Abstract
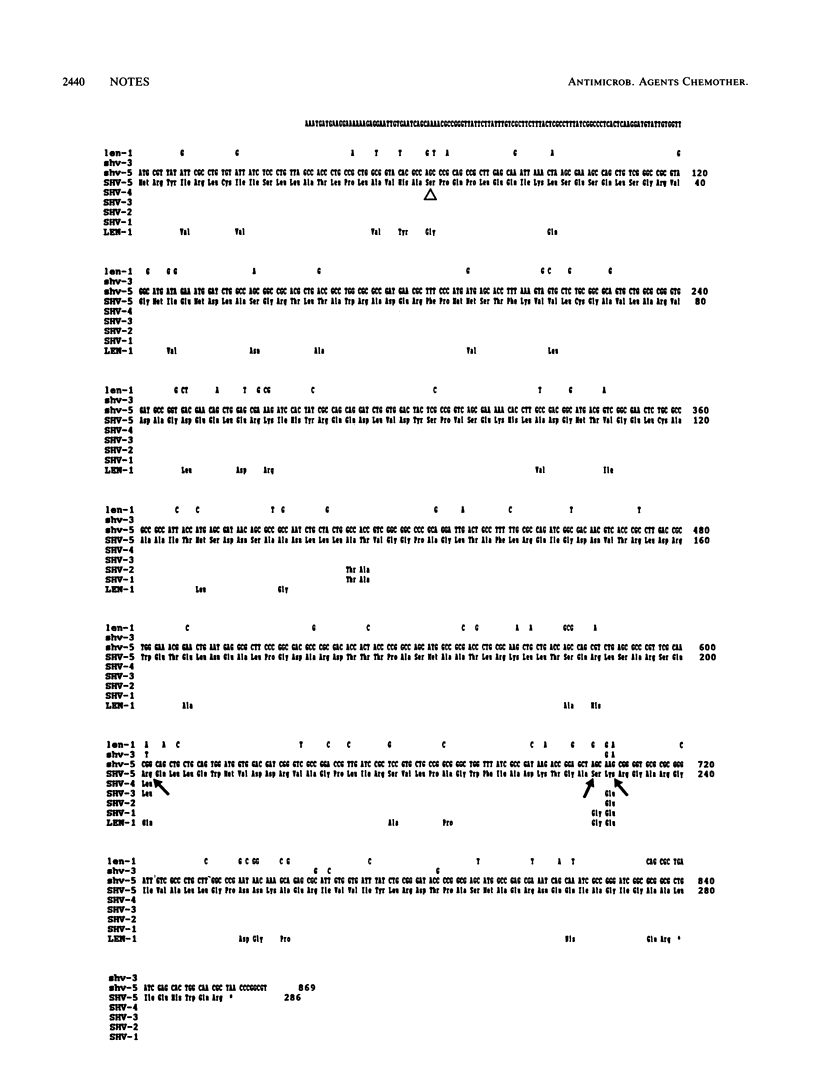
The nucleotide sequence of the SHV-5 beta-lactamase gene, subcloned from a plasmid of Klebsiella pneumoniae, was determined. The amino acid changes thought to be responsible for the extended substrate profile of SHV-5 are Gly----Ser234 and Glu----Lys235. SHV-5 is identical to SHV-4, except for Leu----Arg201, which accounts for the difference in apparent pI of the two enzymes.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Arakawa Y., Ohta M., Kido N., Fujii Y., Komatsu T., Kato N. Close evolutionary relationship between the chromosomally encoded beta-lactamase gene of Klebsiella pneumoniae and the TEM beta-lactamase gene mediated by R plasmids. FEBS Lett. 1986 Oct 20;207(1):69–74. doi: 10.1016/0014-5793(86)80014-x. [DOI] [PubMed] [Google Scholar]
- Barthélémy M., Peduzzi J., Labia R. Complete amino acid sequence of p453-plasmid-mediated PIT-2 beta-lactamase (SHV-1). Biochem J. 1988 Apr 1;251(1):73–79. doi: 10.1042/bj2510073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthélémy M., Péduzzi J., Ben Yaghlane H., Labia R. Single amino acid substitution between SHV-1 beta-lactamase and cefotaxime-hydrolyzing SHV-2 enzyme. FEBS Lett. 1988 Apr 11;231(1):217–220. doi: 10.1016/0014-5793(88)80734-8. [DOI] [PubMed] [Google Scholar]
- Gutmann L., Ferré B., Goldstein F. W., Rizk N., Pinto-Schuster E., Acar J. F., Collatz E. SHV-5, a novel SHV-type beta-lactamase that hydrolyzes broad-spectrum cephalosporins and monobactams. Antimicrob Agents Chemother. 1989 Jun;33(6):951–956. doi: 10.1128/aac.33.6.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzberg O., Moult J. Bacterial resistance to beta-lactam antibiotics: crystal structure of beta-lactamase from Staphylococcus aureus PC1 at 2.5 A resolution. Science. 1987 May 8;236(4802):694–701. doi: 10.1126/science.3107125. [DOI] [PubMed] [Google Scholar]
- Huovinen P., Huovinen S., Jacoby G. A. Sequence of PSE-2 beta-lactamase. Antimicrob Agents Chemother. 1988 Jan;32(1):134–136. doi: 10.1128/aac.32.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarlier V., Nicolas M. H., Fournier G., Philippon A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988 Jul-Aug;10(4):867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- Joris B., Ghuysen J. M., Dive G., Renard A., Dideberg O., Charlier P., Frère J. M., Kelly J. A., Boyington J. C., Moews P. C. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 DD-peptidase family. Biochem J. 1988 Mar 1;250(2):313–324. doi: 10.1042/bj2500313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebe C., Nies B. A., Meyer J. F., Tolxdorff-Neutzling R. M., Wiedemann B. Evolution of plasmid-coded resistance to broad-spectrum cephalosporins. Antimicrob Agents Chemother. 1985 Aug;28(2):302–307. doi: 10.1128/aac.28.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labia R., Morand A., Tiwari K., Sirot J., Sirot D., Petit A. Interactions of new plasmid-mediated beta-lactamases with third-generation cephalosporins. Rev Infect Dis. 1988 Jul-Aug;10(4):885–891. doi: 10.1093/clinids/10.4.885. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossakowska D., Ali N. A., Dale J. W. Oxacillin-hydrolysing beta-lactamases. A comparative analysis at nucleotide and amino acid sequence levels. Eur J Biochem. 1989 Mar 15;180(2):309–318. doi: 10.1111/j.1432-1033.1989.tb14649.x. [DOI] [PubMed] [Google Scholar]
- Nicolas M. H., Jarlier V., Honore N., Philippon A., Cole S. T. Molecular characterization of the gene encoding SHV-3 beta-lactamase responsible for transferable cefotaxime resistance in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1989 Dec;33(12):2096–2100. doi: 10.1128/aac.33.12.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippon A., Labia R., Jacoby G. Extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1989 Aug;33(8):1131–1136. doi: 10.1128/aac.33.8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péduzzi J., Barthélémy M., Tiwari K., Mattioni D., Labia R. Structural features related to hydrolytic activity against ceftazidime of plasmid-mediated SHV-type CAZ-5 beta-lactamase. Antimicrob Agents Chemother. 1989 Dec;33(12):2160–2163. doi: 10.1128/aac.33.12.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirot D., Chanal C., Labia R., Meyran M., Sirot J., Cluzel R. Comparative study of five plasmid-mediated ceftazidimases isolated in Klebsiella pneumoniae. J Antimicrob Chemother. 1989 Oct;24(4):509–521. doi: 10.1093/jac/24.4.509. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]


