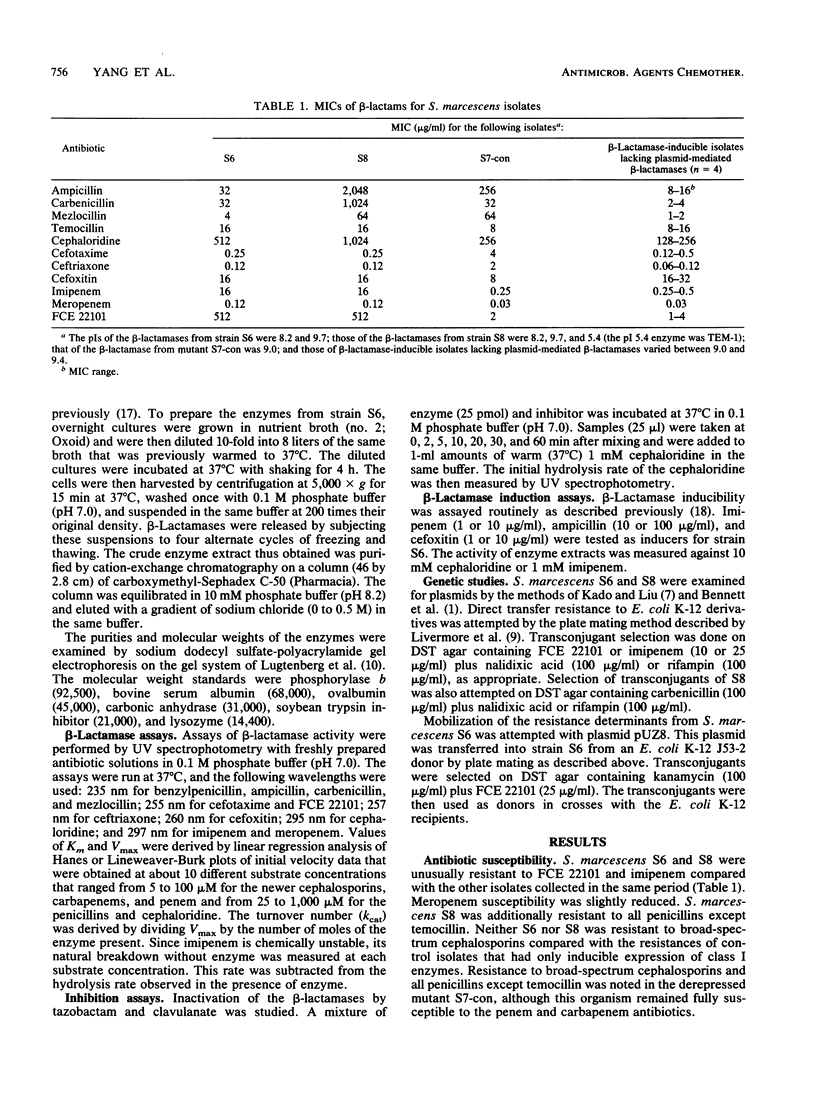
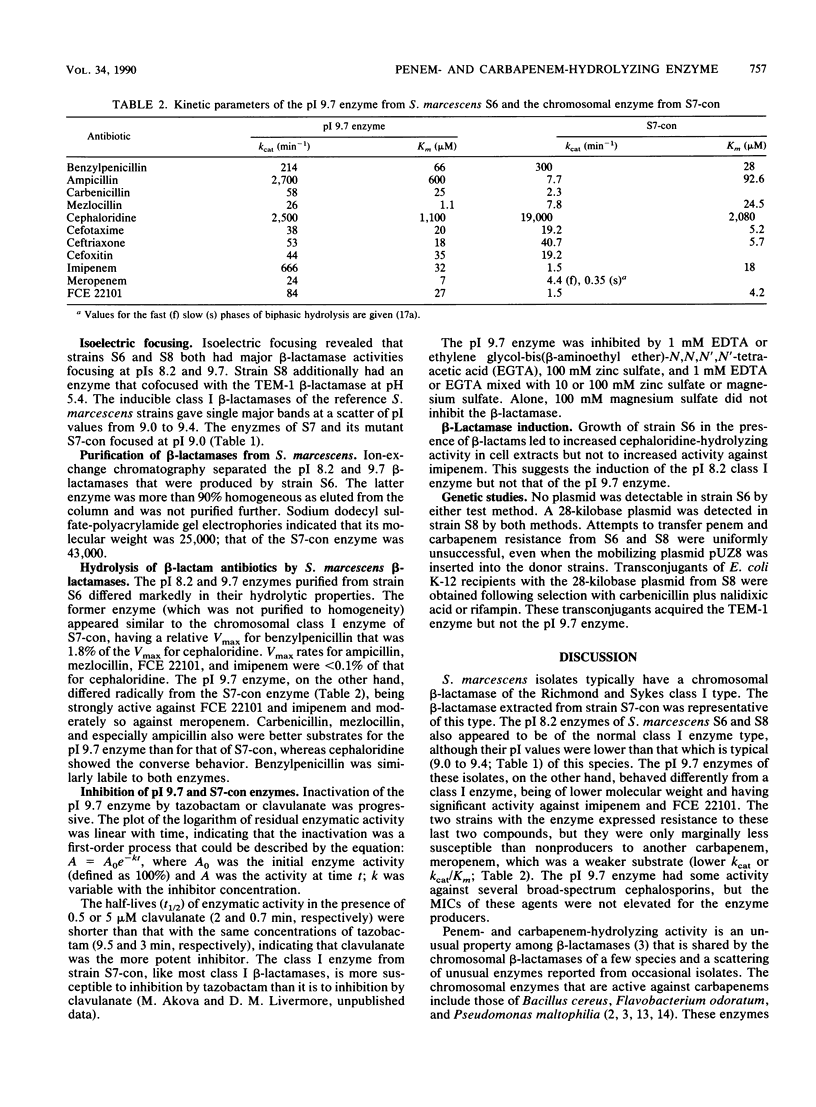
Abstract
Reexamination of Serratia marcescens isolates obtained in 1982 revealed two organisms that were resistant to the penem FCE 22101 (MIC, 512 micrograms/ml) and imipenem (MIC, 16 micrograms/ml) and that had slightly reduced susceptibilities to meropenem (MIC, 0.12 micrograms/ml). MICs of these agents for typical S. marcescens isolates were 1 to 8, 0.25 to 0.5, and 0.03 micrograms/ml, respectively. The two isolates were fully susceptible to broad-spectrum cephalosporins, and only one was highly resistant to ampicillin and carbenicillin (MICs, greater than 1,024 micrograms/ml). Both isolates had beta-lactamases that focused at pIs 8.2 and 9.7. The penicillin-resistant isolate additionally produced the TEM-1 enzyme. The enzymes with pIs of 8.2 and 9.7 were separated by cation-exchange chromatography. The pI 8.2 beta-lactamase was a class I enzyme of the type found in most S. marcescens isolates. It was almost inactive against carbapenems and penems, as was the class I enzyme from another S. marcescens strain. The pI 9.7 enzyme hydrolyzed penems and carbapenems rapidly: kcat (turnover number) values for FCE 22101, imipenem, and meropenem were 3.4, 26, and 1% of the kcat value for cephaloridine, respectively; kcat/Km values were 140, 915, and 150% of the kcat/Km value for cephaloridine, respectively. Otherwise, the pI 9.7 enzyme had predominantly penicillinase activity. It was inhibited more readily by clavulanate than by tazobactam and was inactivated by the chelating agents EDTA and ethylene glycol-bis(beta-aminoethyl ether)-N,N,N',N'-tetraacetic acid. Expression of the pI 9.7 enzyme was not associated with any plasmid, and production was not transferred to Escherichia coli K-12 recipients, even after the mobilizing plasmid pUZ8 was inserted into the S. marcecens donor strains.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennett P. M., Heritage J., Hawkey P. M. An ultra-rapid method for the study of antibiotic resistance plasmids. J Antimicrob Chemother. 1986 Sep;18(3):421–424. doi: 10.1093/jac/18.3.421. [DOI] [PubMed] [Google Scholar]
- Bicknell R., Emanuel E. L., Gagnon J., Waley S. G. The production and molecular properties of the zinc beta-lactamase of Pseudomonas maltophilia IID 1275. Biochem J. 1985 Aug 1;229(3):791–797. doi: 10.1042/bj2290791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. Classification of beta-lactamases: groups 2c, 2d, 2e, 3, and 4. Antimicrob Agents Chemother. 1989 Mar;33(3):271–276. doi: 10.1128/aac.33.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Hedges R. W. Trimethoprim resistance conferred by W plasmids in Enterobacteriaceae. J Gen Microbiol. 1972 Sep;72(2):349–355. doi: 10.1099/00221287-72-2-349. [DOI] [PubMed] [Google Scholar]
- Hedges R. W., Matthew M. Acquisition by Escherichia coli of plasmid-borne beta-lactamases normally confined to Pseudomonas spp. Plasmid. 1979 Apr;2(2):269–278. doi: 10.1016/0147-619x(79)90045-3. [DOI] [PubMed] [Google Scholar]
- Jarlier V., Nicolas M. H., Fournier G., Philippon A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988 Jul-Aug;10(4):867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981 Mar;145(3):1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore D. M., Jones C. S. Characterization of NPS-1, a novel plasmid-mediated beta-lactamase, from two Pseudomonas aeruginosa isolates. Antimicrob Agents Chemother. 1986 Jan;29(1):99–103. doi: 10.1128/aac.29.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore D. M., Pitt T. L., Jones C. S., Crees-Morris J. A., Williams R. J. PSE-4 beta lactamase: a serotype-specific enzyme in Pseudomonas aeruginosa. J Med Microbiol. 1985 Feb;19(1):45–53. doi: 10.1099/00222615-19-1-45. [DOI] [PubMed] [Google Scholar]
- Lugtenberg B., Meijers J., Peters R., van der Hoek P., van Alphen L. Electrophoretic resolution of the "major outer membrane protein" of Escherichia coli K12 into four bands. FEBS Lett. 1975 Oct 15;58(1):254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- Richmond M. H., Sykes R. B. The beta-lactamases of gram-negative bacteria and their possible physiological role. Adv Microb Physiol. 1973;9:31–88. doi: 10.1016/s0065-2911(08)60376-8. [DOI] [PubMed] [Google Scholar]
- Roy C., Foz A., Segura C., Tirado M., Fuster C., Reig R. Plasmid-determined beta-lactamases identified in a group of 204 ampicillin-resistant Enterobacteriaceae. J Antimicrob Chemother. 1983 Nov;12(5):507–510. doi: 10.1093/jac/12.5.507. [DOI] [PubMed] [Google Scholar]
- Saino Y., Kobayashi F., Inoue M., Mitsuhashi S. Purification and properties of inducible penicillin beta-lactamase isolated from Pseudomonas maltophilia. Antimicrob Agents Chemother. 1982 Oct;22(4):564–570. doi: 10.1128/aac.22.4.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Fujii T., Okamoto R., Inoue M., Mitsuhashi S. Biochemical properties of beta-lactamase produced by Flavobacterium odoratum. Antimicrob Agents Chemother. 1985 Apr;27(4):612–614. doi: 10.1128/aac.27.4.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon K., King A., Phillips I. Beta-lactamases with high activity against imipenem and Sch 34343 from Aeromonas hydrophila. J Antimicrob Chemother. 1986 Jan;17(1):45–50. doi: 10.1093/jac/17.1.45. [DOI] [PubMed] [Google Scholar]
- Sirot D., Sirot J., Labia R., Morand A., Courvalin P., Darfeuille-Michaud A., Perroux R., Cluzel R. Transferable resistance to third-generation cephalosporins in clinical isolates of Klebsiella pneumoniae: identification of CTX-1, a novel beta-lactamase. J Antimicrob Chemother. 1987 Sep;20(3):323–334. doi: 10.1093/jac/20.3.323. [DOI] [PubMed] [Google Scholar]
- Yang Y. J., Livermore D. M. Interactions of meropenem with class I chromosomal beta-lactamases. J Antimicrob Chemother. 1989 Sep;24 (Suppl A):207–217. doi: 10.1093/jac/24.suppl_a.207. [DOI] [PubMed] [Google Scholar]
- Yang Y. J., Livermore D. M., Williams R. J. Chromosomal beta-lactamase expression and antibiotic resistance in Enterobacter cloacae. J Med Microbiol. 1988 Mar;25(3):227–233. doi: 10.1099/00222615-25-3-227. [DOI] [PubMed] [Google Scholar]
- Yang Y., Livermore D. M. Interactions of FCE 22101 with class I beta-lactamases. J Antimicrob Chemother. 1989 Mar;23 (Suppl 100):85–94. doi: 10.1093/jac/23.suppl_c.85. [DOI] [PubMed] [Google Scholar]
- Yotsuji A., Minami S., Inoue M., Mitsuhashi S. Properties of novel beta-lactamase produced by Bacteroides fragilis. Antimicrob Agents Chemother. 1983 Dec;24(6):925–929. doi: 10.1128/aac.24.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]


