Abstract
Effective gene therapy depends on the efficient transfer of therapeutic genes and their protein products to target cells. Lentiviral vectors appear promising for virus-mediated gene delivery and long-term expression in nondividing cells. The herpes simplex virus type 1 tegument protein VP22 has recently been shown to mediate intercellular transport of proteins, raising the possibility that it may be helpful in a setting where the global delivery of therapeutic proteins is desired. To investigate the effectiveness of lentiviral vectors to deliver genes encoding proteins fused to VP22, and to test whether the system is sufficiently potent to allow protein delivery from transduced cells in vitro and in vivo, fusion constructs of VP22 and the enhanced green fluorescent protein (EGFP) were prepared and delivered into target cells by using HIV-1-based lentiviral vectors. To follow the spread of VP22-EGFP to other cells, transduced COS-7 cells were coplated with a number of different cell types, including brain choroid plexus cells, human endothelial cells, H9 cells, and HeLa cells. We found that VP22-EGFP fusion proteins were transported from transduced cells to recipient cells and that such fusion proteins accumulated in the nucleus and in the cytoplasm of such cells. To determine the ability to deliver fusion proteins in vivo, we injected transduced H9 cells as well as the viral vector directly into the brain of mice. We present evidence that VP22-EGFP fusion proteins were transported effectively from lentivirus transduced cells in vivo. We also show that the VP22-EGFP fusion protein encoded by the lentivirus is transported between cells. Our data indicate that such fusion proteins are present in the nucleus and in the cytoplasm of neighboring cells. Therefore, lentiviral vectors may provide a potent biological system for delivering genes encoding therapeutic proteins fused to VP22.
A number of obstacles currently limit the effectiveness of gene therapy. One of the most formidable is the delivery of desired genes or proteins to a sufficient number of target cells to elicit a therapeutic response. Recently, a series of virus-encoded and other regulatory proteins were found to possess the ability to cross biological membranes. For example, peptides derived from the Drosophila Antennapedia homeodomain are internalized by cells in culture (1, 2) and conveyed to cell nuclei where they can directly and specifically interfere with transcription (2, 3). The HIV-1 Tat protein was reported to enhance intercellular trafficking in vitro (4, 5). The Tat protein is composed of 86 amino acids and contains a highly basic region and a cysteine-rich region (4). It was found that Tat-derived peptides as short as 11 amino acids are sufficient for transduction of proteins (6, 7). However, the exact mechanism by which the 11-amino acid transduction domain crosses lipid bilayers is poorly understood. Schwarze et al. (8) have recently generated a Tat-β-galactosidase fusion protein that was delivered efficiently into brain tissue and skeletal muscle in vivo. These findings suggest that protein therapies may be successfully developed provided that problems caused by immune response and toxicity that might be associated with long-term expression of novel proteins in vivo can be solved.
The herpes simplex virus type 1 tegument protein VP22 was also reported to exhibit a unique property of effecting intercellular spread. VP22-directed delivery of proteins could be achieved either by transfection of genes encoding VP22 or by exogenous application of a protein extract containing VP22 (9). VP22 is a basic, 38-kDa phosphorylated protein (10) encoded by the viral UL49 gene (11). The transport of VP22 occurs via a mechanism potentially involving actin microfilaments. VP22 is exported from the cytoplasm of expressing cells and imported into neighboring cells where it accumulates in the nucleus (9). These properties aroused interest in VP22 as a delivery vehicle for therapeutic proteins (12). Recent studies suggest that VP22 is distributed to at least three distinct subcellular locations, which were defined as nuclear, diffuse, and cytoplasmic (13). All of the data obtained thus far were based on studies with transfected cells in culture. The delivery of a recombinant fusion protein by a lentiviral vector into the brain in vivo has not been reported. Our goal was to determine whether lentiviral vectors could be used to deliver VP22 fusion genes into mammalian cells and brain tissue and to test whether such genes would be able to effect intercellular protein delivery from infected cells in vitro and in vivo. We prepared recombinant VP22-EGFP fusion constructs that were subsequently delivered into COS-7 cells by using a defective HIV-1-based lentiviral vector. The delivery of such fusion proteins into brain choroid plexus cells, human endothelial cells, HeLa cells, and H9 cells was studied in vitro. The delivery of the fusion protein by lentiviral vector in mouse brain tissue in vivo was also demonstrated.
Materials and Methods
Vector Constructs and Virus Production.
The double-gene HIV-EGFP/HSA vector was described previously (14). The pUL49ep clone encoding VP22 was kindly provided by J. McLauchlan (Institute of Virology, Glasgow, Scotland; ref. 15). The pUL49ep BamHI fragment was cloned in frame to the EGFP coding region present in pEGFP-N1 (CLONTECH). A DNA fragment encoding VP22 fused to EGFP was subsequently subcloned into HIV-EGFP/HSA to yield HIV-VP22-EGFP/HSA. Virus was produced in 293T cells by transient transfection as described (16, 17). Virus stocks were concentrated by ultracentrifugation.
Animals.
Adult mice (57/BL16, 25 g), obtained from Taconic Farms, were maintained in a BSL2/3 animal facility in a temperature and light-controlled room, with food and water available ad libitum. The mice were anesthetized with Avertin solution (Aldrich) i.p. (0.15 ml/10 g body weight) before injection. They were placed in a small-animal stereotactic apparatus fitted to a mouse adaptor with the skull horizontal between lambda and bregma. Following the surgery and injection, the animal's scalp was closed and sterilized before return to the recovery cage. The animal experiment was approved by the Animal Care and Use Committee at the National Institutes of Health.
Cell Culture and Infection.
Human endothelial, brain choroid plexus, HeLa, and COS-7 cell lines were obtained from the American Type Culture Collection. The human H9 cell line was obtained from Dr. Robert Gallo (18) through the AIDS Research and Reference Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. The COS-7, HOS, and HeLa cells were grown in DMEM (GIBCO) containing 10% heat-inactivated FBS. Human endothelial cells were grown in F12 K-medium with 2 mM l-glutamine containing 1.5 g/liter sodium bicarbonate, 100 μg/ml heparin, 30 μg/ml endothelial cell growth supplement (ICN), and 10% FBS. The brain choroid plexus cells were grown in Eagle's minimum essential medium with 0.1 mM nonessential amino acids, 90% Earle's balanced salt solution, and 10% FBS. H9 cells were grown in 80% RPMI 1640 medium (GIBCO) containing 10% FBS, 2× l-glutamine, 0.05 mg/ml gentamicin and 1× Penstrep (GIBCO). Cells were infected in DMEM/FBS containing 4 μg/ml Polybrene for 4–16 h.
Immunofluorescence Analysis and Flow Cytometry.
Approximately 2× 105 COS-7 or HOS cells per well were plated into six-well plates and infected with 0.25 ml of virus. Infected COS-7 cells were trypsinized 24 h after infection and coplated with human endothelial cells, brain choroid plexus cells, and HeLa cells at a ratio of 1:10, then allowed to grow for 3 d. Cells were grown on 12-mm, round coverslips coated with poly-l-lysine (Becton Dickinson) in 12-well culture dishes in 2.2 ml of medium. Cells were fixed with 4% paraformaldehyde in 1× Hanks' balanced salt solution (HSS, GIBCO) containing 2% FBS for 10 min at room temperature. The samples were washed three times with 1× HSS, and blocked with 10% goat serum in 1× HSS for 20 min at room temperature. Monoclonal mouse simian virus 40 T-antigen antibody (Calbiochem) was added at a dilution of 1:100 and the cells were incubated for 60 min at room temperature. The samples were then washed three times with 1× HSS, and incubated with a secondary anti-mouse antibody conjugated with tetramethylrhodamine isothiocyanate (TRITC; Sigma) for 30 min at room temperature. The coverslips were carefully removed after washing three times with 1× HSS, and then mounted on slides for microscopic observation. Statistical evaluation was performed by using a Student's unpaired t test (Statwork, Microsoft). Mean values for the numbers of cells with positive fluorescent staining were determined by averaging values from three experiments.
For immunofluorescence staining of H9 cells, a phycoerythrin-labeled monoclonal anti-IL-2 receptor (IL-2R) antibody (PharMingen) was used. The dish containing transduced COS-7 cells was first washed three times with culture medium. The suspension of H9 cells was then directly transferred onto a monolayer of transduced COS-7 cells. The suspension of H9 cells was collected 3 d after coculturing, and washed three times with 1× HSS solution containing 5% FBS. The resuspended cells were then transferred to a 50-mm tube in which antibody staining (1:100 dilution) was carried out for 30 min on ice. The cells were washed three times with PBS/FBS buffer, and 0.1–0.2 ml of diluted cells (5 × 105) was placed in a Cytospin block. The blocks were centrifuged at 800 rpm for 5 min. After removal from the blocks and fixing in ethanol–glacial acetic acid for 15 min at −20°C, the slides were analyzed by Zeiss Axiophot fluorescence microscopy equipped with a Hamamastu charge-coupled device camera.
For fluorescence-activated cell sorter (FACS) analysis, cells were detached from the plate by using PBS containing 2 mM EDTA 3 d after infection, and then incubated with a phycoerythrin-labeled anti-HSA monoclonal antibody (1:40 dilution) for 30 min on ice. The cells were collected after centrifugation and resuspended in PBS for subsequent FACS analysis.
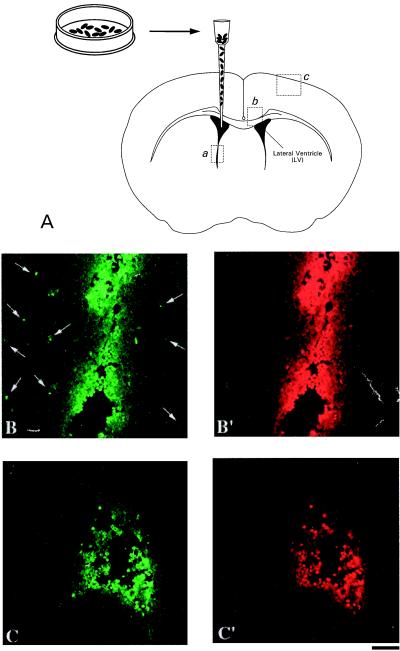
Implantation of Transduced Cells Into Mouse Brain Ventricles.
The animals were divided into two groups (5 animals per group). The first group was implanted with transduced cells previously infected by using the HIV-EGFP/HSA vector. The second group was implanted with cells previously infected by using the HIV-VP22-EGFP/HSA vector. Transduced H9 cells were washed with PBS in 1× HSS containing 0.2% trypsin and subsequently washed two times with PBS in 1× HSS. The cells were then concentrated by centrifugation for implantation. The animal was anesthetized and the head was fastened in the stereotactic apparatus. Injections of transduced cells into the lateral ventricles of the brain were performed at the following coordinates: 0.38 mm to bregma, 0.65 mm to the midline, and 3.0 mm depth (Fig. 5A). Twenty microliters of the transduced cells (106–107 cells per ml) was loaded into an internal cannula needle (23 gauge) with cannula tubing connected to a Hamilton syringe mounted on a microinjection pump (Harvard Apparatus). The cells were delivered into the ventricle of the brain at a rate of 1.0 μl/min.
Figure 5.
Implantation of transduced H9 cells in the lateral ventricle of adult mouse brain. (A) H9 cells previously transduced by using the HIV-VP22-EGFP/HSA vector were injected into the brain ventricle. (B) The transduced cells in the ventricle (position a) and spreading of VP22-EGFP into neighboring tissue (arrows) are illustrated. (C) Implanted cells previously transduced by using the HIV-EGFP/HSA vector lacking VP22 are shown. Implanted cells were visualized by staining using anti IL-2R antibody (B′ and C′, red fluorescence). (Scale bar, 60 μm.)
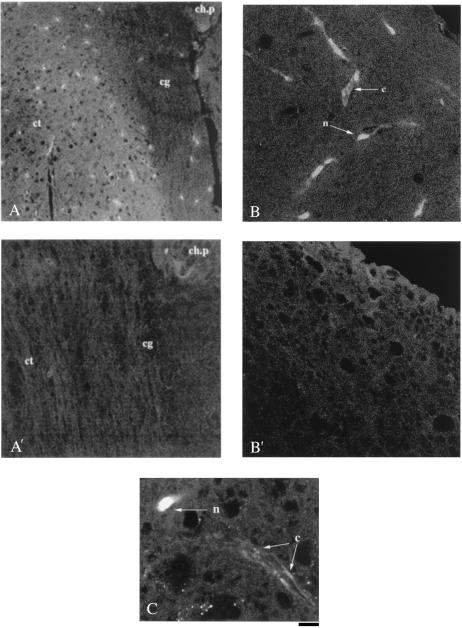
Vector Injection into the Mouse Brain Hippocampus.
Mice were divided into two groups (5 animals per group). The first group was injected with the HIV-EGFP/HSA lentiviral vector. The second group received the HIV-VP22-EGFP/HSA lentiviral vector. The procedure for surgery was as described above using the following coordinates: −2.3 mm to bregma, 1.0 mm to the midline, and 2.0 mm depth (Fig. 6A). Five microliters of concentrated viral vector (corresponding to 3.4 × 106 transducing units of HIV-EGFP/HSA and 1.1 × 106 transducing units of HIV-VP22-EGFP/HSA, respectively) were loaded into an internal cannula needle (C315 × 33) with cannula tubing connected to a Hamilton syringe mounted on a microinjection pump. The viral vector solutions were delivered at a rate of 0.5 μl/min.
Figure 6.
Delivery of VP22-EGFP fusion protein in mouse brain cortex. VP22-EGFP fusion protein (A, green fluorescence) was observed to migrate from the brain cingulum (cg) to the region of brain cortex (ct); the brain section is correlated to position b of the brain anatomic map for cell implantation (see Fig. 5A). The VP22-EGFP fusion proteins were also transported into the nuclei (arrow n) and cytoplasm (arrow c) of brain cortical cells (B and high magnification C); the brain section is correlated to the position c of brain anatomic map (see Fig. 5A). In all adjacent brain sections, the signal for EGFP fluorescence was significantly reduced (A′ and B′) when implanted cells were transduced with HIV-EGFP. ch.p, choroid plexus in ventricle; cc, cingulum; ct, brain cortex. (Scale bar represents 70 μm for A, 50 μm for B, and 35 μm for C.)
Brain Immunofluorescence Assay.
Animals were sacrificed by decapitation 3 weeks after injection and whole brains were carefully removed. The brains were immediately fixed with 4% paraformaldehyde/1% glutaraldehyde for 24 h at 4°C, then washed with PBS in 1× HSS containing 4% sucrose for 2 d at 4°C. The tissues were embedded in O.C.T. (optimum cutting temperature) medium (Tissue-Tek) and frozen in a methanol/dry ice bath. The frozen tissues were sectioned to a thickness of 15 μm per coronal section by using a cryostat (Bright Instrument, Huntingdon, UK) at −18°C. For immunocytochemical detection of implanted cells, the brain sections were washed three times with PBT buffer (PBS in 1× HSS, 0.1% BSA and 0.2% Tween 20), then blocked with 10% goat serum for 15 min. After washing three times with PBT buffer, slides were incubated in the phycoerythrin-labeled monoclonal anti-IL-2R antibody (1:500; PharMingen) for 45 min at room temperature. The slides were washed with PBT buffer and analyzed by using a Zeiss 510 confocal microscope.
Results
Construction of Lentiviral Vectors Encoding VP22-EGFP Fusion Protein.
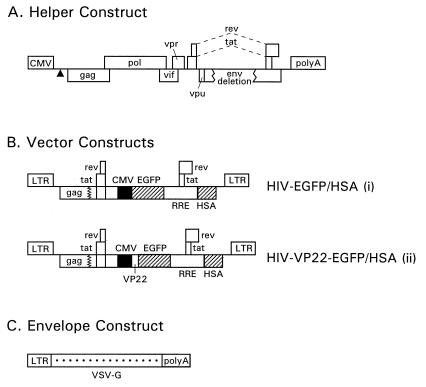
We previously designed pseudotyped, high-titer, replication-defective HIV-1 vector systems to deliver genes into nondividing cells (16). In the present study, we constructed double-gene lentiviral vectors encoding EGFP driven by the human CMV (cytomegalovirus)-IE (immediate early) promoter and the murine HSA driven by the viral long terminal repeat (LTR). One of the vector constructs (HIV-VP22-EGFP/HSA) encodes EGFP fused at its N terminus to the VP22 coding region (15) (Fig. 1B, Lower). A control vector (HIV-EGFP/HSA) (Fig. 1B Upper), expresses unfused EGFP. A three-plasmid expression system consisting of a defective packaging construct (Fig. 1A), a plasmid coding for the vesicular stomatitis virus (VSV) G glycoprotein (Fig. 1C), and the vector constructs shown in Fig.1B were used to generate pseudotyped HIV-1 particles by transient transfection of human embryonic kidney 293T cells.
Figure 1.
HIV-1-based gene transfer systems. (A) Helper (packaging) construct. The triangle symbolizes a deletion affecting the packaging signal between the 5′ splice donor site and the beginning of the gag sequence. The poly(A) site was derived from the bovine growth hormone gene. (B) Transducing vector constructs. The HIV-EGFP/HSA (i) and HIV-VP22 EGFP/HSA (ii) constructs are shown. Boxes interrupted by jagged lines contain partial deletions. CMV, Human CMV-IE promoter. (C) Env expression construct encoding VSV-G. VSV-G expression is driven by the HIV-1 LTR. The poly(A) site was derived from the simian virus 40 late region.
Analysis of Cells Transduced with Double-Gene Vectors Encoding VP22-EGFP Fusion Protein.
Double-gene vectors encoding EGFP and HSA were initially designed to distinguish recipient cells that have taken up the VP22 fusion protein from infected cells delivering the fusion protein. HOS cells infected with these vectors were EGFP-positive as well as HSA-positive by FACS analysis. However, FACS analyses and Northern-blot assay revealed that the number of HSA-positive HOS cells infected with the HIV-VP22-EGFP/HSA vector was notably lower than the number of HSA-positive cells obtained from cultures infected by using HIV-EGFP/HSA vector system (data not shown). These results imply that VP22 somehow affected HSA expression, possibly by down-regulating HSA-specific RNAs.
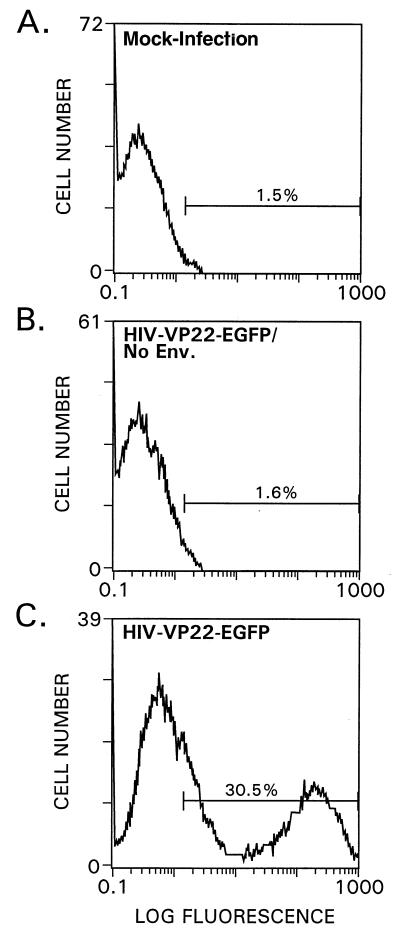
To rule out pseudotransduction events, we prepared vector stocks lacking a viral envelope glycoprotein (Env). FACS analysis revealed that a significant number of EGFP-positive cells were evident in cultures infected by lentiviral vector of HIV-VP22-EGFP/HSA (Fig. 2C), but not for cells infected by the HIV-VP22-EGFP/HSA lacking Env (Fig. 2B). Thus, the transport function of VP22-EGFP fusion protein was abolished when cells were transduced with viral stock lacking an envelope.
Figure 2.
FACS analysis of HOS cells infected with lentiviral vectors encoding VP22-EGFP fusion proteins. Approximately 2 × 105 cells were plated into six-well plates and infected with 0.25 ml of virus. Three days after infection the cells were detached from the plate and were then subjected to FACS analysis. (A) Mock-infected HOS-cells. (B) HOS cells infected by using the HIV-VP22-EGFP/HSA vector lacking viral Env. (C) HOS cells infected by using the HIV-VP22-EGFP/HSA vector.
Intercellular Spread of VP22-EGFP Fusion Proteins from Lentivirus-Transduced Cells.
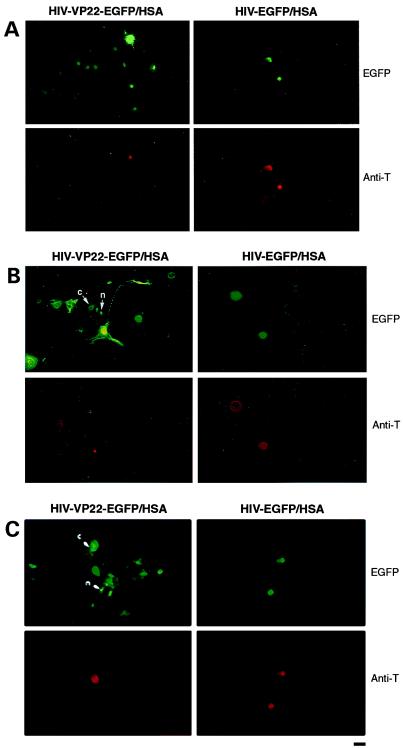
Because the VP22 fusion protein down-regulated the expression of HSA, we adopted a more indirect strategy previously introduced by Elliott and O'Hare (9) for transfected cells. To visualize transduction events involving the movement of VP22-EGFP to neighboring cells, COS-7 cells expressing simian virus 40 T-antigen were infected with the double-gene lentiviral vectors as described above. At 24 h after infection, the cells were coplated with a number of different types of uninfected cells at a ratio of 1:10. The expression of VP22-EGFP fusion proteins in transduced COS-7 cells and the spread of such proteins to uninfected cells, including brain choroid plexus cells (Fig. 3A), human endothelial cells (Fig. 3B), and HeLa cells (Fig. 3C), was investigated by fluorescence microscopy. Fig. 3 (A and B) indicates that the transfer of VP22-EGFP (green fluorescence) into neighboring brain choroid plexus cells and human endothelial cells may be directional, and possibly involve cellular extensions. Furthermore, VP22 fusion proteins in coplated human endothelial cells (Fig. 3B) and HeLa cells (Fig. 3C) were found in both the nucleus and the cytoplasm. The infected COS-7 cells were distinguished from uninfected cells by a monoclonal antibody specific for simian virus 40 T-antigen conjugated to tetramethylrhodamine isothiocyanate (Fig. 3, red fluorescence). The ratio of infected cells to neighboring recipient cells was as follows: 1.3 ± 0.33 to 8.3 ± 0.23 (P < 0.05) for brain choroid plexus cells; 2.0 ± 1.0 to 11.6 ± 2.6 (P < 0.05) for human endothelial cells, and 1.7 ± 0.3 to 10.6 ± 3.0 (P < 0.05) for HeLa cells. The increases in the number of EGFP-positive recipient cells were significant in all three cell lines compared with the number of transduced delivery cells (P < 0.05). EGFP was not transported to neighboring cells when COS-7 cells were infected with a HIV-EGFP/HSA vector lacking the VP22 coding sequence (Fig. 3 A–C Right panels).
Figure 3.
VP22-EGFP fusion protein transport from COS-7 cells infected with HIV–lentiviral vectors to adherent cells. (Left) EGFP-positive cells after coplating of target cells with COS-7 cells previously infected with the HIV-VP22-EGFP/HSA vector. (Right) EGFP-positive cells after coplating of target cells with COS-7 cells previously infected with the HIV-EGFP/HSA vector. Antibody staining for simian virus 40 T-antigen (Anti-T) was tested on the same slides. (A) Brain choroid plexus cells. (B) Human endothelial cells. (C) HeLa cells. VP22-EGFP was found not only in the nucleus (n), but also in the cytoplasm (c) in endothelial cells and in HeLa cells. Approximately 3 × 104 COS-7 cells and 3 × 105 recipient cells were mixed and plated onto cover slips in 12-well plates in 2.2 ml of medium and processed for antibody staining and microscopy 3 d later. (Scale bar, 15 μm.)
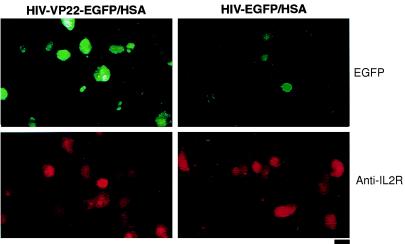
To demonstrate the specificity of VP22-EGFP protein transfer more directly, human H9 cells were used. H9 cells express IL-2Rs (19). These surface receptors can be directly detected by an IL-2R-specific monoclonal antibody. We transferred H9 suspension cells into a culture dish on which transduced COS-7 cells had already adhered and grown for 24 h. Nonadherent cells were removed 3 d later and subjected to fluorescence microscopy. The microscopic analysis presented in Fig. 4 indicates that not only did the H9 cells exhibit binding of a phycoerythrin-labeled IL-2R-specific monoclonal antibody (Fig. 4, red fluorescence), but a significant number of those cells also displayed green fluorescence (Fig. 4, green fluorescence). H9 cells cocultured with COS-7 cells previously infected by the HIV-EGFP/HSA vector displayed red fluorescence but the green fluorescence was greatly reduced and there were no doubly positive cells. The results support the hypothesis that the green fluorescence in H9 cells resulted from the transfer of EGFP mediated by VP22 from the COS-7 cells.
Figure 4.
VP22-EGFP fusion protein transport from COS-7 cells infected with HIV–lentiviral vectors to H9 cells. (Left) EGFP-positive cells after coplating of H9 target cells with COS-7 cells previously infected with the HIV-VP22-EGFP/HSA vector. (Right) EGFP-positive cells after coplating of H9 cells with COS-7 cells previously infected with the HIV-EGFP/HSA vector. Antibody staining for IL-2 receptors on H9 cells (Anti-IL2R) was carried out on the same slides. Approximately 3 × 105 H9 cells were added to a monolayer of infected COS-7 cells and the cells were incubated in 2.2 ml of medium for 3 d at 37°C. Suspended cells were removed and subjected to Cytospin analysis. Cells were visualized by fluorescence microscopy after staining with a phycoerythrin-labeled anti-IL-2R antibody. (Scale bar, 30 μm.)
Lentiviral Vector Delivery of VP22-EGFP Fusion Protein in Mouse Brain.
To determine the capacity to deliver VP22-EGFP from lentivirus-transduced cells in vivo, H9 cells previously infected by lentiviral vectors were injected into the ventricles of brains of mice (Fig. 5A). The results indicate that VP22-EGFP fusion protein had spread into the neighboring tissues from the ventricle (Fig. 5B, green fluorescence), and even as far as the cerebral cortex (Fig. 6A). However, we did not observe such significant transport of EGFP into neighboring tissues (Fig. 5C), nor the cortex when the implanted cells were previously transduced with the HIV-EGFP lentiviral vector lacking VP22 (Fig. 6B). The transplanted H9 cells in brain ventricles were detected by a specific IL-2R antibody (Figs. 5 B′ and C′, red fluorescence).
VP22-EGFP in the cortical region of the mouse brain is present not only in the nuclei of cortical cells (see Fig. 6 B and C, arrow n), but also in the cytoplasm of axons (arrow c).
To further study the delivery of VP22-EGFP fusion protein by lentiviral vector in mouse brain, we injected the viral vectors directly into the pyramidal cell layer in area CA2 of the hippocampus. VP22-EGFP fusion protein was transported throughout the whole pyramidal cell and oriens layers of the hippocampus. Only a local diffusion of EGFP was found when HIV-EGFP/HSA vector lacking a VP22 coding sequence was injected (data not shown).
Discussion
The purpose of this investigation was to determine the efficacy of lentiviral vectors to deliver VP22-EGFP fusion protein to mammalian cells in vitro and to examine the intercellular transport of the corresponding fusion protein in vitro and in vivo. We demonstrated that lentiviral vectors can be used to deliver a gene encoding a VP22-EGFP fusion protein into a mammalian cell such as COS-7 cells. When the infected cells were co-plated with other types of cells, VP22-EGFP was found to migrate to many neighboring cells. An interesting finding is that the transport of VP22-EGFP in cells such as human endothelial cells may be directional, and possibly involves cellular extensions. This intercellular movement suggests cell-to-cell contact during protein transport into this particular cell type from the initial expressing cells. We should not, however, exclude the possibility of extracellular VP22-directed delivery in other cell types. The exact mechanisms involved in the intercellular movement of such a fusion protein are not yet understood. Transport of VP22 fusion protein was reported to occur via a mechanism potentially involving actin microfilaments (20), suggesting that VP22 exhibits a cytoskeletal interaction. Although the numbers of positive recipient cells detected in our analysis were lower than those reported elsewhere (9), the observed transfer was still significant. This discrepancy may, at least in part, be accounted for by the differences in experimental methodology. In our study, cells were trypsinized 24 h after infection with the viral vector before coplating with other cell types as opposed to transfecting or directly microinjecting cells with plasmid DNA (9).
The VP22 fusion protein in the recipient cells was present in both the nucleus and cytoplasm. This discovery is consistent with the findings by Derer et al. (21) that GFP-VP22 fusion protein can be transported into terminally differentiated myotube cell cultures where it preferentially accumulates in the cytoplasm. This predominantly cytoplasmic, rather than nuclear, localization was suggested to be due to the differences in the kinetics of nuclear uptake of the fusion protein. Moreover, Pomeraz and Blaho (13) also found that VP22 exists in the cytoplasm during early herpes simplex virus type 1 infection and later migrates to, and accumulates in, the nucleus. Although most of the VP22 had accumulated in the nucleus at 13 h after infection (13), confocal indirect immunofluorescence detected residual VP22 in the cytoplasm as well.
We have ruled out the possibility of pseudotransduction by including vector stocks lacking a functional Env glycoprotein as negative controls. Also, EGFP lacking the fused VP22 did not appear to be transferred. In addition, we trypsinized the infected cells before coplating with other cell types to eliminate possible contamination from excess virus in the culture solution and on cell surfaces. The immunofluorescence assay was performed 3 days after coplating. Therefore, the data from this study support the concept that intercellular spread of VP22 fusion proteins from cells infected by lentiviral vectors encoding VP22-EGFP is indeed a result of transport involving VP22, and that the VP22-EGFP fusion protein resides in the nucleus and cytoplasm of neighboring cells.
To elicit a sufficient therapeutic response in target tissues in vivo, development of deliverable recombinant fusion protein by viral vectors, such as a lentiviral vector, is needed. To test whether we could improve the global delivery of recombinant proteins in vivo, we constructed a recombinant fusion protein in an HIV-1 based lentiviral vector and examined its delivery in mouse brain. When transduced cells were implanted into the lateral ventricle of the brain, we found VP22-EGFP not only in the tissues immediately surrounding the ventricle, but also in the cerebral cortex. EGFP fluorescence in these brain regions was significantly reduced when cells transduced by a HIV-EGFP/HSA vector lacking VP22 were implanted.
These findings support the view that HIV-1 lentiviral vectors effectively deliver VP22-EGFP fusion proteins from infected target cells to the cytoplasm and nuclei of uninfected cells in vitro and the mouse brain in vivo. VP22-mediated intercellular transport from lentivirus-transduced cells may prove valuable for the delivery of therapeutic proteins. A lentiviral vector coupled with a transport protein such as VP22 may serve as a potent agent in gene therapy applications.
Acknowledgments
We would like to thank Dr. J. McLauchlan for providing the VP22 clone; Dr. Judith Davis for providing technical support at the National Institutes of Health BSL2/3 facility; Drs. Qian Li and Haluk Topaloglu for help with tissue sectioning; Dr. Monique Gelderman for providing the materials for our animal study; Ms. Carolyn Smith for help with confocal microscopy.
Abbreviations
- EGFP
enhanced green fluorescent protein
- FACS
fluorescence-activated cell sorter
- HSA
heat-stable antigen
- VP22
herpes simplex virus type 1 tegument protein
- IL-2R
IL2 receptor
References
- 1.Derossi D, Joliot A H, Chassaing G. J Biol Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- 2.Derossi D, Calvet S, Trembleau A, Brunissen A, Chassaing G, Prochiantz A. J Biol Chem. 1996;271:18188–18193. doi: 10.1074/jbc.271.30.18188. [DOI] [PubMed] [Google Scholar]
- 3.Le Roux I, Duharcourt S, Volovitch M, Prochiantz A, Ronchi E. FEBS Lett. 1995;368:311–314. doi: 10.1016/0014-5793(95)00681-x. [DOI] [PubMed] [Google Scholar]
- 4.Frankel A D, Pabo C O. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 5.Green M, Loewenstein P M. Cell. 1988;55:1179–1188. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- 6.Fawell S, Seery J, Daikh Y, Moore C, Chen L L, Pepinsky B, Barsoum J. Proc Natl Acad Sci USA. 1994;91:664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagahara H, Vocero-Akbani M A, Snyder E, Ho A, Latham G D, Lissy A N, Becker-Hapak M, Ezhevsky A S, Dowdy F S. Nat Med. 1998;4:1449–1452. doi: 10.1038/4042. [DOI] [PubMed] [Google Scholar]
- 8.Schwarze R S, Ho A, Vocero-Akbani A, Dowdy F S. Science. 1999;385:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 9.Elliott G, O'Hare P. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 10.Knopf K W, Kaerner H C. J Gen Virol. 1980;46:405–414. doi: 10.1099/0022-1317-46-2-405. [DOI] [PubMed] [Google Scholar]
- 11.Elliott G D, Meredith D M. J Gen Virol. 1992;73:723–726. doi: 10.1099/0022-1317-73-3-723. [DOI] [PubMed] [Google Scholar]
- 12.Dilber M S, Phelan A, Aints A, Mohamed A J, Elliott G, Edvard Smith S C I, O'Hare P. Gene Ther. 1999;6:12–21. doi: 10.1038/sj.gt.3300838. [DOI] [PubMed] [Google Scholar]
- 13.Pomeranz L, Blaho J. J Virol. 1999;73:6769–6781. doi: 10.1128/jvi.73.8.6769-6781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reiser J, Lai Z, Zhang X Y, Brady R O. J. Virol. 2000. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leslie J, Rixon F J, Mclauchlan J. Virology. 1996;220:60–68. doi: 10.1006/viro.1996.0286. [DOI] [PubMed] [Google Scholar]
- 16.Reiser J, Harmison G, Kluepfel-Stahl S, Brady R O, Karlsson S, Schubert M. Proc Natl Acad Sci USA. 1996;93:15266–15271. doi: 10.1073/pnas.93.26.15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mochizuki H, Schwartz J, Tanaka K, Brady R O, Reiser J. J Virol. 1998;72:8873–8883. doi: 10.1128/jvi.72.11.8873-8883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Popovic M, Sarngadharan G M, Read E, Gallo R C. Science. 1984;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- 19.Gazdar A F, Carney N D, Bunn A P, Russell E K, Jaffe S E, Schechter G P, Guccius J G. Blood. 1980;55:409–417. [PubMed] [Google Scholar]
- 20.Elliott G, O'Hare P. J Virol. 1998;72:6448–6455. doi: 10.1128/jvi.72.8.6448-6455.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derer W, Easwaran H, Knopf C W, Leonhardt H, Cardoso M C. J Mol Med. 1999;77:609–613. doi: 10.1007/s001099900036. [DOI] [PubMed] [Google Scholar]