Abstract
Regulatory changes in genes involved in reproduction are thought to be prime targets for divergence during speciation, since they are expected to play an important role in sexual selection and sexual conflict. We used microarray analysis of RNA from different wild populations of house mouse subspecies (including Mus m. musculus, Mus m. domesticus, and Mus m. castaneus) and from the sister species Mus spretus to test this assumption. A comparison of expression divergence in brain, liver/kidney, and testis shows a major difference in the evolutionary dynamics of testis-related genes. While the comparison between species confirms an excess in divergence in testis genes, we find that all comparisons between subspecies yield only a very small number of genes with significantly different expression levels in the testis. These results suggest that the early phase of the speciation process may not be driven by regulatory changes in genes that are potential targets of sexual selection, and that the divergence in these genes is only established during a later phase of the speciation process.
The process of speciation can rarely be directly observed but has to be inferred from patterns among populations and species. Dating back >100 yr, mechanisms of speciation have been broadly classified as ecological or nonecological (Gulick 1890; Schluter 2000). In the ecological model, divergence in naturally and sexually selected traits is driven by adaptation to different environments. An important nonecological mechanism invokes sexual selection and sexual conflict driving traits in arbitrary directions (Arnqvist and Rowe 2005). Comparisons in secondary sexual traits (Panhuis et al. 2001), in reproductive proteins (Swanson and Vacquier 2002), and in sperm precedence (Price 1997) among species leads to increasing acceptance that these nonecological mechanisms may be very important in speciation.
To date, only a few studies have addressed the question of whether the rapid rate of protein evolution among genes associated with sex and reproduction holds also for the divergence of gene expression and could therefore be a driver of speciation. By comparing the rate of expression change within Drosophila melanogaster as well as between a single D. melanogaster and D. simulans strain, it was shown that genes that are male biased in their expression evolve more rapidly at the expression level than do female-biased or unbiased genes (Meiklejohn et al. 2003; Ranz et al. 2003). A direct comparison of gene expression differences between tissues, including testis as a separate tissue, was done for human–chimpanzee comparisons (Khaitovich et al. 2005). Expression divergence in the testis was found to be higher than for the other tissues considered.
Here we use a similar approach to that of Khaitovich et al. (2005), i.e., a direct comparison of expression differences in testis versus other tissues (brain and liver/kidney). However, we extend the study to two levels of divergence, i.e., divergence between species (Mus musculus with its closest relative Mus spretus), and on the second level, we compare the subspecies of M. musculus among each other. M. musculus and Mus spretus are separated from each other by 1.1–1.5 Myr (Boursot et al. 1993), whereas the different subspecies are assumed to have diverged from each other 500,000–800,000 yr ago (Guenet and Bonhomme 2003). At least three different house mouse subspecies exist that occupy different geographical areas, with a western (M. musculus domesticus), a central (M. musculus musculus), and an eastern house mouse (M. musculus castaneus). These subspecies are partially reproductively isolated from each other as indicated by the presence of hybrid zones that form in areas of secondary contact between them, and by the presence of sterile hybrid males in some experimental crosses (Forejt 1996; Storchova et al. 2004). A fourth, less well characterized subspecies (M. musculus ssp.) occurs close to the center of the assumed origin of the house mouse (throughout northwest India, Iran, and Pakistan).
We find major differences in testis gene expression in the comparisons of the two levels of divergence. This contrasts with divergence in brain and liver/kidney, which is comparable at both taxonomic levels.
Results
We used high-density two-color oligonucleotide (65mers) microarrays designed from Mus musculus to study gene expression differences in animals either caught directly in the wild or animals that were kept under outbreeding conditions for a few generations. Using multiple animals from each population allowed us to study within-population and between-population variation. We focused on comparing expression differences in testis and used brain and liver/kidney for comparison. All data were obtained using a common reference approach (Yang and Speed 2002), using the laboratory strain C57BL/6 as reference.
Identification of differentially expressed genes
We used the program Significance Analysis of Microarray (SAM) (Tusher et al. 2001) to count the number of differentially expressed genes in each tissue for both taxonomic levels, i.e., between M. musculus and M. spretus and among subspecies of M. musculus.
Comparison between species
Together ∼16% of all genes represented on the microarray showed significant differences in expression between M. musculus and M. spretus (a total of 3257 genes). Among these 3257 genes, approximately half were in testis (1666 genes, ∼51%), 1370 (42%) in liver/kidney, and 221 (7%) in brain. In testis and liver/kidney, roughly the same number of genes are up-regulated in the two lineages of Mus (i.e., liver/kidney: 620 genes up-regulated in spretus, 750 up-regulated in musculus; testis: 926 up-regulated in spretus, 740 up-regulated in musculus). In the brain, however, roughly three times as many genes are up-regulated in spretus than in musculus (167 up-regulated in spretus, 54 up-regulated in musculus).
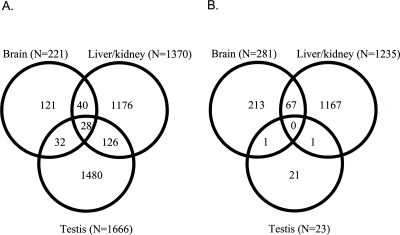
As shown in Figure 1A, we found a relatively small overlap of differentially expressed genes in the different tissues. Only 28 genes (<1%) were differentially expressed in all three tissues. The relative proportions of differentially expressed genes in the different tissues remain the same when we consider only the subset of tissue-specific differentially expressed genes (i.e., testis: 1480 genes, liver/kidney: 1176 genes, brain: 121 genes) (Fig. 1A).
Figure 1.
(A) Venn diagram showing the number of genes differentially expressed between Mus musculus and Mus spretus (FDR < 5%) in the different tissues and the overlap of genes between different tissues. (B) Venn diagram showing the number of genes differentially expressed between M. musculus subspecies (FDR < 5%) in the different tissues and the overlap of genes between different tissues.
Comparison between subspecies
We identified altogether 1539 genes that were differentially expressed in at least one of the subspecies of M. musculus based on results of the SAM multiclass analysis. Among the genes that were identified in the multiclass analysis, only 23 (1.5%) are from testis, 1235 (80.2%) are from liver/kidney, and 281 (18.3%) are from brain (see Fig. 1B). Hence, whereas the numbers of genes differentially expressed in the brain and in the liver/kidney are similar across both comparisons, the number of genes differentially expressed in the testis is strikingly different. This difference in the number of differentially expressed genes in the different tissues is highly significant, when comparing the species and subspecies level (χ22 = 1160.03, P < 0.00001). In the multiclass comparison among musculus subspecies, we found no overlap among differentially expressed genes in all three tissues (Fig. 1B). When only the subset of tissue-specific genes is considered, we find the same proportions of genes differentially expressed in the different tissues (testis: 21 genes, liver/kidney 1167 genes, brain: 213 genes) (Fig. 1B).
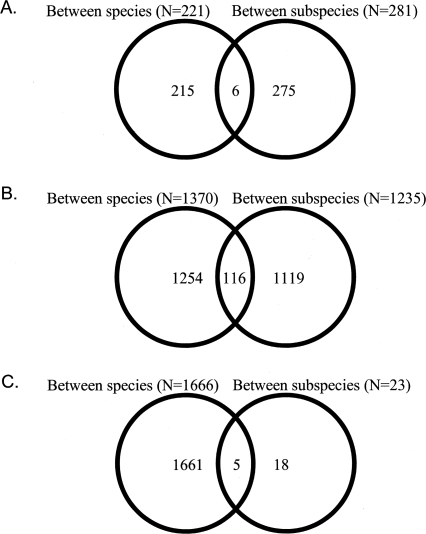
As shown in Figure 2, we found some overlap in differentially expressed genes within a tissue between both levels of divergence. In the brain, six genes (∼2%) were differentially expressed among musculus subspecies as well as between musculus and spretus. In the liver/kidney, the proportion of shared genes is slightly higher (i.e., 116 genes, ∼10%). In the testis, five out of the 23 genes differentially expressed among subspecies of musculus were also differentially expressed between musculus and spretus (Fig. 2). Relative to the 23 genes that were detected among subspecies of musculus, the proportion of shared genes is relatively high (∼30%).
Figure 2.
Venn diagram showing the number and overlap of genes differentially expressed (FDR < 5%) between species and between subspecies. (A) Brain, (B) liver/kidney, (C) testis.
Genome-wide patterns of gene expression change
Differences in expression among tissues can also be measured by using the metric of scaled divergence (Lemos et al. 2005). This method computes a ratio of the between to the within-group components of the variance as estimated from an ANOVA, separately for all genes represented on the microarray. Thus, analyzing scaled divergence measures for all genes allows a genome-wide summary of gene expression divergence in a given tissue, as opposed to an estimate of divergence based on the number of differentially expressed genes. As for the SAM analyses, two levels of divergence were chosen: the between-species and the between-subspecies comparison.
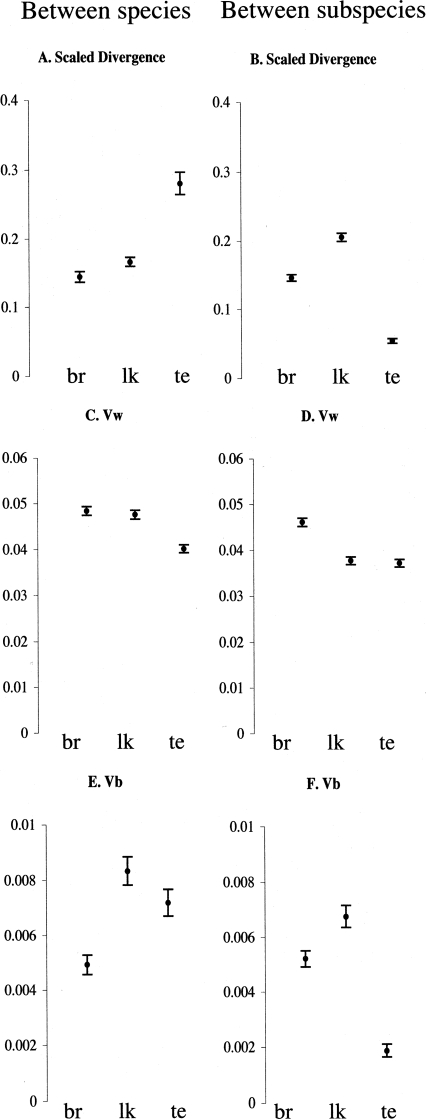
In the between-species comparison, the average (across all genes) scaled divergence is highest in the testis (Fig. 3A), whereas in the between-subspecies comparison, the scaled divergence is highest in the liver/kidney (Fig. 3B). This pattern is consistent with the results obtained from the SAM analyses.
Figure 3.
Mean and 95% confidence interval of scaled divergences, Vw and Vb. (A,C,E) Between Mus spretus and the combined subspecies of Mus musculus. (B,D,F) Between subspecies of M. musculus. br indicates brain; lk, liver/kidney; and te, testis
We further explored the pattern of expression change by decomposing the scaled divergence metric into its variance components, the variance between groups (Vb) and the variance among the individuals within the group (Vw). Their means and 95% confidence intervals are graphically represented in Figure 3C,E,F. The average (across all genes) Vw values vary significantly between tissues (Fig. 3C,D). For both levels of divergence, we find the highest Vw in the brain and the lowest in the testis. The reason for the large variation in gene expression among individuals seen in the brain is unclear but may partly be due to experimental factors, because the complete brain tissue (from olfactory lobe to medulla) is more difficult to extract from the individual than are the liver/kidney and testis; i.e., there may be differences between individuals with respect to different amounts of other tissues included. Vw for liver/kidney is high in the between-species comparison (Fig. 3C) but low in the between-subspecies comparison (Fig. 3D). This can most easily be explained by considering the between-group component of the variance (Vb). In the between-subspecies comparison, Vb is largest in the liver/kidney (Fig. 3F). Because the variance between groups in the subspecies comparison is part of the within-group variance in the species comparison, so Vw in the latter is large.
The Vb components of the total variance in brain and liver/kidney are relatively similar for both levels of divergence (Fig. 3E,F). A large difference between the species and the subspecies comparison is, however, observed in the testis, where the between subspecies Vb is very low but the between species Vb is high.
Validation of the approach
A potential problem with using microarrays that were designed for one species in another closely related (nonfocal) species is that divergence in the sequences that are represented as oligos on the array could confound the measurements of gene expression in the nonfocal species (Gilad et al. 2005). This phenomenon is likely to be more severe the more distantly related the nonfocal species is. Moreover, genes expressed in the testis may evolve more rapidly than do genes expressed in other tissues (Swanson and Vacquier 2002; Good and Nachman 2005). Thus, high sequence divergence in the genes expressed in the testis could be erroneously interpreted as high divergence in gene expression in the testis.
Several lines of evidence suggest that this possible artifact is unlikely to substantially affect the results. First, if high divergence in gene expression in the testis between the species is driven by high DNA sequence evolution of testis expressed genes, we would expect that most of the identified genes differentially expressed between musculus and spretus would show a decrease in expression in spretus (because the presence of mismatches between oligo sequence and cDNA sequence would result in less efficient binding and therefore lower signal intensity). This is not the case, with slightly more genes showing up-regulation in spretus. Second, musculus and spretus are very closely related and show only 1%–2% sequence divergence in noncoding (intergenic) regions (Galtier et al. 2004; B. Harr, unpubl.). Our microarrays consist of relatively long oligos (65mers) so we would expect only marginal effects of sequence divergence on hybridization efficiency in M. spretus. Assuming that DNA sequence constraint is approximately three times higher in 3′UTRs of genes (most oligos on the chip target the 3′ UTR) compared with intergenic regions (D.J. Gaffney and P.D. Keightley, in prep.), we would expect an average of 0.3 SNPs within a given 65mer (i.e., 0.015/ 3 × 65 = 0.3, assuming 1.5% intergenic divergence between the species) that could affect the hybridization characteristic of the oligo in the two different species.
Nevertheless, to rule out differences in hybridization intensity, we performed hybridizations of genomic DNA from M. spretus against the genomic DNA of a M. musculus reference. These results show (see Supplemental material) that, while we cannot rule out that a small number of differentially expressed genes between species could be affected by sequence changes in the oligo sequence on the chip, this effect is not predominant in the testis and therefore cannot explain the accumulation of testis-specific differences in the between-species comparison but not in the between-subspecies comparison.
Chromosomal location of candidate genes
A compilation of the chromosomal position of the identified differentially expressed genes from the SAM analyses is given in Supplemental Table 1. An expected 3.1% of genes should be located on the X chromosome given the chromosomal affiliation of the complete set of genes represented on the microarray. Between the two species of Mus, the observed proportion of X-linked over autosomal genes among the differentially expressed genes is 2.7% (brain), 2.8% (liver/kidney), and 2.5% (testis), which is lower than expected (although not significantly so). Among subspecies of musculus, we find evidence for an overrepresentation of X-chromosomal genes among the differentially expressed genes in the brain (χ2-test, df = 1, P = 0.0078). A surplus of X-chromosomal genes relative to autosomal ones is also found for the divergence among subspecies in the testis (8.7%), but this difference is not statistically significant, due to a very small sample size (Supplemental Table 1).
Functional annotation of differentially expressed genes
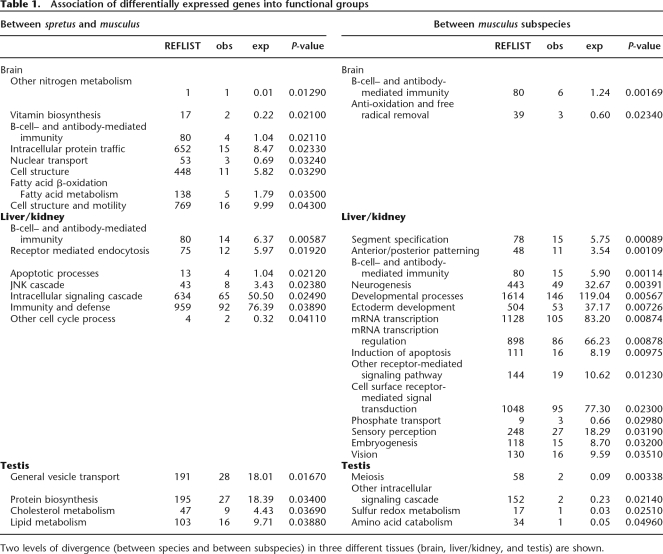
We determined the functional annotations of all differentially expressed genes identified in the SAM analyses to identify overrepresented gene categories. Table 1 lists all cases for which significantly more genes than expected by chance were found for a given annotated biological process. There is little consistency with respect to the species/subspecies comparisons concerning the functional categories that are overrepresented for the individual tissues. The only category that is shared among both levels of divergence and also among different tissues is the group of genes related to humoral immunity. None of the functional categories were significantly overrepresented when P-values were Bonferroni corrected. Thus, these results do not yield a clear hint toward functional processes that would show a particular high rate of divergence.
Table 1.
Association of differentially expressed genes into functional groups
Two levels of divergence (between species and between subspecies) in three different tissues (brain, liver/kidney, and testis) are shown.
Discussion
Under the assumption of neutral divergence of gene expression patterns, the within-species (i.e., between subspecies) divergence should be reflected in the between-species pattern. This is, however, not what we observe. In particular, we find a large surplus of testis-related changes in the between-species comparison relative to the between-subspecies comparison. This result is independent of the analysis method and holds if expression divergence is described in terms of the number of differentially expressed genes (i.e., the SAM method) or the genome-wide analysis of the scaled divergence metric (Lemos et al. 2005).
We can rule out phenotypic plasticity as a cause of the expression differences. Although two of the subspecies in the experiment (M. m. domesticus and M. m. musculus) were held in a common environment for only 3–5 d after collection in the wild, individuals of M. m. castaneus and M. m. ssp. and Mus spretus were derived from stocks that have been maintained in the laboratory for at least one generation. Hence, the animals were kept under laboratory conditions for different time intervals and yet show all a similar overrepresentation of genes, implying that the differences can be attributed to genetic divergence, rather than plasticity.
We also tested whether the same overrepresentation of testis genes between species relative to between subspecies would also be seen when individual subspecies are compared to M. spretus in a pairwise manner. This is indeed the case. Based on SAM, we find in all pairwise comparisons between M. spretus and a single M. musculus subspecies, many more genes that are significantly differentially expressed in the testis than in the liver/kidney (data not shown). Moreover, an overrepresentation of liver/kidney and deficiency of testis-related genes was observed in all pairwise comparisons among individual subspecies (data not shown). Thus, the enrichment of testis-related expression changes between species seems to be a general effect and cannot be ascribed to a single (or a subset of) subspecies or an artifact of pooling over subspecies in the between species analysis.
Khaitovich et al. (2005) showed that testis gene expression evolves especially fast between humans and chimpanzees compared with other tissues such as brain, liver, kidney, and heart, which all evolve at a relatively similar rate. This is in accord with our finding, but evolution below the species level has not been studied in this system. While different tissues have not been contrasted in Drosophila, rates of divergence of sex-biased genes are different between D. melanogaster and D. simulans than they are between two populations of D. melanogaster (Lemos et al. 2005). Thus, as suggested by Lemos et al. (2005), the time scale of the comparison is relevant to the understanding of how within-group variation is converted into between-group divergence. This is in line with our observation of contrasting patterns of gene expression evolution in the testis at and below the species level.
Many studies highlight the peculiarity of the X chromosome in adaptation and species differentiation. Polymorphism studies in Drosophila (Begun and Whitley 2000; Andolfatto 2001), humans (Payseur et al. 2002), and mice (J.F. Baines and B. Harr, in prep.) have found reduced levels of polymorphism on X chromosomes relative to autosomes. This has been interpreted as evidence for a higher rate of adaptive evolution on the X chromosome relative to the autosomes due to recessive beneficial mutations (Charlesworth et al. 1987). The X chromosome is also peculiar with respect to its “male-biased” gene content (i.e., genes that are more strongly expressed in males than in females): Male-biased genes are deficient on the X chromosome in Drosophila (Parisi et al. 2003) and also in mice, at least if genes expressed relatively late in spermatogenesis are considered (Khil et al. 2004). Inactivation of the X chromosome in male meiosis seems to be a universal driving force for this X chromosome “demasculinization” (Khil et al. 2004). Thus, while we would expect to find genes that are involved in male meiosis to be depleted on the X chromosome, we would expect genes without this function to be enriched on the X chromosome.
In our data we do not find much evidence that differentially expressed genes associate predominantly with the X chromosome or the autosomes in any of the tissues assayed. The only comparison for which we detect a significant effect is for the genes differentially expressed among subspecies in the brain where a surplus of X-linked genes was observed. Interestingly, a recent study found the brain to be enriched for X-linked sexual dimorphic genes in M. musculus (Yang et al. 2006). It is well known that sexually dimorphic genes evolve more rapidly on the gene expression level than unbiased (i.e., sexually monomorphic) genes (for a review, see Ranz and Machado 2006). Further studies will be needed to determine whether the surplus of X-linked genes among the differentially expressed genes in the brain is driven by unusual sexual dimorphic selection pressures.
One reason for the absence of a significant effect of the X chromosome in most comparisons could be that most of the identified differentially expressed genes are downstream targets of major regulatory changes, that themselves might be depleted (in the case of genes involved in male meiosis) or enriched on the X chromosome (for genes that do have no function in male meiosis). Further studies that investigate in detail the regulatory pathways and timing and space of the identified differentially expressed genes in different tissues would be needed to clarify this.
Among the list of annotated genes that show a change in expression among species or subspecies, we found only one common theme, i.e., humoral immune system–related genes. This is in contrast to a recent study in primates, where an excess of transcription factors with increased expression specifically in the human lineage was found (Gilad et al. 2006), and to studies in Drosophila, where a surplus of enzymatic functions among the differentially expressed genes has been described (Rifkin et al. 2003). Being involved in host–pathogen interaction, immune system related genes have been shown to be common targets of directional selection (Tanaka and Nei 1989; Schlenke and Begun 2003) and thus represent likely candidate genes for functional divergence. However, the inconsistency of the results from the different systems, as well as the marginal statistical significance, suggests that the question of which biological processes are most affected by expression divergence is not yet resolved.
The common finding that the male reproductive system is rapidly evolving is supported by our study, but only for the between-species comparison. The low divergence of testis gene expression in the within-species comparison (i.e., among the subspecies) was unexpected, given that divergence in brain and liver/kidney is similar in both comparisons. One explanation might be that testis-related differences only arise at a late stage in the divergence between populations or after speciation is complete. The subspecies of house mouse show partial reproductive isolation but meet in hybrid zones (Boursot et al. 1993). One can speculate that genes that are favored under sexual selection rapidly move across such zones, homogenizing the subspecies, as has been shown for one Y-linked haplotype (Boissinot and Boursot 1997).
Methods
Animal material
Unrelated male mice have been collected in the wild in the Czech Republic (M. m. musculus) and in Germany (M. m. domesticus). Animals were captured in live traps and transferred to the laboratory, where they were kept individually under common laboratory conditions between 3 and 5 d. Six males from both subspecies of similar age (judged based on their adult body weight) were selected for further experiments. Three male mice from the outgroup M. spretus were also included in the analysis. These were F1 offspring derived from three pairs of wild caught unrelated mice collected in Spain near Madrid in August 2004. Six male individuals from the central subspecies M. m. ssp. and three male individuals from subspecies M. m. castaneus with similar age and body weight as individuals collected in the wild were selected. M. m. ssp. and M. m. castaneus had been kept between two and 10 generations in the Laboratoire Génome Populations Interactions Adaptation in Montpellier under standard conditions. The reference strain C57BL/6 was obtained from the Charles River Laboratories, Germany. All animals were sacrificed using CO2, and tissues were excised and immediately snap frozen in liquid nitrogen. Frozen tissues were stored at –80°C not >1 wk. Exact geographic origins of the animals used are given in Supplemental Table 2.
Microarrays
The mouse OligoLibrary by Sigma-Genosys/Compugen was duplicate-spotted on Schott/Nexterion Slides H using a Biorobotics MicroGrid II Spotter (Genomic Solutions). The library consists of 21,997 5′-C6 amino modified 65-mers with 21,766 unique transcripts and 231 controls. The oligos represent all of the mouse genes associated with public mRNA sequences found in GenBank release 126 (October 2001). Each array consists of 46,128 spots, representing the OligoLibrary in duplicate, plus external controls and reference spots. After removal of those spots that showed inconsistencies in spotting quality across arrays, 40,656 oligos remained viable.
Sample preparation
Tissue samples were homogenized in TRIzol using an electric homogenizer. RNA was extracted following the manufacturer’s protocol. Briefly, after resuspension of the RNA in DEPC-H2O, the RNA was precipitated in 4 M LiCl for storage at −80°C.
Twenty micrograms of each total RNA was reverse transcribed using the FairPlay Microarray Labeling Kit (Stratagene) and labeled by incorporation of Alexa Fluor 555 and Alexa Fluor 647 (Molecular Probes), respectively, according to the manufacturer’s protocols. Half of the labeled cDNA was used for hybridization.
Hybridization
The two labeled cDNA samples to be compared were combined in 65 μL of 5× SSC and competitively hybridized to a microarray under a coverslip (Implen LifterSlip 24×60l) for 16 h at 42°C in an Advalytix SlideBooster. Slides were washed in graded SSC/SDS and spun dry.
Data acquisition
The arrays were scanned with a GeneTAC LS IV (Genomic Solutions) confocal laser scanner. Scanner settings were adjusted individually for every channel and microarray. Images were analyzed using Spotfinder from the TIGR institute (Saeed et al. 2003) and the extracted intensities were stored as tab-delimited .mev files. The arrays were normalized using block-wise LOWESS and SD regularization (Yang et al. 2002) as described in Quackenbush (2002).
Experimental design
Microarray analysis was performed on three different tissue samples. For each individual, labeled cDNA from total RNA of the whole brain and testis tissue was individually hybridized to the chip, whereas RNA from liver and kidney was pooled in equivalent amounts and hybridized jointly. We employed the common reference design with experimental samples labeled with Alexa Fluor 647 and the reference sample labeled with Alexa Fluor 555. To obtain enough reference RNA for each of the tissues, we extracted RNA from nine animals of the reference inbreed strain C57BL/6 and pooled the RNA’s tissue-wise. Thus, for each tissue the exact same reference RNA was used for all hybridizations. The experimental samples were hybridized together with the reference sample. Since we are using a common reference design and all reference samples are labeled with the same dye, differences in the amount of label per amount of RNA (i.e., “dye bias”) should affect all hybridizations similarly.
Altogether we performed 72 hybridizations, i.e., six individuals × three tissue samples for each of the subspecies musculus, domesticus, and ssp. and three tissue samples for each of the castaneus individuals CTP and CIM and CTA. For M. spretus, nine hybridizations were performed (three tissue samples from each of the tissues brain, testis and liver/kidney).
Data processing and statistical analysis
For the cDNA hybridizations, we calculated the log2-transformed ratio of the normalized signal intensity of the “sample” channel relative to the “reference” channel for each gene. Log2 ratios for the duplicate spots were averaged. These values were submitted to the Gene Expression Omnibus (GSE2597).
Gene expression differences between M. musculus and M. spretus
We used SAM (Tusher et al. 2001) in a two-class unpaired comparison to identify genes differentially expressed between the house mouse subspecies and M. spretus. This analysis was performed for each tissue independently. In SAM, the relative difference (d(i)) in gene expression is compared to the distribution of d(i) following random permutation (200 permutations) of the sample categories. For each d(i), a certain proportion of all genes in the permutation set (control set) will be found to be “significant” by chance, and this parameter is then used to calculate a “false discovery rate” (FDR) (Reiner et al. 2003). Only genes with a FDR <5% were selected as differentially expressed in this “between species” comparison.
Gene expression differences between the M. musculus subspecies
To identify the genes that are differentially expressed in at least one subspecies, we used the multiclass option in SAM and a false discovery rate <5%. Genes identified by the multiclass analysis include (1) genes where all house mouse subspecies differ significantly from each other or (2) genes where combinations of two subspecies differ from each other or (3) genes that are differentially expressed only in one subspecies relative to all other subspecies.
Scaled divergence
The scaled divergence metric is basically a measure of the rate of gene expression divergence in that it compares the between-species to the within-species components of the variance, as determined from the mean squares (MS) from an ANOVA (Lemos et al. 2005).The scaled divergence is defined as Δt = Vb/Vw, where t is the total number of generations separating the two lineages, Vb is the between-species component of variance, and Vw is the within-species component of variance. Vb is calculated as Vb = (MSb − MSw)/n0, where MSb and MSw are the between-population and within-population MS from an ANOVA, and n0 is the average sample size. Therefore, the between-population component of variance is already corrected for the within-population component, since MSw is subtracted from MSb. Two different group assignments were used, which reflected the two different levels of divergence. The first grouping assorts all individuals from all musculus subspecies into one group and all spretus individuals into a second group, yielding an ANOVA model with only two levels. The second grouping comprised only the subspecies of house mouse in a four-level one-way ANOVA. Statistical analyses were performed using the statistical language R and Perl scripts.
Identification of functional categories
The gene list for the between subspecies and the between species analysis obtained from SAM were submitted to PANTHER (http://www.pantherdb.org), which identifies overrepresented functional categories among the significant regions (Mi et al. 2005). Significance is established by comparing the gene list with a reference list (in this case the full gene content that was represented on our microarray) by means of a binomial test (Cho and Campbell 2000).
Acknowledgments
We are thankful to R. Rottscheidt and T. Bayer for catching Mus spretus; B. Bimova, E. Bozikova, J. Pialek, A. Orth, F. Bonhomme, S. Kipp, and S. Ihle for providing wild mice; and T. Price and H. Tang for statistical advice. We also thank T. Price for helpful comments on the manuscript. The work has been funded by an Emmy Noether fellowship by the DFG to B.H., a Volkswagen Stiftung grant to D.T., and a DFG grant to L.E.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5683806
References
- Andolfatto P. Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 2001;18:279–290. doi: 10.1093/oxfordjournals.molbev.a003804. [DOI] [PubMed] [Google Scholar]
- Arnqvist G., Rowe L., Rowe L. Sexual conflict. Princeton University Press; Princeton, NJ: 2005. [Google Scholar]
- Begun D.J., Whitley P., Whitley P. Reduced X-linked nucleotide polymorphism in Drosophila simulans. Proc. Natl. Acad. Sci. 2000;97:5960–5965. doi: 10.1073/pnas.97.11.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S., Boursot P., Boursot P. Discordant phylogeographic patterns between the Y chromosome and mitochondrial DNA in the house mouse: Selection on the Y chromosome? Genetics. 1997;146:1019–1034. doi: 10.1093/genetics/146.3.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursot P., Auffray J.C., Brittondavidian J., Bonhomme F., Auffray J.C., Brittondavidian J., Bonhomme F., Brittondavidian J., Bonhomme F., Bonhomme F. The evolution of house mice. Annu. Rev. Ecol. Syst. 1993;24:119–152. [Google Scholar]
- Charlesworth B., Coyne J.A., Barton N., Coyne J.A., Barton N., Barton N. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 1987;130:113–146. [Google Scholar]
- Cho R.J., Campbell M.J., Campbell M.J. Transcription, genomes, function. Trends Genet. 2000;16:409–415. doi: 10.1016/s0168-9525(00)02065-5. [DOI] [PubMed] [Google Scholar]
- Forejt J. Hybrid sterility in the mouse. Trends Genet. 1996;12:412–417. doi: 10.1016/0168-9525(96)10040-8. [DOI] [PubMed] [Google Scholar]
- Galtier N., Bonhomme F., Moulia C., Belkhir K., Caminade P., Desmarais E., Duquesne J.J., Orth A., Dod B., Boursot P., Bonhomme F., Moulia C., Belkhir K., Caminade P., Desmarais E., Duquesne J.J., Orth A., Dod B., Boursot P., Moulia C., Belkhir K., Caminade P., Desmarais E., Duquesne J.J., Orth A., Dod B., Boursot P., Belkhir K., Caminade P., Desmarais E., Duquesne J.J., Orth A., Dod B., Boursot P., Caminade P., Desmarais E., Duquesne J.J., Orth A., Dod B., Boursot P., Desmarais E., Duquesne J.J., Orth A., Dod B., Boursot P., Duquesne J.J., Orth A., Dod B., Boursot P., Orth A., Dod B., Boursot P., Dod B., Boursot P., Boursot P. Mouse biodiversity in the genomic era. Cytogenet. Genome Res. 2004;105:385–394. doi: 10.1159/000078211. [DOI] [PubMed] [Google Scholar]
- Gilad Y., Rifkin S.A., Bertone P., Gerstein M., White K.P., Rifkin S.A., Bertone P., Gerstein M., White K.P., Bertone P., Gerstein M., White K.P., Gerstein M., White K.P., White K.P. Multi-species microarrays reveal the effect of sequence divergence on gene expression profiles. Genome Res. 2005;15:674–680. doi: 10.1101/gr.3335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y., Oshlack A., Smyth G.K., Speed T.P., White K.P., Oshlack A., Smyth G.K., Speed T.P., White K.P., Smyth G.K., Speed T.P., White K.P., Speed T.P., White K.P., White K.P. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature. 2006;440:242–245. doi: 10.1038/nature04559. [DOI] [PubMed] [Google Scholar]
- Good J.M., Nachman M.W., Nachman M.W. Rates of protein evolution are positively correlated with developmental timing of expression during mouse spermatogenesis. Mol. Biol. Evol. 2005;22:1044–1052. doi: 10.1093/molbev/msi087. [DOI] [PubMed] [Google Scholar]
- Guenet J.L., Bonhomme F., Bonhomme F. Wild mice: An ever-increasing contribution to a popular mammalian model. Trends Genet. 2003;19:24–31. doi: 10.1016/s0168-9525(02)00007-0. [DOI] [PubMed] [Google Scholar]
- Gulick J.T. Unstable adjustments as affected by isolation. Nature. 1890;42:28–29. [Google Scholar]
- Khaitovich P., Hellmann I., Enard W., Nowick K., Leinweber M., Franz H., Weiss G., Lachmann M., Pääbo S., Hellmann I., Enard W., Nowick K., Leinweber M., Franz H., Weiss G., Lachmann M., Pääbo S., Enard W., Nowick K., Leinweber M., Franz H., Weiss G., Lachmann M., Pääbo S., Nowick K., Leinweber M., Franz H., Weiss G., Lachmann M., Pääbo S., Leinweber M., Franz H., Weiss G., Lachmann M., Pääbo S., Franz H., Weiss G., Lachmann M., Pääbo S., Weiss G., Lachmann M., Pääbo S., Lachmann M., Pääbo S., Pääbo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- Khil P.P., Smirnova N.A., Romanienko P.J., Camerini-Otero R.D., Smirnova N.A., Romanienko P.J., Camerini-Otero R.D., Romanienko P.J., Camerini-Otero R.D., Camerini-Otero R.D. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat. Genet. 2004;36:642–646. doi: 10.1038/ng1368. [DOI] [PubMed] [Google Scholar]
- Lemos B., Meiklejohn C.D., Caceres M., Hartl D.L., Meiklejohn C.D., Caceres M., Hartl D.L., Caceres M., Hartl D.L., Hartl D.L. Rates of divergence in gene expression profiles of primates, mice, and flies: Stabilizing selection and variability among functional categories. Evolution Int. J. Org. Evolution. 2005;59:126–137. [PubMed] [Google Scholar]
- Meiklejohn C.D., Parsch J., Ranz J.M., Hartl D.L., Parsch J., Ranz J.M., Hartl D.L., Ranz J.M., Hartl D.L., Hartl D.L. Rapid evolution of male-biased gene expression in Drosophila. Proc. Natl. Acad. Sci. 2003;100:9894–9899. doi: 10.1073/pnas.1630690100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Lazareva-Ulitsky B., Loo R., Kejariwal A., Vandergriff J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M.J., Lazareva-Ulitsky B., Loo R., Kejariwal A., Vandergriff J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M.J., Loo R., Kejariwal A., Vandergriff J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M.J., Kejariwal A., Vandergriff J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M.J., Vandergriff J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M.J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M.J., Guo N., Muruganujan A., Doremieux O., Campbell M.J., Muruganujan A., Doremieux O., Campbell M.J., Doremieux O., Campbell M.J., Campbell M.J., et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panhuis T.M., Butlin R., Zuk M., Tregenza T., Butlin R., Zuk M., Tregenza T., Zuk M., Tregenza T., Tregenza T. Sexual selection and speciation. Trends Ecol. Evol. 2001;16:364–371. doi: 10.1016/s0169-5347(01)02160-7. [DOI] [PubMed] [Google Scholar]
- Parisi M., Nuttall R., Naiman D., Bouffard G., Malley J., Andrews J., Eastman S., Oliver B., Nuttall R., Naiman D., Bouffard G., Malley J., Andrews J., Eastman S., Oliver B., Naiman D., Bouffard G., Malley J., Andrews J., Eastman S., Oliver B., Bouffard G., Malley J., Andrews J., Eastman S., Oliver B., Malley J., Andrews J., Eastman S., Oliver B., Andrews J., Eastman S., Oliver B., Eastman S., Oliver B., Oliver B. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur B.A., Cutter A.D., Nachman M.W., Cutter A.D., Nachman M.W., Nachman M.W. Searching for evidence of positive selection in the human genome using patterns of microsatellite variability. Mol. Biol. Evol. 2002;19:1143–1153. doi: 10.1093/oxfordjournals.molbev.a004172. [DOI] [PubMed] [Google Scholar]
- Price C.S.C. Conspecific sperm precedence in Drosophila. Nature. 1997;388:663–666. doi: 10.1038/41753. [DOI] [PubMed] [Google Scholar]
- Quackenbush J. Microarray data normalization and transformation. Nat. Genet. (Suppl.) 2002;32:496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- Ranz J.M., Machado C.A., Machado C.A. Uncovering evolutionary patterns of gene expression using microarrays. Trends Ecol. Evol. 2006;21:29–37. doi: 10.1016/j.tree.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Ranz J.M., Castillo-Davis C.I., Meiklejohn C.D., Hartl D.L., Castillo-Davis C.I., Meiklejohn C.D., Hartl D.L., Meiklejohn C.D., Hartl D.L., Hartl D.L. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- Reiner A., Yekutieli D., Benjamini Y., Yekutieli D., Benjamini Y., Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- Rifkin S.A., Kim J., White K.P., Kim J., White K.P., White K.P. Evolution of gene expression in the Drosophila melanogaster subgroup. Nat. Genet. 2003;33:138–144. doi: 10.1038/ng1086. [DOI] [PubMed] [Google Scholar]
- Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Braisted J., Klapa M., Currier T., Thiagarajan M., Klapa M., Currier T., Thiagarajan M., Currier T., Thiagarajan M., Thiagarajan M., et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Schlenke T.A., Begun D.J., Begun D.J. Natural selection drives Drosophila immune system evolution. Genetics. 2003;164:1471–1480. doi: 10.1093/genetics/164.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D. The ecology of adaptive radiation. Oxford University Press; Oxford, UK: 2000. [Google Scholar]
- Storchova R., Gregorova S., Buckiova D., Kyselova V., Divina P., Forejt J., Gregorova S., Buckiova D., Kyselova V., Divina P., Forejt J., Buckiova D., Kyselova V., Divina P., Forejt J., Kyselova V., Divina P., Forejt J., Divina P., Forejt J., Forejt J. Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm. Genome. 2004;15:515–524. doi: 10.1007/s00335-004-2386-0. [DOI] [PubMed] [Google Scholar]
- Swanson W.J., Vacquier V.D., Vacquier V.D. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Nei M., Nei M. Positive Darwinian selection observed at the variable-region genes of immunoglobulins. Mol. Biol. Evol. 1989;6:447–459. doi: 10.1093/oxfordjournals.molbev.a040569. [DOI] [PubMed] [Google Scholar]
- Tusher V.G., Tibshirani R., Chu G., Tibshirani R., Chu G., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.H., Speed T., Speed T. Design issues for cDNA microarray experiments. Nat. Rev. Genet. 2002;3:579–588. doi: 10.1038/nrg863. [DOI] [PubMed] [Google Scholar]
- Yang Y.H., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P., Lin D.M., Peng V., Ngai J., Speed T.P., Peng V., Ngai J., Speed T.P., Ngai J., Speed T.P., Speed T.P. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Schadt E.E., Wang S., Wang H., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J., Schadt E.E., Wang S., Wang H., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J., Wang S., Wang H., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J., Wang H., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J., Ingram-Drake L., Drake T.A., Lusis A.J., Drake T.A., Lusis A.J., Lusis A.J. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]