Abstract
The exon/intron structure of eukaryotic genes differs extensively across species, but the mechanisms and relative rates of intron loss and gain are still poorly understood. Here, we used whole-genome sequence alignments of human, mouse, rat, and dog to perform a genome-wide analysis of intron loss and gain events in >17,000 mammalian genes. We found no evidence for intron gain and 122 cases of intron loss, most of which occurred within the rodent lineage. The majority (68%) of the deleted introns were extremely small (<150 bp), significantly smaller than average. The intron losses occurred almost exclusively within highly expressed, housekeeping genes, supporting the hypothesis that intron loss is mediated via germline recombination of genomic DNA with intronless cDNA. This study constitutes the largest scale analysis for intron dynamics in vertebrates to date and allows us to confirm and extend several hypotheses previously based on much smaller samples. Our results in mammals show that intron gain has not been a factor in the evolution of gene structure during the past 95 Myr and has likely been restricted to more ancient history.
Reconstructing the evolutionary history of spliceosomal introns remains one of the most fervently debated topics in eukaryotic evolution (Roy and Gilbert 2006). The long-standing debate over the introns-early versus introns-late hypotheses (Stoltzfus 1994; de Souza et al. 1998) contrasts the ideas of introns either originating in the early RNA world or evolving from an expansion of group II self-splicing introns in an early eukaryotic ancestor (Cavalier-Smith 1991). Understanding the natural history of introns is essential to understanding their function: Are introns simply selfish DNA elements that have been maintained in large genomes akin to retrotransposons, or do they serve a function, such as promoting recombination (Comeron and Kreitman 2000) and alternative splicing (AS) (Kim et al. 2004), resulting in increased proteome diversity and complexity?
Evolutionary investigations of the dynamics of intron gains and losses are generally hampered by the limited availability of high-quality data on the sequence and structure of gene orthologs from diverse species. To date, we have been unable to utilize the entire gene complements of most organisms in question, and the data sets commonly used range from hundreds (Rogozin et al. 2003) to at most a thousand genes (Roy et al. 2003) or several thousand introns (Nielsen et al. 2004).
Here, we make use of the complete, high-quality genomic sequences of four mammalian species—human, mouse, rat, and dog—to investigate intron gain and loss dynamics in mammals. We utilize a gene mapping technique to map annotated reference human genes onto genome-wide, multispecies sequence alignments, allowing us to investigate the predicted intron-exon boundaries of 152,146 introns within 17,242 autosomal genes. A recent study that considered a much smaller number of mammalian genes (Roy et al. 2003) uncovered six differences in intron positions between human and rodents, and suggested that there is no evidence for intron gain, and a very slow rate of intron loss in mammals. Here, we detect >100 cases of intron loss and still no evidence for any intron gain during mammalian evolution. Our large sample size allows us to determine the relative rates of intron losses in mammalian lineages and characterize the types of introns and genes that appear susceptible to loss, providing us with new insight regarding the mechanisms of intron deletion.
Results
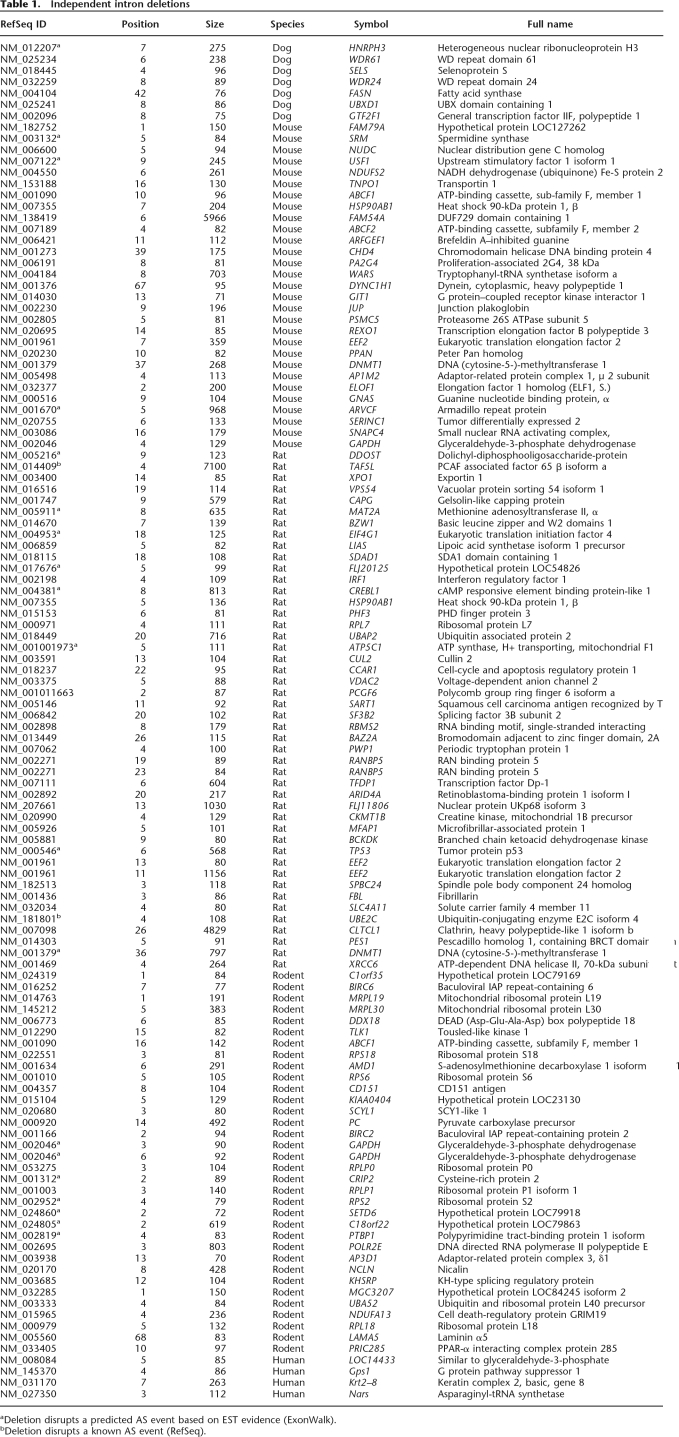
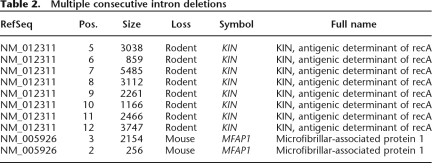
We used the mapping of annotated human exon–intron boundaries onto the mouse, rat, and dog genomes to detect changes in gene architecture that occurred during the evolution of the four mammalian species. This approach makes use of the highest quality gene annotation (17,242 human genes), but it allows us only to detect either intron loss events that occurred in rodent and dog or intron gain events that occurred in the human lineage. Thus, we also employed the reverse approach: mapping known mouse genes onto mouse/human whole-genome alignments. The latter strategy results in a slightly smaller data set (16,068 mouse genes) but allows us to detect intron losses in the human and intron gains in the mouse genome. The results of the combined analyses are listed in Tables 1 and 2. The name and symbol correspond to the human RefSeq gene where the loss/gain event occurred, except for the events in human, where the symbol refers to the mouse gene. The length for dog, mouse, and rat events is the length of the corresponding intron in human. For human events, the length corresponds to the mouse intron. We classified the results into isolated events (Table 1), i.e., those where a single intron gain/loss event (or multiple nonconsecutive events) occurred in a gene, and concerted events (Table 2), where the change involved multiple successive introns from the same gene. We propose that the single and multiple events may be mediated by slightly different mechanisms (see Discussion), and the two classes were henceforth analyzed separately.
Table 1.
Independent intron deletions
Table 2.
Multiple consecutive intron deletions
We were able to uncover a total of 120 isolated changes: four occurring in human, 29 in mouse, 46 in rat, 34 in the rodent lineage prior to the mouse/rat divergence, and seven in dog. Remarkably, all of the changes were consistent with a loss, rather than a gain of an intron; i.e., for each case of a deletion of an intron relative to the reference gene structure (either mouse or human), the annotated intron was present in an earlier diverged organism. The loss of each intron was verified by using dog as the outgroup for changes occurring in human, mouse, or rat and by using chicken as the outgroup for changes occurring in dog.
Figure 1 shows an example of an intron deletion event occurring in mouse displayed in the vertebrate MultiZ alignment track of the UCSC Genome Browser. This case illustrates common misalignments close to the splice sites, which is the reason we allowed for a 25-bp margin of error in the distance between exon edges in the target species during the search for intron loss and gain (see Methods). To confirm each gain/loss event, we extracted the original genomic sequences from the assemblies and used ClustalW to realign the reference species intron and 100 bp of flanking upstream and downstream sequences with the homologous target species region. The data for all the alignments are available in Supplemental data online. Our analysis shows that at least 117 of the detected intron losses are exact. The remaining three cases are also likely to be exact losses but fall into regions of relatively poor quality genomic sequence and require single base insertion/deletion events in the alignments.
Figure 1.
An example of intron loss in the mouse ortholog of the human DYNC1H1 gene, visualized in the UCSC Genome Browser display of multispecies alignments. Uppercase, boxed sequences correspond to exons. Note that the alignment is inexact at the splice sites, resulting in an artifactual 3-bp intron length in mouse, which necessitates an approximate search strategy (described in Methods), and realignment of sequences using an appropriate parameter choice in order to confirm all candidate intron deletions. Realigned sequences of the introns and neighboring exons for all 120 cases of intron loss are provided in Supplemental data.
Rates of intron loss/gain
We find a very low rate of intron loss throughout the mammalian evolution and no evidence for intron gain. Based on the total number of donor/acceptor splice site pairs identified in the alignments (146,964, 141,942, 146,727, and 124,474 for mouse, rat, dog, and human, respectively), we determined the rates for intron loss per million years per intron as follows: 5.32 × 10−6 for the mouse–rat common ancestor, 6.58 × 10−6 for mouse, 1.08 × 10−5 for rat, 5.30 × 10−7 for dog, and 4.28 × 10−7 for human. These estimates assume that human and dog lineages diverged 95 Mya, human and rodent 75 Mya (Waterston et al. 2002), and mouse and rat 30 Mya (Nei et al. 2001; Springer et al. 2003). In order to assess whether the rates are proportional to generation time, we multiplied these rates by the age of sexual maturity of each organism (1⁄6, 1⁄3, 3, and 12 yr for mouse, rat, dog, and human) and normalized the resulting figures, so that the rate for human is equal to one. (Note that we are making a somewhat simplistic assumption that the ages behaved proportionally during the evolution of each lineage). We obtain ratios of 0.21, 0.70, 0.31, and 1 for mouse, rat, dog, and human, respectively. It appears that generation time is not the only factor affecting the rate of intron loss. Other possible factors include the relative activity of reverse transcriptase within each lineage. We note that the rat genome has a higher density of LINE elements, which encode their own reverse transcriptase, than the mouse genome (data not shown), possibly resulting in higher amounts of reverse-transcribed cDNA available for recombination.
Projected sizes of deleted introns
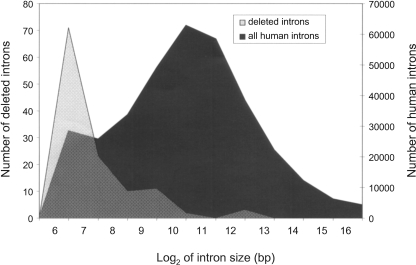
One of the most striking characteristics distinguishing the deleted introns is their extremely small size. The mean size of a human intron is 6259 bp, while the deleted cases were on average 355 bases long (in human). Figure 2 illustrates the difference in projected size distribution of deleted introns and that of all introns. The difference in the distributions is highly statistically significant (Student’s t-test assuming unequal variances, T = −57.3, df = 208, two-tailed P < 10−10). Most of the deleted introns (81 out of 120) are <150 bases. We further investigated five cases of unusual intron deletions that exceeded 1000 bp in length (5968, 7100, 1030, 1158, and 4380 nucleotides in genes FAM54A, TAF5L, FLJ11806, EEF2, and CLTCL1, respectively). Four of those cases occurred in the rat lineage and one in mouse. We identified the corresponding intron in the closest relative (mouse, in case the loss occurred in rat, and vice versa) and observed that the introns in the closest relative were actually considerably shorter than in human (984, 4902, 224, 134, and 79 bases, respectively) and hence were likely to be short at the time of their deletion. This suggests that the size of the intron must be an important factor affecting the underlying molecular mechanism of deletion.
Figure 2.
Log2(size) distribution of all introns (black) versus deleted introns (gray). The deleted introns are unusually short and much shorter than the human genome average.
Intron phases
Introns can be classified as phase 0 (inserted between two codons), phase 1 (after the first base of a codon), or phase 2 (after the second base). We examined the phase distribution of the 116 deleted introns present in human from Table 1 and compared it to the phase distribution of all introns from the RefSeq data set. The proportions for the deleted introns were 0.52, 0.26, and 0.22 for phases 0, 1, and 2, respectively, while the ratios for the genome average were 0.46, 0.32, and 0.22. The distribution of phases of deleted introns did not differ significantly from the expected (χ2 = 2.24, df = 2, P = 0.33). Intron deletions in mammals appear to occur randomly with respect to their phase. This finding contrasts with some earlier observations of phase 0–biased intron loss (Lynch 2002; Roy and Gilbert 2005b).
Positions of deleted introns within genes
We defined the relative position of each intron within a gene as the position, n, of the intron (measured from the 5′ end of the CDS; i.e., for the first CDS intron, n = 1), divided by the total number of CDS introns, N. In order to obtain a symmetrical distribution centered about 1⁄2, we subtracted 1⁄2 from the numerator. Hence, the adjusted relative position, (n − 1⁄2)/N has a range between 1/(2N) and 1 − 1/(2N), and an expectation of 1⁄2. We used a χ2 test to compare the proportion of deletions in the 5′ half of the gene versus the 3′ half. We found that the positions of deleted introns were significantly skewed toward the 3′ half of each gene (χ2 = 7.76, df = 1, P = 0.0053). Seventy-three of the intron losses occurred closer to the 3′ end of genes, compared with 43 that were closer to the 5′ end.
Splice site characteristics
We examined the distributions of bases around both splice sites and compared them with the distributions for all introns. We found that the consensus at the 5′ splice site was not significantly different from the control. However, at the 3′ splice site, the two positions following the acceptor AG dinucleotide had a significantly greater frequency of the bases G (χ2 = 3.82, df = 1, P = 0.05) and T (χ2 = 4.93, df = 1, P = 0.03), respectively. Since the GT at the 3′ splice site is very often preceded by an AG, this stronger consensus sequence may have served to promote a recombination event occurring between the two splice sites, leading to the deletion of the intron.
Expression patterns and ontology of genes undergoing intron loss
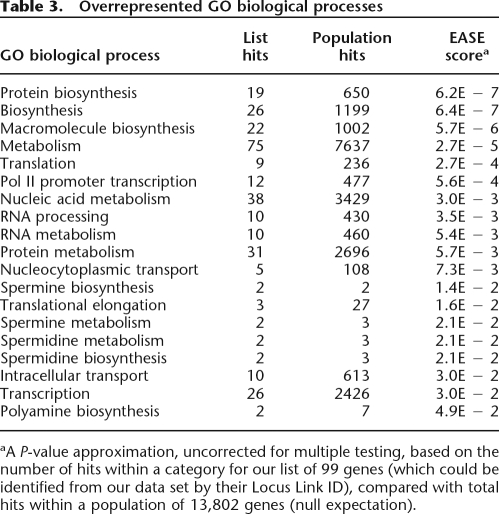
We used the EASE (Hosack et al. 2003) interface to classify our genes into GO categories (biological process) and characterize the types of genes that undergo intron deletion events. EASE calculates overrepresentation statistics for each GO category using an EASE score, which approximates a P-value by using the upper bound of the distribution of Jackknife Fisher exact probabilities. In Table 3, we list the most overrepresented biological processes (EASE score < 0.05). We note that most of the genes with intron deletions are involved in biosynthesis, metabolism, translation, transcription, and RNA processing. All of the overrepresented categories correspond to ubiquitous housekeeping functions, suggesting that intron deletion events occur predominantly in genes that are both highly expressed and expressed in the germline. In order to further confirm this hypothesis, we utilized microarray expression data available from SymAtlas (Su et al. 2002) to determine the expression intensities and breadths of the candidate genes. Since germline gene expression levels are not known, we used averaged gcRMA (Robust Multichip Average with GC correction) expression over all tissues as a proxy of germline expression (Majewski 2003) and compared the averages of the intron-deleted sample to all genes. The average gcRMA expression level was of 952 overall, and significantly higher, 9560, for the genes with intron deletion (Student’s t = −4.3, df = 108, P = 3.58 × 10−5). In order to study the breadth of expression, we used MASS 5.0 present/absent calls from >300 tissues and cell lines and determined the fraction of tissues where expression was positively detected (present). Again, we compared the expression breadth for genes with intron deletions (0.54) to that of all genes (0.26) and found a highly significant difference (t = −6.9, df = 92, P = 7.15 × 10−10). Thus intron deletions occur preferentially in genes with housekeeping functions, which have experimentally been determined to be both highly and broadly expressed.
Table 3.
Overrepresented GO biological processes
aA P-value approximation, uncorrected for multiple testing, based on the number of hits within a category for our list of 99 genes (which could be identified from our data set by their Locus Link ID), compared with total hits within a population of 13,802 genes (null expectation).
Genes experiencing frequent intron loss: GAPDH
We performed a detailed analysis of the GAPDH gene, where we found evidence of multiple, independent intron losses occurring in mouse, human, and rat. GAPDH is a known, very highly expressed housekeeping gene, which supports the hypothesis that expression in the germline is essential for intron loss. We extracted genomic DNA and mRNA GAPDH sequences for 15 vertebrate species and used multiple sequence alignment to reconstruct the intron/exon structure of the gene in each species (Fig. 3). A Dollo parsimony approach (assuming a single appearance of the derived character—intron) suggests that there were no gain events throughout vertebrates but numerous losses, including several independent losses of the same intron (intron 9 of the ancestral gene). The result also suggests that the phenomenon of intron loss in vertebrates (at least within this gene) may be accelerated in the mammalian branch.
Figure 3.
The evolution of the intron–exon structure of the GAPDH gene throughout the vertebrate phylogeny. The numbers on the branches indicate the inferred deletion events. The introns are numbered according to their position within the coding sequence of the ancestral gene.
Does intron loss disrupt AS?
We identified two cases of intron losses disrupting known (RefSeq-confirmed) AS events that alter the predicted amino acid sequence of the gene (for details, see footnote to Table 1). We also detected 20 losses that disrupt predicted (EST-based) events alternatively processed in human. If intron losses occurred randomly, without any regard to preserving AS, the expected number, based on all the RefSeq introns used in this analysis is a disruption of four known events and 17 predicted events. It is unexpected that the number of observed losses that disrupt predicted AS events is actually slightly greater than the null expectation. However, since our ability to predict AS events is highly dependent on the availability of mRNAs and ESTs, and the set of genes undergoing intron losses is extremely highly and broadly expressed, there is likely to be a bias in the annotation of the deleted sample. That is, because of their high expression levels, genes experiencing intron loss have deeper EST coverage and are better annotated with respect to AS than the genome average.
In view of the annotation bias, it is difficult to conclude whether the disrupted predicted AS events are truly functional or constitute an artifact of deep EST coverage and the presence of inadvertent splicing errors. It is also possible that, since AS may be only weakly conserved across species (Pan et al. 2005), a predicted disruption of AS in humans may have no effect on AS in the species where the deletion occurred.
Discussion
We identify >100 cases of intron loss in the four examined mammalian species. Our approach, based on mapping of known human genes to whole-genome sequence alignments of multiple species, allows us to utilize the annotation information from well-studied model species, such as human and mouse, and predict gene structure in other, relatively poorly annotated species. Using our method, we recover all six intron deletion events detected in a smaller scale study (Roy et al. 2003) and extend previous conclusions regarding the patterns of intron loss in mammals. There are several remarkable characteristics of our data set: (1) losses appear to occur almost exclusively for small introns; (2) essentially all of our examples of loss are consistent with an exact deletion event; (3) the loss events are biased toward the 3′ ends of genes, but can be found at all positions; (4) genes that are associated with intron loss events are generally highly expressed and have housekeeping functions; (5) the rate of intron loss is related to (but not fully explained by) the generation time of the organisms and follows a pattern similar to spontaneous mutation; and (6) all of the differences in gene structure are consistent with intron loss events—no detectable intron insertions have occurred in human or mouse since the divergence of their lineages. Below, we discuss some implications of these findings.
Mechanism of intron loss
It has been suggested that intron loss may be mediated either by genomic deletion events or recombination of the genomic locus with a reverse-transcribed, processed mRNA molecule of the gene (Logsdon Jr. et al. 1998). Our analysis suggests that at least 98% (and possibly all) of the observed deletions are exact. In addition, we do not find any evidence for inexact deletions, which would retain a small part of the intron or remove parts of neighboring exons. It has been argued (Roy et al. 2003) that random genomic deletion events would be unlikely to always result in exact intron losses. This is even more evident in our large data set. It would be extremely unlikely that, if intron loss were generally mediated by random deletions, we would not recover any cases of inexact losses. Even in the presence of purifying selection against such potentially deleterious events, it seems plausible that some minor insertion/deletions of the boundary sequence, particularly ones that do not alter the reading frame, would be evolutionarily neutral. Thus the exact character of the detected intron loss events supports the latter model, i.e., recombination with an intronless cDNA of the gene.
The small projected size of the introns provides another insight into the mechanism of loss. It is well documented that genetic recombination events occur less frequently in the presence of mismatches, insertions, or deletions within the recombining substrates (Majewski and Cohan 1999). We propose that in the cases of intron loss, recombination with cDNA is much more likely if the introns are small, resulting in a high relative effective proportion of sequence identity.
We also find that genes susceptible to intron loss tend to be involved in housekeeping functions and expressed at relatively high levels. Again, high level of expression most likely results in relatively high levels of reverse-transcribed copies of the gene, leading to an increased probability of recombination. A similar effect has been demonstrated for the frequency of processed pseudogenes (Zhang et al. 2003). Furthermore, in order for the recombination events to result in intron losses that are transmitted to the next generation and have a chance to increase in frequency in the population, the loss events must occur in the germline, as opposed to somatic cells. Thus, germline expression of the gene would be an essential condition for intron loss. In accordance with this prediction, we find that our intron-deleted data set is highly enriched in housekeeping (ubiquitously expressed) genes. Thus both the expression levels and the expression patterns of the genes support the recombination-mediated model of intron loss.
Finally, we find that the position of the lost introns is significantly biased toward the 3′ ends of the genes. This is in accordance with recent studies of lower eukaryotes (Sverdlov et al. 2004; Roy and Gilbert 2005b) and again supports intron loss being mediated by recombination, since reverse transcription of the mRNA is believed to occur preferentially from the 3′ end (Weiner et al. 1986). However, this result may also reflect the bias in distribution of intron sizes (first introns are generally longer and more difficult to remove by recombination) and selective pressures against deleting potentially regulatory regions, which may be present close to the 5′ termini of genes (Majewski and Ott 2002).
Multiple intron losses in single genes
We identify several cases of multiple intron loss in single genes, which we classify as two distinct types of events. The first, concerted type, is exemplified by a loss of multiple successive introns: eight successive introns of the HsKin17 gene in the rodent lineage (before the rat–mouse divergence) and two introns of the MFAP1 gene. The introns lost in the concerted events are generally considerably longer than those involved in the usual single-loss cases. We propose that this type of rearrangement event occurs as a result of recombination with nearly complete cDNA, whereas in the more typical cases the recombining cDNAs may be incomplete or fragmented. The overall greater available length of substrate involved in the concerted losses may promote recombination despite the presence of long unmatched intronic regions.
The second, punctuated type of multiple losses, involves the more typical short introns and is exemplified by the GAPDH and EEF2 genes. Those genes exhibit evidence of several independent intron losses, occurring either in the same species, or distinct lineages. A similar phenomenon has been previously observed in the white gene in insects (Krzywinski and Besansky 2002). For our most extreme case, GAPDH (seven introns within the human CDS), we manually examined the genomic species alignments of other mammalian and vertebrates species, which we had not considered in the genome-wide study. We found evidence for three independent losses of the ancestral intron 9 (dog, primate/rodent lineage, possum), and a total of nine losses that occurred in this gene in mammals (Fig. 3). The fish, frog, chicken, elephant, and cow have retained the same ancestral structure. GAPDH is a known, extremely highly expressed housekeeping gene, often used as a control in gene expression studies due to its consistent elevated expression in all cell types. Consistent with general observations for highly expressed genes, GAPDH has multiple annotated pseudogenes (Kent et al. 2003; Zhang et al. 2003), further supporting the hypothesis that high levels of potentially reverse-transcribed mRNA in germ cells lead to an elevated probability of not only retrotransposition but also intron loss.
Selection favoring intron loss?
The preferential intron loss in highly expressed housekeeping genes is also consistent with selection for transcription efficiency favoring the resulting short transcript (Castillo-Davis et al. 2002). While the selection pressure and the increased likelihood of recombination in highly transcribed genes are not mutually exclusive and, in theory, may both contribute to the association of intron loss and expression levels, it seems unlikely that selection is a major force responsible for the observed intron losses. Most of the deleted introns are extremely short (∼100 bp), while much longer introns are present in the corresponding genes and had not been deleted. Selection alone would favor the loss of longer introns. In the example of the GAPDH gene, a loss of an 82-bp intron from a 3783-bp transcript would result in only a very modest 2% decrease in the time of transcription. In comparison, loss of the first intron fully contained within the CDS (∼1700 bp) could result in a 45% reduction. We propose that the availability of cDNA and the length of unmatched intronic sequences in the recombining strands are the primary limiting factors in the process of intron loss. Once the gene conversion event occurs, selection may be an additional force increasing the probability of fixation of such events. However, because of low effective population sizes, genetic drift, rather than selection (Jeffares et al. 2006; Roy and Gilbert 2006), is much more likely to be the main determinant of fixation rates in mammals.
Rates of intron loss and gain
The rate of intron loss in mammals appears extremely slow. The fastest genome-wide rate, in the rat lineage is approximately one intron loss per 1.53 Myr. This corresponds to one intron loss per 217,644 Myr, per intronic site (based on 141,942 introns identified in the human/rat comparison). We note that the rates are not clocklike and appear to be dependent on the generation time of each lineage: rodent > dog > human, but they are also likely to be affected by other factors, such as the effective population size (in the absence of selection, the rate of fixation of intron losses is expected to be inversely proportional to the population size). At the current rate, it would take >1012 yr for the human genome to shed half of its introns. Hence, intron loss/gain does not appear to be a major factor in mammalian evolution.
Since we have not detected any cases of intron gain, we estimate the process to proceed considerably slower than intron loss. Our approach would allow us to detect intron gain events occurring in the mouse/rodent lineage, after its divergence from the human lineage. Since no cases of intron gain (and 63 losses) were found, the genome-wide rate of gain is at least 60-fold slower than the rate of loss. Although some claims to intron gain in mammals have been reported (Veeramachaneni and Makalowski 2005), more recent analyses (Shepelev and Fedorov 2006), including ours, do not confirm such findings. Our observations support the premise that modern introns are evolutionarily inert and, having expanded through early eukaryotic genomes (or being inherited through earlier yet ancestors), have been gradually, albeit very slowly, disappearing within mammalian lineages over at least the past 100 Myr.
Our results in mammals bear similarity to two recent studies of intron dynamics. Cho et al. (2004)investigated closely related nematode species and found frequent and recurrent intron loss and a much slower (fivefold to 10-fold) rate of putative intron gain. Roy and Hartl (2006) studied two relatively closely related Plasmodium species and found a very slow rate of intron loss and possibly no gain during ∼100 Myr of evolution. On the other hand, these results contrast with estimates from fungi (Nielsen et al. 2004) and more distantly related eukaryotic clades (Fedorov et al. 2002; Rogozin et al. 2003; Babenko et al. 2004; Roy and Gilbert 2005c; Yoshihama et al. 2006), which suggest prevalence of intron gain over intron loss over periods >200 Myr.
The combined data on intron dynamics implies that introns have not been actively proliferating during the past 100 Myr of evolution in any of the studied species. Most of inferred intron gains must have occurred significantly earlier. However, ours and other studies in closely related species show a pattern of recurrent intron loss, particularly in highly expressed housekeeping genes that are most likely to recombine with their reverse transcribed cDNA. Because of their high degree of conservation at both sequence and functional levels, such genes are the ones most often used in gene structure comparisons (Rogozin et al. 2003; Yoshihama et al. 2006). The process of loss is likely to be much more accelerated in organisms with high reproductive rates and large population sizes where selection for reduced transcript length, rather than genetic drift, will lead to fixation of losses in populations. It is also likely to be rapid in species with short introns, where recombination with intronless substrates is more efficient. In the presence of recurrent parallel loss, studies using parsimony methods and large evolutionary distances (Rogozin et al. 2003; Coghlan and Wolfe 2004; Yoshihama et al. 2006) will certainly underestimate the rate of loss and overestimate the rate of intron gains. Even maximum likelihood approaches (Nguyen et al. 2005; Roy and Gilbert 2005a; Csuros 2006) are not fully immune to this error and will overestimate the gain rate particularly if they fail to account for the rate variation among sites (Yang 1996), which we have shown to occur in the case of intron loss. While we are still very far from resolving the debate over the age and origin of spliceosomal introns, our analysis suggests that many studies may mistake parallel intron losses for gains, and that as a result most introns may be significantly older than we currently believe.
Methods
DNA sequences and interspecies alignments
We used the RefSeq annotation of the human genomic sequence to extract coding sequences of human genes (Hinrichs et al. 2006). Only the sequences which could be in silico translated into their predicted protein were retained. This strategy resulted in a high confidence, nonredundant data set of 17,242 human autosomal genes, containing 152,146 distinct introns within their coding sequence. We based our analysis on the four available highest-quality mammalian genome assemblies: human (hg17), mouse (mm7), rat (rn3), and dog (canFam2). We mapped the well-annotated human genes onto the genome-wide alignments present within the 17-way MultiZ (Blanchette et al. 2004) alignment tracks in order to determine the intron–exon structures in the target species. We considered only introns that were flanked by coding, or partially coding, exons, since noncoding UTR sequences are poorly conserved (and often not conserved) among species and provide poor anchors for detecting splice sites within alignments. We also performed the reverse analysis by mapping a set of 16,068 mouse RefSeq genes (129,336 CDS introns) onto the mouse vs. human genomic sequence alignments.
We used the following criteria to detect intron loss events in the target sequence (or gain in the reference sequence): (1) for each reference species intron, we identified the positions of both the donor and acceptor splice sites within the MultiZ alignment; (2) within the target species, we flagged an intron as potentially lost if the distance between the donor and acceptor sites was lower than a predetermined cutoff of 25 bp. The latter condition was necessary since alignments are often imperfect at the exon–intron boundary (Fig. 1). In particular, especially in the case of intron loss events, the last 2 bp of an exon, which have an AG consensus, tend to align with the downstream intronic acceptor site (also AG), but more serious misalignments are also common. Nevertheless, allowing a margin of 25 bp did not introduce any false––positive results (as manually verified in the final curated results), since sequences <25 bp cannot be efficiently spliced in mammals (Lim and Burge 2001) and correspond to imperfect alignments, rather than actual introns. Using the above first pass search criteria, we identified 623 cases of potential intron loss/gain.
Since the genome assemblies and the resulting alignment contain numerous sequencing, assembly, and alignment artifacts, all potential intron loss events were further filtered based on the quality of the underlying alignment. In the process of constructing the BLASTZ alignments, gaps in the sequences may be filled in using secondary (non-syntenic) sequences. This significantly increases the proportion of aligned sequences but also results in an increased probability of introducing alignment errors. Thus, only potential intron loss cases that mapped to the highest confidence, top, syntenic, long (encompassing at least two neighboring genes) alignment nets (Kent et al. 2003) were retained for further analysis. Cases occurring in genes that were aligned to multiple or nonsyntenic portions of target genomes, which could potentially constitute alignments to duplicate genes or pseudogenes, were rejected. This strategy resulted in 157 cases of intron loss/gain, of which 35 occurred in both rat and mouse, for a total of 122 events.
For all the candidates, we extracted the sequence of 100 bp flanking the intronic site from the genomic sequence assembly and used ClustalW (Thompson et al. 1994) with high gap opening penalty (80) and low gap extension penalty (zero) to align it to the human intron-containing sequence and visualize the detailed evidence for intron loss. After performing some minor supervised adjustments, mainly correcting the misalignment of the terminal AG of an upstream exon with the downstream acceptor site (see above), this allowed us to confirm the deletion events and demonstrate that essentially all of the events are cases of exact deletion, with no alteration to the coding sequence. All of the 120 isolated intron loss/gain events were successfully validated using the sequence alignments (Supplemental data).
Characterization of genes involved in loss events
In order to functionally classify the genes involved in intron loss events, we used the EASE (Hosack et al. 2003) interface to the Gene Ontology annotation. We identified the GO categories with the highest support—lowest EASE score—for overrepresentation by the genes within our list, compared with all known genes.
In order to approximate expression levels and expression breadth of the genes, we used microarray expression data from SymAtlas (Su et al. 2002). Although the relevant variable is the expression level in the germline, this information is currently not available. As a proxy for gene expression levels, we used the mean values of gcRMA summaries across all tissues studied. Note that because of developmental history of germ cells, testes- and ovary-specific expression levels may not be the appropriate indicator of germline expression, and a global average expression may provide a better estimate (Majewski 2003), particularly in the case of housekeeping genes. As an estimate of expression breadth, we used the present/absent calls from the MASS 5.0 summaries and, for each gene, calculated the percentage of tissues where expression was detected.
Intron loss and AS
In order to study the relationship of intron loss and AS, we cross-referenced the set of lost intron positions with alternative gene isoforms present in the RefSeq data set. We identified all the introns where the deletion in the target species disrupts a known AS event in human. For example, deletion of an intron would prevent alternative usage of the adjacent exons (cassette events), as well as alternative (cryptic) splice site usage of the adjacent splice sites. We further limited the AS events of interest to only those that altered the predicted amino acid sequence of the gene. In order to obtain a background genome-wide estimate for the probability of any intron loss disrupting AS, we also determined how many introns from our entire input data set border alternatively spliced exons that would be disrupted by a deletion.
While the RefSeq set of genes is manually curated and highly accurate, it contains relatively few alternatively spliced isoforms. Hence we also analyzed predicted AS events from the ExonWalk annotation of the UCSC Database. Briefly, the ExonWalk program merges EST and cDNA evidence together to predict full-length isoforms, including alternative transcripts. To predict transcripts that are biologically functional, rather than the result of technical or biological noise, ExonWalk requires that every intron and exon be: (1) present in cDNA libraries of another organism (i.e., also present in mouse), (2) have three separate cDNA GenBank entries supporting it, or (3) be evolving like a coding exon as determined by the Exoniphy program (Siepel and Haussler 2004). Once the transcripts are predicted an open reading frame finder is used to find the best open reading frame. Transcripts that are targets for nonsense mediated decay are filtered. We further filtered out all predicted transcripts that did not begin with an ATG and did not end with a stop codon.
Acknowledgments
This research was supported by funds from the Canadian Institute of Health Research and the Canada Research Chairs program.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5703406
References
- Babenko V.N., Rogozin I.B., Mekhedov S.L., Koonin E.V., Rogozin I.B., Mekhedov S.L., Koonin E.V., Mekhedov S.L., Koonin E.V., Koonin E.V. Prevalence of intron gain over intron loss in the evolution of paralogous gene families. Nucleic Acids Res. 2004;32:3724–3733. doi: 10.1093/nar/gkh686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Rosenbloom K., Clawson H., Green E.D., Clawson H., Green E.D., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis C.I., Mekhedov S.L., Hartl D.L., Koonin E.V., Kondrashov F.A., Mekhedov S.L., Hartl D.L., Koonin E.V., Kondrashov F.A., Hartl D.L., Koonin E.V., Kondrashov F.A., Koonin E.V., Kondrashov F.A., Kondrashov F.A. Selection for short introns in highly expressed genes. Nat. Genet. 2002;31:415–418. doi: 10.1038/ng940. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Intron phylogeny: A new hypothesis. Trends Genet. 1991;7:145–148. [PubMed] [Google Scholar]
- Cho S., Jin S.W., Cohen A., Ellis R.E., Jin S.W., Cohen A., Ellis R.E., Cohen A., Ellis R.E., Ellis R.E. A phylogeny of Caenorhabditisreveals frequent loss of introns during nematode evolution. Genome Res. 2004;14:1207–1220. doi: 10.1101/gr.2639304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan A., Wolfe K.H., Wolfe K.H. Origins of recently gained introns in Caenorhabditis. Proc. Natl. Acad. Sci. 2004;101:11362–11367. doi: 10.1073/pnas.0308192101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J.M., Kreitman M., Kreitman M. The correlation between intron length and recombination in Drosophila. Dynamic equilibrium between mutational and selective forces. Genetics. 2000;156:1175–1190. doi: 10.1093/genetics/156.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csuros M. On the estimation of intron evolution. PLoS Comput. Biol. 2006;2:e84. e83. doi: 10.1371/journal.pcbi.0020084. author reply. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza S.J., Long M., Klein R.J., Roy S., Lin S., Gilbert W., Long M., Klein R.J., Roy S., Lin S., Gilbert W., Klein R.J., Roy S., Lin S., Gilbert W., Roy S., Lin S., Gilbert W., Lin S., Gilbert W., Gilbert W. Toward a resolution of the introns early/late debate: Only phase zero introns are correlated with the structure of ancient proteins. Proc. Natl. Acad. Sci. 1998;95:5094–5099. doi: 10.1073/pnas.95.9.5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov A., Merican A.F., Gilbert W., Merican A.F., Gilbert W., Gilbert W. Large-scale comparison of intron positions among animal, plant, and fungal genes. Proc. Natl. Acad. Sci. 2002;99:16128–16133. doi: 10.1073/pnas.242624899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Diekhans M., Furey T.S., Harte R.A., Hsu F., Furey T.S., Harte R.A., Hsu F., Harte R.A., Hsu F., Hsu F., et al. The UCSC Genome Browser Database: Update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosack D.A., Dennis G., Jr., Sherman B.T., Lane H.C., Lempicki R.A., Dennis G., Jr., Sherman B.T., Lane H.C., Lempicki R.A., Sherman B.T., Lane H.C., Lempicki R.A., Lane H.C., Lempicki R.A., Lempicki R.A. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffares D.C., Mourier T., Penny D., Mourier T., Penny D., Penny D. The biology of intron gain and loss. Trends Genet. 2006;22:16–22. doi: 10.1016/j.tig.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Kent W.J., Baertsch R., Hinrichs A., Miller W., Haussler D., Baertsch R., Hinrichs A., Miller W., Haussler D., Hinrichs A., Miller W., Haussler D., Miller W., Haussler D., Haussler D. Evolution’s cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Klein R., Majewski J., Ott J., Klein R., Majewski J., Ott J., Majewski J., Ott J., Ott J. Estimating rates of alternative splicing in mammals and invertebrates. Nat. Genet. 2004;36:915–916. 916–917. doi: 10.1038/ng0904-915. author reply. [DOI] [PubMed] [Google Scholar]
- Krzywinski J., Besansky N.J., Besansky N.J. Frequent intron loss in the white gene: A cautionary tale for phylogeneticists. Mol. Biol. Evol. 2002;19:362–366. doi: 10.1093/oxfordjournals.molbev.a004091. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Burge C.B., Burge C.B. A computational analysis of sequence features involved in recognition of short introns. Proc. Natl. Acad. Sci. 2001;98:11193–11198. doi: 10.1073/pnas.201407298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon J.M., Jr., Stoltzfus A., Doolittle W.F., Stoltzfus A., Doolittle W.F., Doolittle W.F. Molecular evolution: Recent cases of spliceosomal intron gain? Curr. Biol. 1998;8:R560–R563. doi: 10.1016/s0960-9822(07)00361-2. [DOI] [PubMed] [Google Scholar]
- Lynch M. Intron evolution as a population-genetic process. Proc. Natl. Acad. Sci. 2002;99:6118–6123. doi: 10.1073/pnas.092595699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J. Dependence of mutational asymmetry on gene-expression levels in the human genome. Am. J. Hum. Genet. 2003;73:688–692. doi: 10.1086/378134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J., Cohan F.M., Cohan F.M. DNA sequence similarity requirements for interspecific recombination in Bacillus. Genetics. 1999;153:1525–1533. doi: 10.1093/genetics/153.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J., Ott J., Ott J. Distribution and characterization of regulatory elements in the human genome. Genome Res. 2002;12:1827–1836. doi: 10.1101/gr.606402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Xu P., Glazko G., Xu P., Glazko G., Glazko G. Estimation of divergence times from multiprotein sequences for a few mammalian species and several distantly related organisms. Proc. Natl. Acad. Sci. 2001;98:2497–2502. doi: 10.1073/pnas.051611498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H.D., Yoshihama M., Kenmochi N., Yoshihama M., Kenmochi N., Kenmochi N. New maximum likelihood estimators for eukaryotic intron evolution. PLoS Comput. Biol. 2005;1:e79. doi: 10.1371/journal.pcbi.0010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen C.B., Friedman B., Birren B., Burge C.B., Galagan J.E., Friedman B., Birren B., Burge C.B., Galagan J.E., Birren B., Burge C.B., Galagan J.E., Burge C.B., Galagan J.E., Galagan J.E. Patterns of intron gain and loss in fungi. PLoS Biol. 2004;2:e422. doi: 10.1371/journal.pbio.0020422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Bakowski M.A., Morris Q., Zhang W., Frey B.J., Hughes T.R., Blencowe B.J., Bakowski M.A., Morris Q., Zhang W., Frey B.J., Hughes T.R., Blencowe B.J., Morris Q., Zhang W., Frey B.J., Hughes T.R., Blencowe B.J., Zhang W., Frey B.J., Hughes T.R., Blencowe B.J., Frey B.J., Hughes T.R., Blencowe B.J., Hughes T.R., Blencowe B.J., Blencowe B.J. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 2005;21:73–77. doi: 10.1016/j.tig.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Rogozin I.B., Wolf Y.I., Sorokin A.V., Mirkin B.G., Koonin E.V., Wolf Y.I., Sorokin A.V., Mirkin B.G., Koonin E.V., Sorokin A.V., Mirkin B.G., Koonin E.V., Mirkin B.G., Koonin E.V., Koonin E.V. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr. Biol. 2003;13:1512–1517. doi: 10.1016/s0960-9822(03)00558-x. [DOI] [PubMed] [Google Scholar]
- Roy S.W., Gilbert W., Gilbert W. Complex early genes. Proc. Natl. Acad. Sci. 2005a;102:1986–1991. doi: 10.1073/pnas.0408355101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S.W., Gilbert W., Gilbert W. The pattern of intron loss. Proc. Natl. Acad. Sci. 2005b;102:713–718. doi: 10.1073/pnas.0408274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S.W., Gilbert W., Gilbert W. Rates of intron loss and gain: Implications for early eukaryotic evolution. Proc. Natl. Acad. Sci. 2005c;102:5773–5778. doi: 10.1073/pnas.0500383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S.W., Gilbert W., Gilbert W. The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat. Rev. Genet. 2006;7:211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- Roy S.W., Hartl D.L., Hartl D.L. Very little intron loss/gain in Plasmodium: Intron loss/gain mutation rates and intron number. Genome Res. 2006;16:750–756. doi: 10.1101/gr.4845406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S.W., Fedorov A., Gilbert W., Fedorov A., Gilbert W., Gilbert W. Large-scale comparison of intron positions in mammalian genes shows intron loss but no gain. Proc. Natl. Acad. Sci. 2003;100:7158–7162. doi: 10.1073/pnas.1232297100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepelev V., Fedorov A., Fedorov A. Advances in the Exon-Intron Database (EID) Brief. Bioinform. 2006;7:178–185. doi: 10.1093/bib/bbl003. [DOI] [PubMed] [Google Scholar]
- Siepel A., Haussler D., Haussler D. Combining phylogenetic and hidden Markov models in biosequence analysis. J. Comput. Biol. 2004;11:413–428. doi: 10.1089/1066527041410472. [DOI] [PubMed] [Google Scholar]
- Springer M.S., Murphy W.J., Eizirik E., O’Brien S.J., Murphy W.J., Eizirik E., O’Brien S.J., Eizirik E., O’Brien S.J., O’Brien S.J. Placental mammal diversification and the cretaceous-tertiary boundary. Proc. Natl. Acad. Sci. 2003;100:1056–1061. doi: 10.1073/pnas.0334222100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus A. Origin of introns—early or late. Nature. 1994;369:526–527. 527–528. doi: 10.1038/369526b0. author reply. [DOI] [PubMed] [Google Scholar]
- Su A.I., Cooke M.P., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Cooke M.P., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Vega R.G., Sapinoso L.M., Moqrich A., Sapinoso L.M., Moqrich A., Moqrich A., et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sverdlov A.V., Babenko V.N., Rogozin I.B., Koonin E.V., Babenko V.N., Rogozin I.B., Koonin E.V., Rogozin I.B., Koonin E.V., Koonin E.V. Preferential loss and gain of introns in 3′ portions of genes suggests a reverse-transcription mechanism of intron insertion. Gene. 2004;338:85–91. doi: 10.1016/j.gene.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J., Higgins D.G., Gibson T.J., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeramachaneni V., Makalowski W., Makalowski W. DED: Database of Evolutionary Distances. Nucleic Acids Res. 2005;33:D442–D446. doi: 10.1093/nar/gki094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwala R., Ainscough R., Alexandersson M., An P., Ainscough R., Alexandersson M., An P., Alexandersson M., An P., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Weiner A.M., Deininger P.L., Efstratiadis A., Deininger P.L., Efstratiadis A., Efstratiadis A. Nonviral retroposons: Genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu. Rev. Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Yang Z. Maximum-likelihood models for combined analyses of multiple sequence data. J. Mol. Evol. 1996;42:587–596. doi: 10.1007/BF02352289. [DOI] [PubMed] [Google Scholar]
- Yoshihama M., Nakao A., Nguyen H.D., Kenmochi N., Nakao A., Nguyen H.D., Kenmochi N., Nguyen H.D., Kenmochi N., Kenmochi N. Analysis of ribosomal protein gene structures: Implications for intron evolution. PLoS Genet. 2006;2:e25. doi: 10.1371/journal.pgen.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Harrison P.M., Liu Y., Gerstein M., Harrison P.M., Liu Y., Gerstein M., Liu Y., Gerstein M., Gerstein M. Millions of years of evolution preserved: A comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13:2541–2558. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]