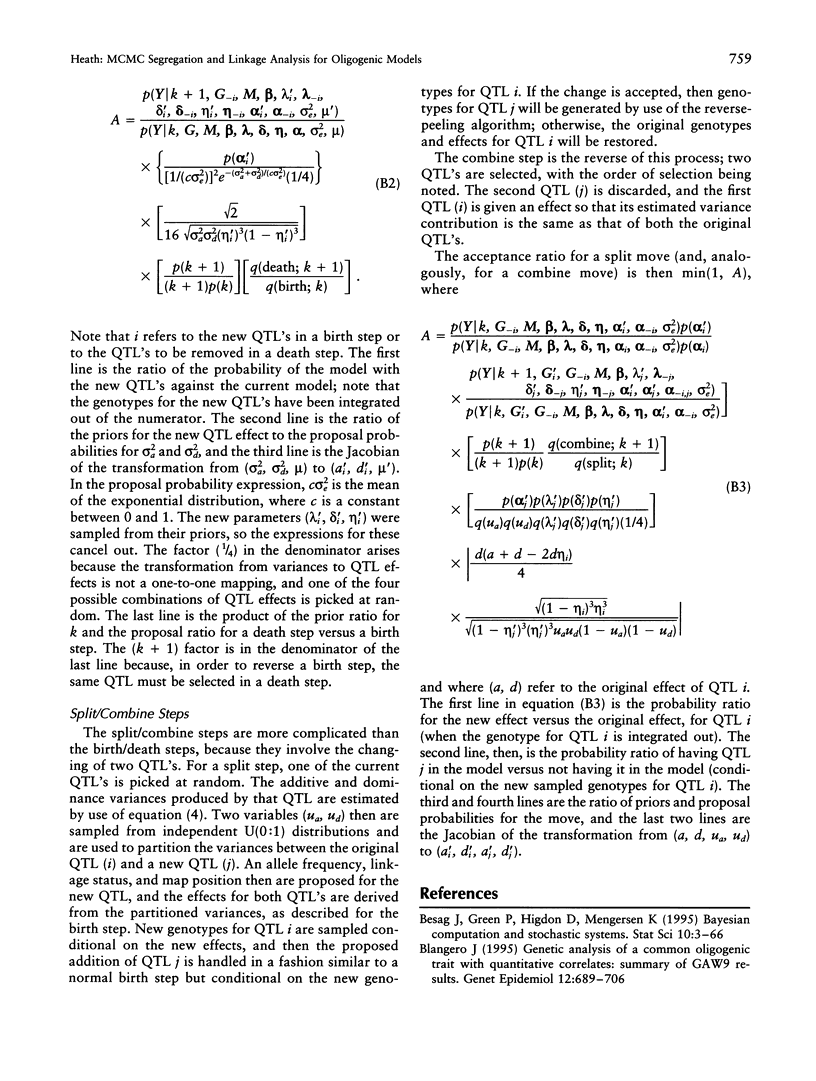
Abstract
A new method for segregation and linkage analysis, with pedigree data, is described. Reversible jump Markov chain Monte Carlo methods are used to implement a sampling scheme in which the Markov chain can jump between parameter subspaces corresponding to models with different numbers of quantitative-trait loci (QTL's). Joint estimation of QTL number, position, and effects is possible, avoiding the problems that can arise from misspecification of the number of QTL's in a linkage analysis. The method is illustrated by use of a data set simulated for the 9th Genetic Analysis Workshop; this data set had several oligogenic traits, generated by use of a 1,497-member pedigree. The mixing characteristics of the method appear to be good, and the method correctly recovers the simulated model from the test data set. The approach appears to have great potential both for robust linkage analysis and for the answering of more general questions regarding the genetic control of complex traits.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blangero J. Genetic analysis of a common oligogenic trait with quantitative correlates: summary of GAW9 results. Genet Epidemiol. 1995;12(6):689–706. doi: 10.1002/gepi.1370120628. [DOI] [PubMed] [Google Scholar]
- Dizier M. H., Bonaïti-Pellié C., Clerget-Darpoux F. Conclusions of segregation analysis for family data generated under two-locus models. Am J Hum Genet. 1993 Dec;53(6):1338–1346. [PMC free article] [PubMed] [Google Scholar]
- Elston R. C., Stewart J. A general model for the genetic analysis of pedigree data. Hum Hered. 1971;21(6):523–542. doi: 10.1159/000152448. [DOI] [PubMed] [Google Scholar]
- Guo S. W., Thompson E. A. A Monte Carlo method for combined segregation and linkage analysis. Am J Hum Genet. 1992 Nov;51(5):1111–1126. [PMC free article] [PubMed] [Google Scholar]
- Haley C. S., Knott S. A. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity (Edinb) 1992 Oct;69(4):315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- Kruglyak L., Daly M. J., Lander E. S. Rapid multipoint linkage analysis of recessive traits in nuclear families, including homozygosity mapping. Am J Hum Genet. 1995 Feb;56(2):519–527. [PMC free article] [PubMed] [Google Scholar]
- Lander E. S., Green P. Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Thompson E., Wijsman E. Achieving irreducibility of the Markov chain Monte Carlo method applied to pedigree data. IMA J Math Appl Med Biol. 1993;10(1):1–17. doi: 10.1093/imammb/10.1.1. [DOI] [PubMed] [Google Scholar]
- MacCluer J. W., Blangero J., Dyer T. D., Kammerer C. M. Simulation of a common oligogenic disease with quantitative risk factors. GAW9 problem 2: the answers. Genet Epidemiol. 1995;12(6):707–712. doi: 10.1002/gepi.1370120629. [DOI] [PubMed] [Google Scholar]
- Ott J. Computer-simulation methods in human linkage analysis. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4175–4178. doi: 10.1073/pnas.86.11.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satagopan J. M., Yandell B. S., Newton M. A., Osborn T. C. A bayesian approach to detect quantitative trait loci using Markov chain Monte Carlo. Genetics. 1996 Oct;144(2):805–816. doi: 10.1093/genetics/144.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan N., Thomas A. On the irreducibility of a Markov chain defined on a space of genotype configurations by a sampling scheme. Biometrics. 1993 Mar;49(1):163–175. [PubMed] [Google Scholar]
- Stephens D. A., Smith A. F. Bayesian inference in multipoint gene mapping. Ann Hum Genet. 1993 Jan;57(Pt 1):65–82. doi: 10.1111/j.1469-1809.1993.tb00887.x. [DOI] [PubMed] [Google Scholar]
- Thomas A. Approximate computation of probability functions for pedigree analysis. IMA J Math Appl Med Biol. 1986;3(3):157–166. doi: 10.1093/imammb/3.3.157. [DOI] [PubMed] [Google Scholar]


