Abstract
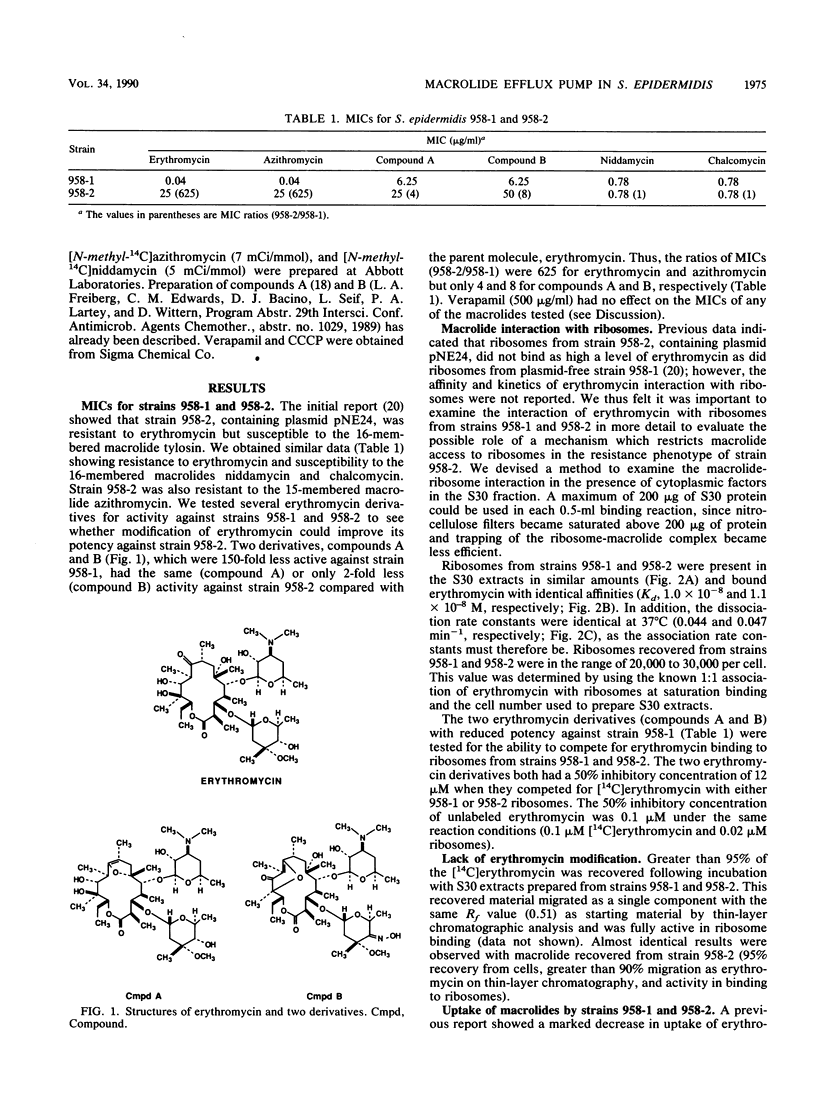
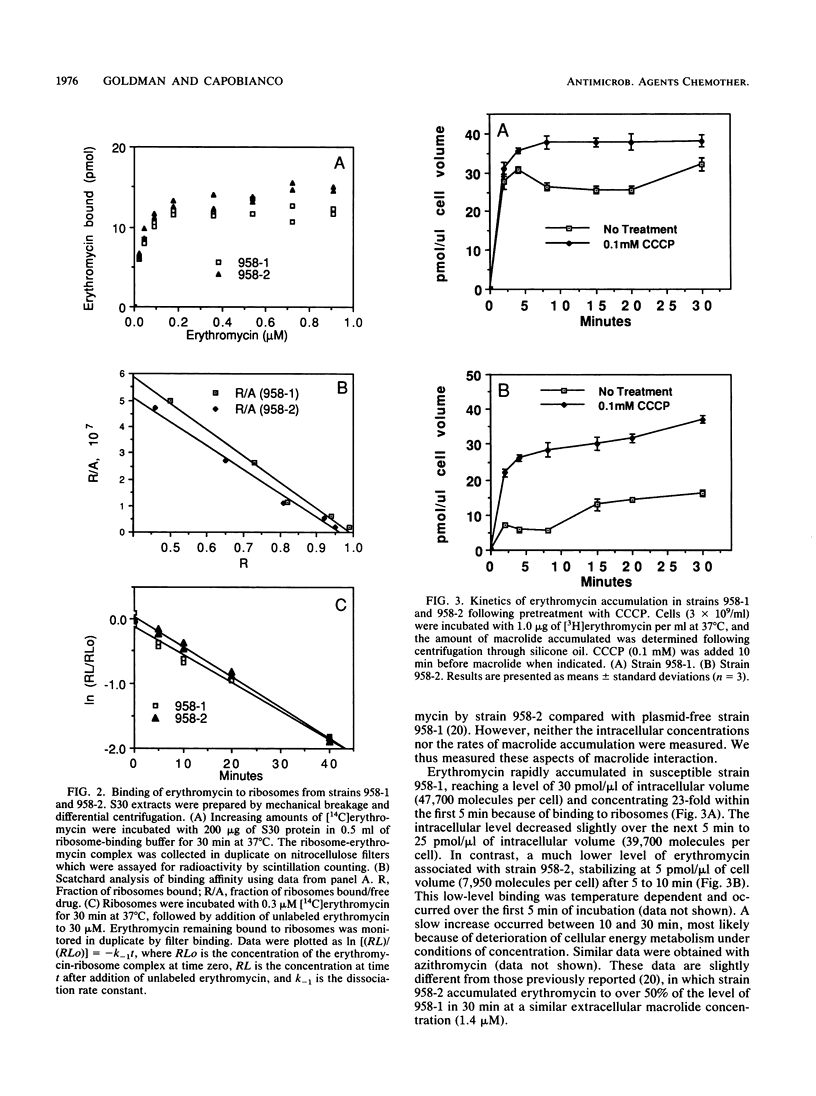
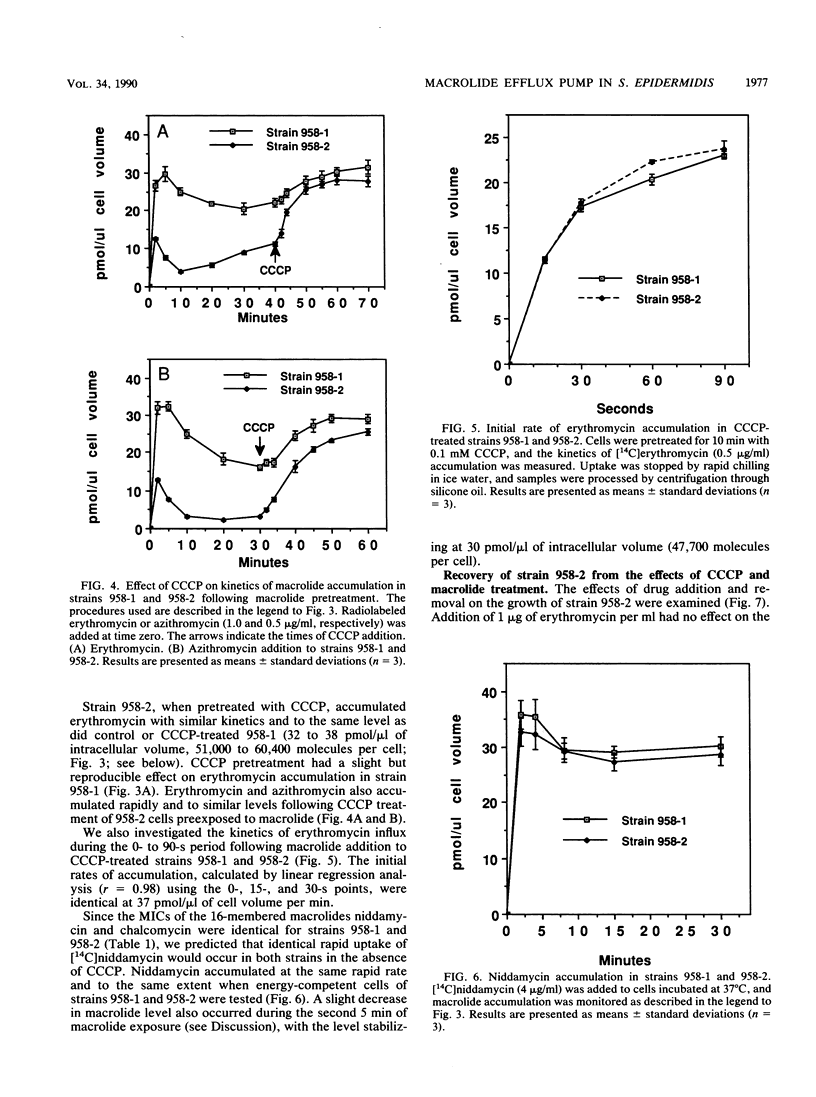
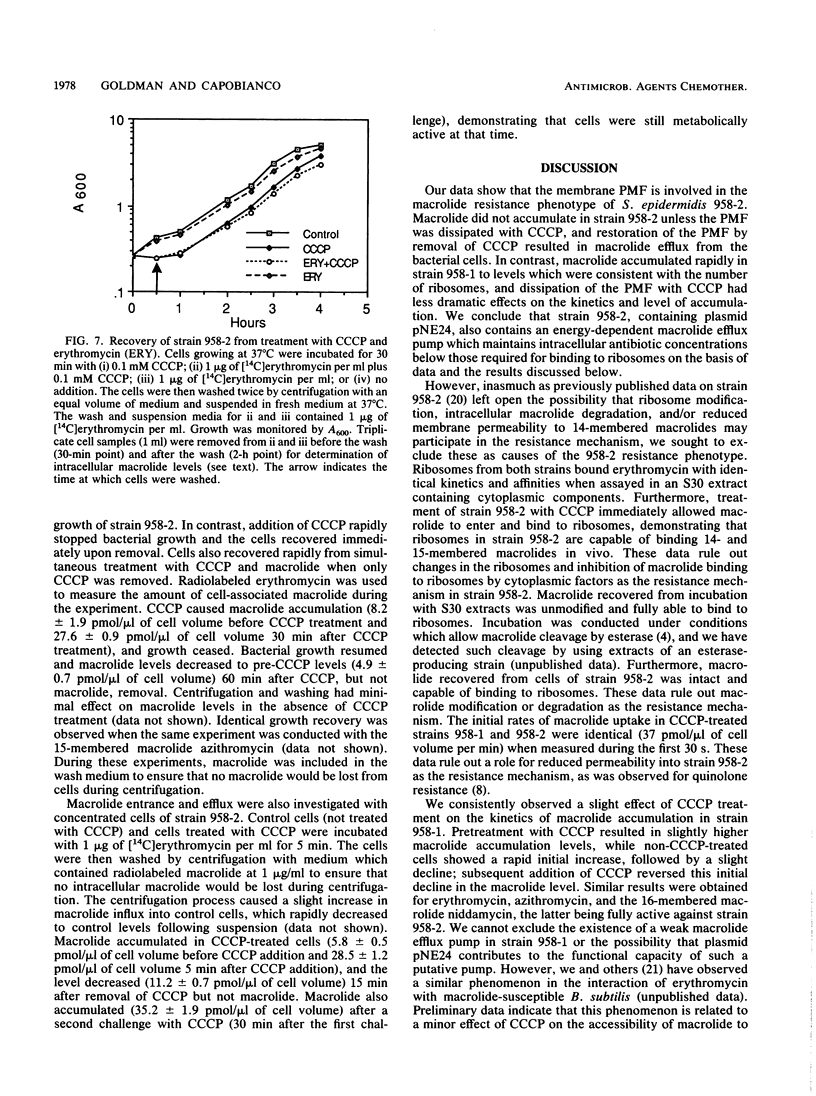
We have elucidated a new mechanism for bacterial resistance to the 14-membered macrolides oleandomycin and erythromycin and the 15-membered macrolide azithromycin. Plasmid pNE24, previously isolated from a clinical specimen of Staphylococcus epidermidis, was characterized as causing resistance to 14-membered but not 16-membered macrolides by a mechanism suggested to involve reduced antibiotic permeation of bacterial cells (B. C. Lampson, W. von David, and J. T. Parisi, Antimicrob. Agents Chemother. 30:653-658, 1986). Our recent investigations have demonstrated that S. epidermidis 958-2 containing plasmid pNE24 also contains an energy-dependent macrolide efflux pump which maintains intracellular antibiotic concentrations below those required for binding to ribosomes. Thus, when strain 958-2 was pretreated with the inhibitor carbonyl cyanide m-chlorophenylhydrazone (CCCP), macrolide accumulated at the same rate and to the same extent as in CCCP-treated or untreated control cells lacking plasmid pNE24 (strain 958-1). In contrast, macrolide did not accumulate in energy-competent strain 958-2 but did accumulate to levels equal to those of ribosomes immediately following CCCP addition. Furthermore, intracellular macrolide was excreted and bacteria resumed growth when CCCP but not macrolide was removed from the growth medium. As expected, the 16-membered macrolide niddamycin accumulated to the same level in energy-competent strains 958-1 and 958-2 at the same rapid rate. Macrolide incubated with lysates prepared from both strains or recovered from cells of strain 958-2 was unmodified and bound to ribosomes from strains 958-1 and 958-2 with identical affinities and kinetics, thus precluding a role for ribosome or drug alteration in the resistance mechanism. We conclude that the presence of plasmid pNE24 results in specific energy-dependent efflux of 14- and 15-membered macrolides.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arthur M., Andremont A., Courvalin P. Distribution of erythromycin esterase and rRNA methylase genes in members of the family Enterobacteriaceae highly resistant to erythromycin. Antimicrob Agents Chemother. 1987 Mar;31(3):404–409. doi: 10.1128/aac.31.3.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur M., Brisson-Noël A., Courvalin P. Origin and evolution of genes specifying resistance to macrolide, lincosamide and streptogramin antibiotics: data and hypotheses. J Antimicrob Chemother. 1987 Dec;20(6):783–802. doi: 10.1093/jac/20.6.783. [DOI] [PubMed] [Google Scholar]
- Barre J., Fournet M. P., Zini R., Deforges L., Duval J., Tillement J. P. In vitro [3H]-erythromycin binding to Staphylococcus aureus. Biochem Pharmacol. 1986 Mar 15;35(6):1001–1004. doi: 10.1016/0006-2952(86)90090-0. [DOI] [PubMed] [Google Scholar]
- Barthélémy P., Autissier D., Gerbaud G., Courvalin P. Enzymic hydrolysis of erythromycin by a strain of Escherichia coli. A new mechanism of resistance. J Antibiot (Tokyo) 1984 Dec;37(12):1692–1696. doi: 10.7164/antibiotics.37.1692. [DOI] [PubMed] [Google Scholar]
- Birmingham V. A., Cox K. L., Larson J. L., Fishman S. E., Hershberger C. L., Seno E. T. Cloning and expression of a tylosin resistance gene from a tylosin-producing strain of Streptomyces fradiae. Mol Gen Genet. 1986 Sep;204(3):532–539. doi: 10.1007/BF00331036. [DOI] [PubMed] [Google Scholar]
- Capobianco J. O., Doran C. C., Goldman R. C. Mechanism of mupirocin transport into sensitive and resistant bacteria. Antimicrob Agents Chemother. 1989 Feb;33(2):156–163. doi: 10.1128/aac.33.2.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capobianco J. O., Goldman R. C. Erythromycin and azithromycin transport into Haemophilus influenzae ATCC 19418 under conditions of depressed proton motive force (delta mu H). Antimicrob Agents Chemother. 1990 Sep;34(9):1787–1791. doi: 10.1128/aac.34.9.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. P., McMurry L. M., Hooper D. C., Wolfson J. S., Levy S. B. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob Agents Chemother. 1989 Aug;33(8):1318–1325. doi: 10.1128/aac.33.8.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dette G. A., Knothe H., Kaula S. Modifying effects of pH and temperature on (14C)erythromycin uptake into Staphylococcus aureus--relation to antimicrobial activity. Zentralbl Bakteriol Mikrobiol Hyg A. 1987 Jul;265(3-4):393–403. [PubMed] [Google Scholar]
- Dubnau D. Translational attenuation: the regulation of bacterial resistance to the macrolide-lincosamide-streptogramin B antibiotics. CRC Crit Rev Biochem. 1984;16(2):103–132. doi: 10.3109/10409238409102300. [DOI] [PubMed] [Google Scholar]
- Duval J. Evolution and epidemiology of MLS resistance. J Antimicrob Chemother. 1985 Jul;16 (Suppl A):137–149. doi: 10.1093/jac/16.suppl_a.137. [DOI] [PubMed] [Google Scholar]
- Fierro J. F., Hardisson C., Salas J. A. Involvement of cell impermeability in resistance to macrolides in some producer streptomycetes. J Antibiot (Tokyo) 1988 Jan;41(1):142–144. doi: 10.7164/antibiotics.41.142. [DOI] [PubMed] [Google Scholar]
- Goldman R. C., Fesik S. W., Doran C. C. Role of protonated and neutral forms of macrolides in binding to ribosomes from gram-positive and gram-negative bacteria. Antimicrob Agents Chemother. 1990 Mar;34(3):426–431. doi: 10.1128/aac.34.3.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman R. C., Kadam S. K. Binding of novel macrolide structures to macrolides-lincosamides-streptogramin B-resistant ribosomes inhibits protein synthesis and bacterial growth. Antimicrob Agents Chemother. 1989 Jul;33(7):1058–1066. doi: 10.1128/aac.33.7.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofsli E., Nissen-Meyer J. Effect of erythromycin and tumour necrosis factor on the drug resistance of multidrug-resistant cells: reversal of drug resistance by erythromycin. Int J Cancer. 1989 Mar 15;43(3):520–525. doi: 10.1002/ijc.2910430330. [DOI] [PubMed] [Google Scholar]
- Kuo M. S., Chirby D. G., Argoudelis A. D., Cialdella J. I., Coats J. H., Marshall V. P. Microbial glycosylation of erythromycin A. Antimicrob Agents Chemother. 1989 Dec;33(12):2089–2091. doi: 10.1128/aac.33.12.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurath P., Jones P. H., Egan R. S., Perun T. J. Acid degradation of erythromycin A and erythromycin B. Experientia. 1971 Apr 15;27(4):362–362. doi: 10.1007/BF02137246. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Weisblum B. Altered methylation of ribosomal RNA in an erythromycin-resistant strain of Staphylococcus aureus. Proc Natl Acad Sci U S A. 1971 Apr;68(4):856–860. doi: 10.1073/pnas.68.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson B. C., von David W., Parisi J. T. Novel mechanism for plasmid-mediated erythromycin resistance by pNE24 from Staphylococcus epidermidis. Antimicrob Agents Chemother. 1986 Nov;30(5):653–658. doi: 10.1128/aac.30.5.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J. C., Putterman M. Accumulation in gram-postive and gram-negative bacteria as a mechanism of resistance to erythromycin. J Bacteriol. 1968 Mar;95(3):1111–1117. doi: 10.1128/jb.95.3.1111-1117.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura S., Namiki S., Shibata M., Muro T., Sawada J. Studies on the antibiotics from Streptomyces spinichromogenes var. kujimyceticus. V. Some antimicrobial characteristics of kujimycin A and kujimycin B against macrolide resistant staphylococci. J Antibiot (Tokyo) 1970 Sep;23(9):448–460. doi: 10.7164/antibiotics.23.448. [DOI] [PubMed] [Google Scholar]
- Pestka S. Binding of [14C]erythromycin to Escherichia coli ribosomes. Antimicrob Agents Chemother. 1974 Oct;6(4):474–478. doi: 10.1128/aac.6.4.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath L. D., Lorian V., Gerstein D., Loder P. B., Finland M. Enhancing effect on alkalinization of the medium on the activity of erythromycin against gram-negative bacteria. Appl Microbiol. 1968 Sep;16(9):1288–1292. doi: 10.1128/am.16.9.1288-1292.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner R., Cundliffe E., Schmidt F. J. Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J Biol Chem. 1983 Oct 25;258(20):12702–12706. [PubMed] [Google Scholar]
- Taber H. W., Mueller J. P., Miller P. F., Arrow A. S. Bacterial uptake of aminoglycoside antibiotics. Microbiol Rev. 1987 Dec;51(4):439–457. doi: 10.1128/mr.51.4.439-457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai P. C., Wallace B. J., Davis B. D. Selective action of erythromycin on initiating ribosomes. Biochemistry. 1974 Oct 22;13(22):4653–4659. doi: 10.1021/bi00719a029. [DOI] [PubMed] [Google Scholar]
- Walter A., Gutknecht J. Monocarboxylic acid permeation through lipid bilayer membranes. J Membr Biol. 1984;77(3):255–264. doi: 10.1007/BF01870573. [DOI] [PubMed] [Google Scholar]
- Wiley P. F., Baczynskyj L., Dolak L. A., Cialdella J. I., Marshall V. P. Enzymatic phosphorylation of macrolide antibiotics. J Antibiot (Tokyo) 1987 Feb;40(2):195–201. doi: 10.7164/antibiotics.40.195. [DOI] [PubMed] [Google Scholar]
- Zalacain M., Cundliffe E. Methylation of 23S rRNA caused by tlrA (ermSF), a tylosin resistance determinant from Streptomyces fradiae. J Bacteriol. 1989 Aug;171(8):4254–4260. doi: 10.1128/jb.171.8.4254-4260.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]


