Abstract
DsRed, a brilliantly red fluorescent protein, was recently cloned from Discosoma coral by homology to the green fluorescent protein (GFP) from the jellyfish Aequorea. A core question in the biochemistry of DsRed is the mechanism by which the GFP-like 475-nm excitation and 500-nm emission maxima of immature DsRed are red-shifted to the 558-nm excitation and 583-nm emission maxima of mature DsRed. After digestion of mature DsRed with lysyl endopeptidase, high-resolution mass spectra of the purified chromophore-bearing peptide reveal that some of the molecules have lost 2 Da relative to the peptide analogously prepared from a mutant, K83R, that stays green. Tandem mass spectrometry indicates that the bond between the alpha-carbon and nitrogen of Gln-66 has been dehydrogenated in DsRed, extending the GFP chromophore by forming —C⩵N—C⩵O at the 2-position of the imidazolidinone. This acylimine substituent quantitatively accounts for the red shift according to quantum mechanical calculations. Reversible hydration of the C⩵N bond in the acylimine would explain why denaturation shifts mature DsRed back to a GFP-like absorbance. The C⩵N bond hydrolyses upon boiling, explaining why DsRed shows two fragment bands on SDS/PAGE. This assay suggests that conversion from green to red chromophores remains incomplete even after prolonged aging.
In the preceding companion paper (1), we showed that the red fluorescent protein DsRed cloned from coral by Matz et al. (2) has impressive brightness and stability against pH changes, denaturants, and photobleaching, and that it can be mutated to even longer wavelengths of excitation and emission. However, DsRed also shows slow and complex kinetics of maturation, proceeding via a green fluorescent protein (GFP)-like green intermediate to the final red species. DsRed also proves to be at least an obligate tetramer, two of which may weakly associate into an octamer (1). The present paper examines one of the most important fundamental unsolved questions about DsRed, namely the structure of the red chromophore and the mechanism by which it attains such long wavelengths. Matz et al. (2) speculated that “an additional autocatalytic reaction, presumably inhibited in GFP, takes place during fluorophore maturation and leads to the extension of its conjugated π-system.” In an accompanying commentary, Tsien (3) raised the possibility that an amine side chain might attack the terminal carbonyl of the GFP chromophore to form an iminium cation somewhat analogous to that in rhodopsin. More recently, Terskikh et al.** suggested instead that “the additional reaction is another cyclization within the backbone rather than attachment of a group derived from a neighboring side chain.” All of these suggestions have been of generic reaction types rather than specific proposals of a detailed structure for the red chromophore.
Our strategy relied critically on a point mutant, K83R, which remains almost entirely arrested at the green stage of maturation (1) and provides a reference standard for that intermediate. Lysyl endopeptidase proteolysis, isolation of the chromophore-bearing peptides by HPLC, and high-resolution mass spectrometry show that DsRed, unlike K83R, has undergone an additional dehydrogenation of the αC-N bond of Gln-66, which extends the GFP chromophore by two strongly electron-withdrawing double bonds. The resulting acylimine substituent quantitatively ac-counts for the red shift by detailed quantum chemical calculations. It also explains why the DsRed chromophore shifts back to a GFP-like absorbance spectrum upon protein denaturation and why harsher treatments cleave DsRed into two fragments.
Materials and Methods
Mass Spectral Analysis of LysC Digests.
Approximately 2 nmol of His-tagged DsRed protein or its K83R mutant (1) was evaporated to near dryness in vacuo then resuspended in 3 μl of 6 M guanidinium chloride and heated to 80°C for 30 s. The DsRed solution, which turned yellow after this treatment, was cooled to room temperature. To this cooled solution was added 1.0 μl of 0.33 M hydrochloric acid (to neutralize the digestion buffer to pH 8.6) and 10 μl LysC endoprotease (Wako Biochemicals, Osaka) (0.05 μg/μl in 100 mM Tris, pH 9.2) (4). The digest was allowed to proceed for 22 h at 37oC, and then quenched with 40 μl trifluoroacetic acid (0.1% vol/vol) before injection onto an HPLC column (C8 300-Å Zorbax column, 1 mm i.d. by 150-mm length). Peptide fractions were collected and their masses were analyzed with an Applied Biosystems QSTAR quadrupole time-of-flight mass spectrometer.
Mass Spectral Analysis After Mild Trypsinolysis.
One microliter of a 50 μg/ml solution of trypsin in pH 7.5 Tris buffer was added to 20 μl samples containing 50 μg of either DsRed or the K83R mutant. The tubes then were incubated overnight at room temperature. The ratio of fluorescent protein to trypsin was 1,000:1 by weight, which was sufficient to quantitatively cleave away the 36 N-terminal amino acid residues, which included all of the His tag through to the first arginine in the polypeptide sequence. The C terminus remained intact in this treatment. The trimmed protein samples (5 μg per injection) were desalted by reverse-phase HPLC using a Michrom (Auburn, CA) UMA model narrow-bore chromatograph. The column was 1 mm i.d. by 50-mm length, with a stationary phase of C18 modified polystyrene-divinylbenzene beads, from which the protein was eluted after 5 min of the gradient program (50 μl/min, 20–68% B, where A = 10% acetonitrile, B = 90% acetonitrile, 0.1% trifluoroacetic acid). The fraction was manually collected and immediately injected into the mass spectrometer interface, a Hewlett–Packard Mass Engine 5989B, in which the electrospray was continuously supplied by a constant flow of 10 μl/min of 50% methanol, 1% acetic acid. The resulting mass spectra were averaged and deconvolved by using the Hewlett–Packard hpchemstation programs supplied with the instrument control software.
Computational Methods.
All calculations were carried out by using the gaussian 98 (5) and gamess (6) software packages, running on the hardware facilities at the San Diego Supercomputer Center. The molecular structures were determined by using a variety of levels of theory to establish self-consistency in terms of basis sets as well as effects of dynamic correlation. This level then was used for the remaining analogues. Hybrid density functional theory (HDFT) methods were thus primarily used. The HDFT methods used Becke's three-parameter hybrid exchange functional (7) in combination with the nonlocal correlation provided by the Lee-Parr expression (8, 9) that contains both local and nonlocal terms, B3LYP. The 6–31G+(d,p) (10) split valence basis set was used for all fully unrestrained geometry optimizations, and the 6–31++G(d,p) basis was used for additional HDFT time-dependent analysis. These basis sets include one set of six d polarization functions on all heavy atoms, and one set of p polarization functions on hydrogen atoms. In addition, diffuse functionality was included, (+) and (++), to more adequately accommodate the negative charge of these molecules. These levels of theory have been previously shown by us and others to be reliable for structural determination in these types of compounds. Excited-state energy computations were performed by using both Zerner's modified intermediate neglect of differential overlap (ZINDO) method (11, 12) as well as the more rigorous time-dependent HDFT (TDHDFT) method (13), both performed at the optimized HDFT geometry using the basis set specifications indicated above.
Results
Chromophore Structure Revealed by Mass Spectra.
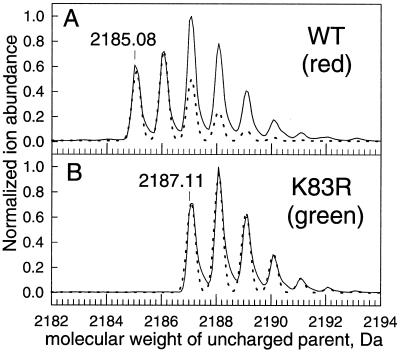
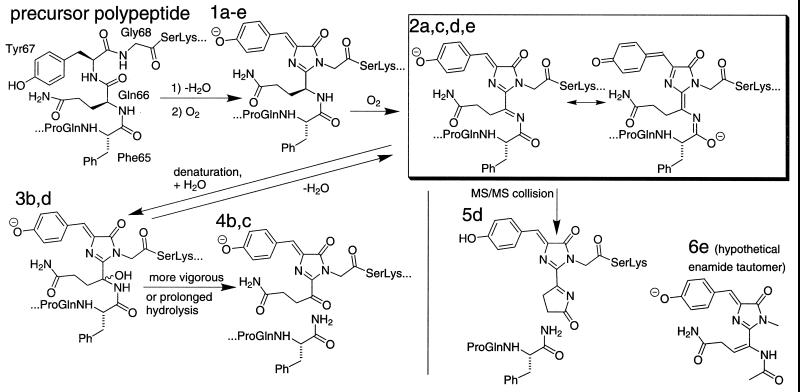
Mature DsRed and its green K83R mutant were separately digested by LysC, a lysyl endopeptidase. The peptide containing the chromophore absorbing at 360 nm was isolated by HPLC as described in Materials and Methods. The mass spectrum of the K83R chromopeptide (Fig. 1B) showed a monoisotopic mass of 2187.11. This mass is exactly what would be expected from cyclization (loss of H2O) and oxidation (O2-mediated loss of H2) of Gln-66–Tyr-67–Gly-68 (Fig. 2, precursor polypeptide) to form a p-hydroxybenzylideneimidazolidinone (Fig. 2, structure 1a) just as in GFP, followed by LysC cleavage after lysines 50 and 70 to give the fragment GGPLPFAWDILSPQF-[green chromophore]-SK. The mass spectrum of the wild-type chromopeptide showed a mixture of the 2187.11-Da species plus a congener having lost an additional 2 Da, with approximately equal amplitudes (Fig. 1A). Loss of 2 Da immediately suggested an additional dehydrogenation, for which the most plausible location would be within the remnants of Gln-66 to form an acylimine, as shown in structure 2 of Fig. 2. Table 1 lists the optical and mass spectral properties of each structure in up to five different contexts, the intact (a) or denatured (b) protein with the N-terminal polyhistidine tag, with that tag removed (c), the HPLC-purified LysC proteolytic fragment derived from residues 51 to 70 (d), and a computational model (e) where the aliphatic linkages to the rest of the protein have been replaced by methyls. Thus the 2187.11- and 2185.08-Da species are interpreted as structures 1d and 2d, respectively.
Figure 1.
Mass spectra (solid lines) of HPLC-purified chromopeptides from LysC digests of denatured DsRed (A) and green mutant K83R (B). The dashed lines indicate the theoretical mass spectra for structures 2d and 1d considering the natural abundances of 13C, 2H, 15N, 17O, and 18O. The mass scale is for the uncharged parent species, deconvoluted from the +2 to +4 species (A) or +2 to +3 species (B). WT, Wild type.
Figure 2.
Summary scheme for conversion of immature green form (1) to mature red form of DsRed (2) and reactions after denaturation of the latter. Letter suffixes denote different contexts for the same chromophore: (a) intact or (b) denatured protein with the N-terminal polyhistidine tag, (c) with that tag removed, (d) the HPLC-purified LysC proteolytic fragment derived from residues 51–70, and (e) a computational model where the aliphatic linkages to the rest of the protein have been replaced by methyls. Table 1 enumerates the interconversions and properties of the protein species and fragments.
Table 1.
DsRed species formed during maturation, denaturation, and degradation
| Structure | Produced by | Mass or optical properties |
|---|---|---|
| 1a (nondenatured) | First stage of maturation (K83R goes no further) | Exc 475, em 500 nm |
| 1b (denatured, full length) | Denaturation of 1a (especially K83R) | 37 kDa by SDS/PAGE |
| 1c (trimmed with trypsin) | Mild trypsinolysis of 1a | 25648 Da (K83R) found 25651.4 Da calculated |
| 1d (LysC fragment) | Complete digestion of 1a with LysC | 2187.11 Da found 2187.08 Da calculated |
| 1e (computational model) | Truncation of Phe-65 & Gly-68 to methyls | Exc 456 nm (ZINDO) or 445 nm (TDHDFT) |
| 2a (nondenatured) | Second stage of maturation | Exc 558, em 583 nm |
| 2c (trimmed with trypsin) | Mild trypsinolysis of 2a, dehydration in electrospray | 25622 ± 4 Da found 25621.4 Da calculated |
| 2d (LysC fragment) | Complete digestion of 2a with LysC to form 3d, dehydration in electrospray | 2185.08 Da found 2185.06 Da calculated |
| 2e (computational model) | Truncation of Phe-65 & Gly-68 to methyls | Exc 563 nm (ZINDO) or 531 nm (TDHDFT) |
| 3b (denatured, hydrated) | Mild denaturation of 2a | Abs 383 nm (neutral pH) Abs 452 nm (alkaline pH) |
| 3d (LysC fragment) | Complete digestion of 2a with LysC to form 3d | |
| 4b (denatured, cleaved) | Boiling of 2a, especially at extreme pH | 15 kDa + 22 kDa fragments by SDS/PAGE |
| 4c (trimmed with trypsin) | Same as for 2c | 7149 ± 3 Da found 7150.2 Da calculated 18488 ± 3 Da found 18489.1 Da calculated |
| 5d (LysC fragment after cyclization in MS/MS) | Cyclization of 2d in MS/MS and cleavage between Ser-62 and Pro-63 | 542.22 Da found 542.21 Da calculated 389.20 Da found 389.21 Da calculated |
| 5d (internal fragment ProGlnPhe-amide) | ||
| 6e (computational model for enamide tautomer of 2e) | Truncation of Phe-65 and Gly-68 to methyls | Exc 490 nm (ZINDO) or 493 nm (TDHDFT) |
Mild trypsinolysis removes the N-terminal polyhistidine tag and residues 1, 1a, and 2 (MetValArg) of DsRed. LysC cuts at many lysines but only the chromophore-containing fragment from residues 51 to 70 was isolated by HPLC and analyzed by mass spectrometry. Masses are stated for the neutral parent species before charging with 1 or more protons. See Fig. 2. for corresponding structures of the chromophore regions.
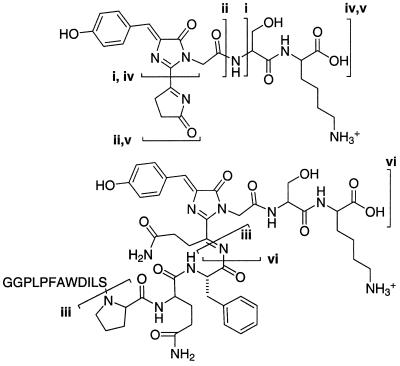
Both species were separately subjected to further fragmentation in tandem mass spectrometry, which showed that the 2185.08-Da but not the 2187.11-Da species gave rise to prominent peaks at 542.2 and 390.2 Da. These species (5d in Fig. 2 and Table 1) are explained by intramolecular attack of the side-chain amide of Gln-66 on the acylimine and cleavage between Ser-62 and Pro-63. Many other carboxyl-terminal chromopeptide fragments, i.e., “y” ions, also showed the loss of 2 Da relative to the corresponding fragments from the K83R chromopeptide (Table 2 and Fig. 3).
Table 2.
Major peaks in secondary mass spectrum derived from collisional fragmentation of the DsRed chromopeptide ion (parent mass 2185.08) and the corresponding ion from the K83R mutant (parent mass 2187.11)
| Assignment | Calculated m/z | Observed m/z | SD | Calculated −observed m/z | Relative amplitude |
|---|---|---|---|---|---|
| Distinctive ions in secondary MS derived from DsRed chromopeptide ion | |||||
| x4c18(i) | 246.088 | 246.120 | .015 | −0.032 | 14 |
| y5b18-NH3(ii) | 310.083 | 310.076 | .013 | 0.007 | 9 |
| PQF + 17 y8c15(iii) | 390.214 | 390.208 | .010 | 0.006 | 13 |
| x4c18(iv) | 462.199 | 462.198 | .013 | 0.001 | 22 |
| (y8-2H)2+ | 466.717 | 466.724 | .006 | −0.007 | 8 |
| (y9-2H)2+ | 510.233 | 510.236 | .004 | −0.003 | 6 |
| y5-2H-NH3(v) | 543.220 | 543.224 | .011 | −0.004 | 100 |
| y5-2H (vi) | 560.247 | 560.256 | .014 | −0.009 | 11 |
| (y10-2H)2+ | 566.775 | 566.788 | .012 | −0.013 | 2 |
| (y11-2H)2+ | 623.317 | 623.320 | .004 | −0.003 | 1 |
| (y12-2H)2+ | 680.831 | 680.830 | .017 | 0.001 | 2 |
| y8-2H | 932.427 | 932.457 | .023 | −0.030 | 25 |
| y9-2H | 1019.459 | 1019.485 | .014 | −0.026 | 10 |
| y10-2H | 1132.543 | 1132.546 | .023 | −0.003 | 3 |
| Distinctive ions in secondary MS derived from K83R chromopeptide ion | |||||
| y82+ | 467.725 | 467.723 | .006 | 0.002 | 17 |
| y92+ | 511.241 | 511.240 | .004 | 0.001 | 11 |
| y102+ | 567.783 | 567.790 | .026 | −0.007 | 3 |
| y112+ | 624.325 | 624.349 | .035 | −0.024 | 1 |
| y122+ | 681.839 | 681.860 | .016 | −0.021 | 3 |
| y8 | 934.442 | 934.461 | .026 | −0.019 | 30 |
| y9 | 1021.474 | 1021.506 | .007 | −0.032 | 19 |
| y10 | 1134.558 | 1134.556 | .039 | 0.002 | 6 |
Separate wild-type and K83R mutant peptide digests were prepared and the resulting spectra were obtained on three different days. Fig. 3 gives the structures of the ions assigned as i–vi. C-terminal fragment ions (yn species, where n denotes the peptide bond cleaved counting from the C terminus of the peptide at Lys-70) already include the dehydration and dehydrogenation necessary to generate the K83R or GFP-like chromophore 1; yn − 2H indicates the corresponding ion with one additional dehydrogenation. Twenty-nine b-type ions and internal ions are omitted because they did not include the residues modified by the chromophore and therefore were observed in both of the secondary mass spectra. The ions that did include the chromophore generally differed from their calculated mass-to-charge by less than .01 m/z, within the measurement error.
Figure 3.
Proposed structures for the more complex fragments of Table 1. Note that iii and v are the same as the two fragments in structure 5d of Fig. 2.
Theoretical Calculations of Spectra.
The proposed acylimine would extend the GFP-like chromophore by two electron-withdrawing double bonds, which would intuitively explain a considerable red shift. To obtain more quantitative and objective estimates, computations were performed on the immature green form of the chromophore 1e, the proposed red species 2e, and the enamide tautomer 6e. The peptide backbone was truncated at the α carbons of Phe-65 and Gly-68, so that those aliphatic carbons were modeled as methyl groups. Ground-state optimizations indicated that the acylimine 2e is more stable than 6e by 7 kcal/mol, presumably because structures of generic structure 2 permit more extensive delocalization of the chromophore through the N⩵C—C⩵N—C⩵O moiety. Greater delocalization in 2 is confirmed by lesser alternation in bond lengths in 2e compared with 6e. Simple acylimines with enolizable hydrogens are normally less stable than their enamide tautomers (14), so it was important to check whether our computational methods could reproduce this preference or whether they were biased in favor of acylimines. Ground-state computations performed at the same level of theory as that of the protein fragments predicted the acylimine AcN⩵C(Me)—CH2Me to be 1.6 kcal/mol less stable than the enamide AcNH—C(Me)⩵CH—Me. Thus our computations predict the correct tautomer for a small model enamide. Presumably the superior resonance possible in an amide dominates when additional conjugation as in 2e is impossible.
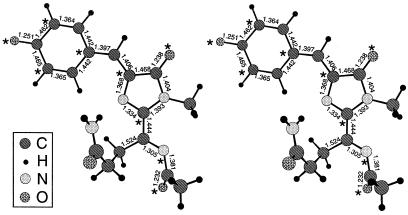
The theoretically predicted ground-state structure is shown in Fig. 4. The conjugated π-system making up the chromophore is quite planar, with the surprising exception of the terminal oxygen of the acylimine. The dihedral angle of the C⩵N—C⩵O was computed to be 48°, and recalculations with this dihedral angle forced to zero predicted such a conformation to be energetically less favorable. Perhaps the C⩵O tilts out of the plane to achieve greater conjugation with the lone pair of electrons on the adjacent N. Another interesting feature of the computed structure is the pronounced alternation in bond lengths within the phenolate ring and the nearly equal lengths of the two bonds connecting the phenolate to the imidazolidinone ring. These bond lengths suggest that in this portion of the molecule, the benzenoid and quinonoid resonance forms shown for 2 in Fig. 2 make roughly equal contributions.
Figure 4.
Computed ground-state structure of the DsRed chromophore 2e, shown as a stereo pair for viewing with uncrossed eyes. Symbols for C, H, N, and O are explained in the Inset. Relevant bond lengths are given in angstroms. Starred and unstarred atoms, respectively, indicate where maxima and nodes of the highest occupied molecular orbital should be located according to basic molecular orbital theory (15), in agreement with the far more detailed calculations presented in Fig. 7.
Fig. 7, which is published as supplemental material on the PNAS website, www.pnas.org, illustrates the highest occupied and lowest unoccupied molecular orbitals for structure 2e. As predicted by the lowest-level theory of odd alternate hydrocarbons (15), the highest occupied molecular orbital is a nonbonding orbital with its maxima roughly on the starred atoms, whereas its nodes are near the unstarred atoms in Fig. 4. Several of these maxima and nodes approximately swap places in the lowest unoccupied molecular orbital.
From the optimized structures of 1e, 2e, and 6e, both ZINDO and TDHDFT computations were done to predict the excitation wavelengths. For 1e, the two values were 456 nm and 445 nm, respectively, slightly lower than the observed values of 475 nm in the intact green form 1a and quite close to the 452 nm for denatured protein 1b. For 2e, the predictions were 563 nm and 531 nm, and for 6e, 490 nm and 493 nm, respectively. The experimental value of 558 nm for DsRed agrees much better with 2e than with 6e. A fairly close analog of 6e with an ethylenic double bond linked to the 2-position of the imidazolidinone was synthesized by McCapra et al. (16) and measured to absorb at 445 nm. In general, ZINDO more closely reproduced the experimental wavelengths than did TDHDFT.
Behavior of DsRed upon Denaturation.
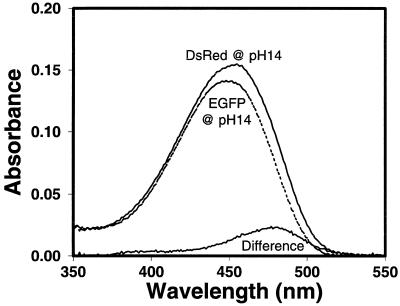
Acylimines are normally considered unstable species prone to nucleophilic attack. After denaturation of the protective protein shell, the initial step should be addition of water across the C⩵N bond to form an N-acylcarbinolamine as in 3 (Fig. 2), but more vigorous conditions should lead to irreversible hydrolysis (17) and cleavage of the peptide backbone at residue 66 as in 4. These predictions explained two previously puzzling observations. The first is that the absorbance of denatured DsRed is almost identical to that of denatured GFP, i.e., that the red shift instantly disappears upon denaturation. Thus when DsRed and GFP are separately diluted into neat DMSO, they both absorb maximally at 383 nm and only differ in relative amounts of 280-nm absorbance, as one would expect because DsRed has three times as many tryptophans per molecule as GFP. Alternatively, wild-type and K83R DsRed are denatured by raising the pH above 12, after which they absorb maximally at 452 nm (Fig. 5, solid line). For comparison, alkali-denatured GFP (Fig. 5, dashed line) peaks at 446 nm, but all three have similar extinction coefficients near 4 × 104 M−1⋅cm−1. Our interpretation is that upon denaturation, the red chromophore 2 hydrates to 3, whose conjugated system is identical to the p-hydroxybenzylideneimidazolidinone (1) of GFP or immature DsRed. The pH-dependent shift between 383 and 446 nm is known to be caused by ionization of the phenolic group (4). The residual slight discrepancy of 5 nm between the spectra of base-denatured DsRed (wild type or K83R) and base-denatured GFP can be attributed to a fluorescent component within the DsRed that still absorbs and excites maximally at 475 nm (see difference spectrum in Fig. 5) and emits at 500 nm even at pH 14. Evidently the immature green form is more alkali-resistant than the mature red form. By contrast GFP is negligibly fluorescent at such high pH. When the alkaline solution of DsRed was heated, both the absorbance and fluorescence disappeared.
Figure 5.
Absorbance spectra of DsRed (solid line, λmax 451.5 nm) and enhanced GFP (EGFP) (dashed line, λmax 446 nm), both 3.9 μM by BCA protein assay, denatured in 1 M NaOH. The difference between the DsRed and EGFP spectra also is indicated.
Fragmentation of DsRed.
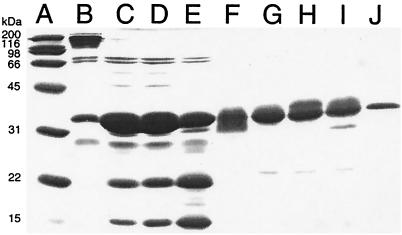
The other puzzling observation explained by the acylimine structure is that DsRed boiled before SDS/PAGE always showed two fragment bands of apparent masses 15 and 22 kDa (Fig. 6, lane C). The extent of cleavage increased when the sample was preboiled at pH 13 or 1, especially the latter (Fig. 6, lanes D and E), but did not increase beyond the extent shown in lane E even upon more prolonged boiling. No fragmentation was seen with GFP samples or the permanently green mutant K83R (Fig. 6, lanes F–J), indicating that only red protein samples containing acylimine undergo cleavage. A more accurate estimate of the molecular weights of the two fragments and the site of cleavage was obtained by electrospray mass spectrometry, after very mild trypsin digestion of wild-type or K83R DsRed to remove the N-terminal polyhistidine tag and the initial residues Met-Val-Arg. The green mutant K83R remained full length as shown by a mass within experimental error of the 25,651 Da predicted for 1c. Wild-type protein gave peaks at 18,488 and 7,149 Da, corresponding nicely to the two fragments (4c in Fig. 2) resulting from the expected hydrolysis of the C⩵N bond. Also a peak at mass 25,622 Da was observed corresponding to uncleaved material, although the experimental uncertainty of ±4 Da was too large to decide between structures 1c and 2c, i.e., to provide independent confirmation of dehydrogenation. Mass spectrometry clearly gave more accurate masses, but SDS/PAGE was probably more reliable for estimating the relative proportions of cleaved vs. uncleaved protein.
Figure 6.
Oligomerization and cleavage of DsRed monitored by electrophoresis in a 15% polyacrylamide gel. Lane A: Broad range protein standard (Bio-Rad); molecular masses in kDa are indicated. Lane B: Unboiled DsRed (the band with molecular mass > 100 kDa was brightly red fluorescent before Coomassie staining). Lane C: Boiled DsRed. Lane D: DsRed boiled in 0.1 M KOH. Lane E: DsRed boiled in 0.1 M HCl. Lane F: Unboiled enhanced yellow fluorescent protein (EYFP) (the band with apparent molecular mass ≈ 30 kDa was brightly yellow fluorescent before Coomassie staining). Lane G: Boiled EYFP. Lane H: EYFP boiled in 0.1 M KOH. Lane I: EYFP boiled in 0.1 M HCl. Lane J: Boiled DsRed K83R. The two bands at ≈70 kDa in nearly every lane are non-GFP impurities we sometimes observe in protein preps from JM109 Escherichia coli.
Is O2 Required for Green to Red Conversion?
The proposed dehydrogenation of Gln-66 requires an oxidant to carry away the two hydrogens. The most obvious candidate would be molecular oxygen just as is used to generate the p-hydroxybenzylideneimidazolidinone 1 from amino acids (18, 19). Another possibility would be the hydrogen peroxide that is presumably generated by that first oxidation (19). To test whether the green-to-red maturation requires O2, a freshly prepared DsRed sample with an approximately maximal green fraction was deoxygenated by addition of glucose, glucose oxidase, and catalase in a sealed cuvette under argon. After 13 h the green species had declined considerably without any of the normal increase in red fluorescence. Likewise, a 2-μl aliquot of concentrated protein diluted in 1 ml of degassed buffer and sealed into an ampule developed red fluorescence at a much lower rate than that of an air-saturated control. These results suggest that O2 is indeed required. However, it is still possible that de-aeration inhibits development of red fluorescence partly by preventing fresh conversion of precursor polypeptide to the green intermediate 1a as well as subsequent conversion to red 2a. In separate experiments we tested whether H2O2 could be the secondary oxidant, but exposure to 0.3 M H2O2 for 1 h had no obvious effect, whereas 1.6 M H2O2 for 16 h degraded both species but particularly the red. Thus excess H2O2 seems if anything deleterious.
Discussion
Our interpretation for the structure of the DsRed chromophore and its mechanisms of formation and degradation is shown in Fig. 2 and Table 2, which explain an array of disparate observations. (i) The most indicative are the mass spectra (Fig. 1) showing that the chromopeptide prepared from DsRed (2d) contains a major component 2 Da lighter than the chromopeptide prepared from a mutant that stays green (1d). (ii) Maturation from green (1a) to red (2a) is a slow step (half-life ca. 1 day) that probably requires O2 but does not produce peaks with intermediate wavelengths. If the red shift had been caused by the summation of multiple noncovalent interactions accumulating during slow protein folding, one might have expected to see such intermediate wavelength spectra. (iii) When the protective protein shell around the DsRed chromophore is denatured, the acylimine should be prone to addition of water across the C⩵N bond as in 3b, which would interrupt the conjugation. Therefore the UV-visible absorbance should immediately revert to a spectrum very similar to that of denatured GFP, as indeed observed. Upon entry into the electrospray mass spectrometer inlet at pH 2.2, the strongly dehydrating conditions would remove H2O to restore the acylimine (2c, 2d) seen in the mass spectra (Fig. 1). (iv) Upon harsher treatment such as boiling, especially at extreme pHs, the carbinolamide (3b, 3d) should hydrolyze and split the polypeptide chain into two fragments between Phe-65 and the oxidized remnant of Gln-66 (4b, 4c), just as seen in Fig. 6 (lanes C–E) and in mass spectra of DsRed not treated with LysC. Because the permanently green mutant K83R does not progress beyond 1 and lacks the hydrolyzable acylimine, it does not break into two fragments. (v) Theoretical calculations confirm that the two extra double bonds, both polarized to attract electron density from the phenolate anion at the other end of the continuous conjugation, are indeed sufficient to explain the red shift in 2a or 2e, whereas the tautomeric enamide 6e would give a much smaller shift. Although we doubt that native DsRed contains either an enamide as in 6 or a ketone as in 4, the interesting possibility remains that such species might spontaneously occur among the many other fluorescent proteins with different colors (2, 20).
Several lines of evidence agree that even after prolonged maturation, DsRed still contains a substantial amount of GFP-like green chromophore (1a) in addition to the red species 2a. Mass spectra of the LysC chromopeptide from DsRed consistently show both species 1d and 2d. Likewise the full-length (≈35 kDa) protein in Fig. 6 that resisted cleavage by boiling at extreme pHs probably represents the fraction of molecules that had not yet formed acylimine. Both the mass spectra and the gels suggest that even after prolonged aging, DsRed may contain structures 1 and 2 in about 1:1 ratio. If so, the true extinction coefficient of the intact red monomers would be about 150,000 M−1⋅cm−1. Optical spectroscopy also confirms the admixture of green species among the red, in that both the absorbance and excitation spectra of DsRed retain a peak near 475 nm (see figure 2 of ref. 1), which is not attributable to the vibrational shoulder shown by other fluorescent proteins, and which is more alkali-stable than the red species (Fig. 5). Emission near 500 nm is very low in mature DsRed, because of fluorescence resonance energy transfer to the red species, but such green emission increases markedly upon photobleaching the latter (D.A. Zacharias and R.Y.T., unpublished observations), similarly to the photobleaching of immature DsRed discussed in the previous paper (1). We do not know why the green-to-red conversion stops well short of 100%, nor whether individual tetramers consist of a stochastic vs. a deterministic composition of green vs. red monomers. The latter question would be best solved by spectroscopy of single tetramers.
Our proposed structure (2 in Fig. 2) was formulated without reliance on any x-ray crystallographic structure, in which the most obvious sign of dehydrogenation of the remnants of Gln-66 should be a change from tetrahedral to planar geometry for the α-carbon. However, the presence of a roughly equimolar amount of nondehydrogenated chromophores could well obscure the change in bond angles, unless one specifically fitted the electron density as a superposition of two structures, or if the tetramer is an ordered rather than stochastic mixture of green and red monomers. A crystal structure would be the most efficient way to determine the cis vs. trans configurations of many single and double bonds, which could make a noticeable difference to the spectrum.
Many of the results of the present paper and its companion (1) are summarized in Table 3 as a comparison with the corresponding properties of GFP and its mutants. We see the following three challenges as the most important areas where DsRed needs improvement before it becomes generally useful as a fusion partner in cell biology: suppressing its obligate aggregation and increasing both the speed and completeness of maturation to the desired red form. Even though the green form does not show up in the emission spectrum because of fluorescence resonance energy transfer within the tetramer, its excitation peak will contribute spectral cross-talk in experiments using GFP as a separate label or fluorescence resonance energy transfer donor. Furthermore, if tetramerization were suppressed, then any residual immature green form would indeed contribute to the emission. The excellent brightness and stability of DsRed and many potential uses for a long-wavelength fluorescent protein provide ample justification for major efforts to remedy these remaining deficiencies.
Table 3.
Comparison of major properties of GFP and its mutants (19) vs. DsRed and its K83M mutant (ref. 1 and this work)
| GFP [and its mutants] | DsRed [and K83M] | |
|---|---|---|
| Source | Bioluminescent jellyfish Aequorea victoria | Nonbioluminescent coral Discosoma spp. |
| Excitation wavelengths, nm | 395, 470; [360–516] | 475, 558; [564] |
| Emission wavelength, nm | 504; [442–529] | 583; [602] |
| Fluorescence quantum yield | 0.8; [up to 0.7] | 0.7; [0.44] |
| Photobleach quantum yield | 3 × 10−6 (EGFP), 5 × 10−5 (EYFP Q69K) | 7 × 10−7 |
| Molar extinction coefficient, 103 M−1 ⋅ cm−1 | 25–30, 10–14; [up to 95] | 75 (average per subunit); [not determined] |
| Native size | 238 aa, 27 kDa | 225 aa, 26 kDa |
| State of aggregation | Mostly monomeric, weak dimerization (Kd ∼ 100 μM) | Obligate tetramer, possible weak dimerization (Kd ∼ 39 μM) of tetramers |
| Chromophore formed by: | Cyclization (dehydration) and one oxidation (dehydrogenation) | Cyclization (dehydration) and two oxidations (dehydrogenations) |
| Maturation time scale | Minutes to hours | Hours to days |
| Extent of maturation | Complete | Incomplete |
| pH sensitivity | Medium; [moderate to high] | Low |
EGFP, enhanced GFP (F64L/S65T); EYFP, enhanced yellow fluorescent protein (Q69K, S65G/V68L/Q69K/S72A/T203Y).
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NS27177) and Howard Hughes Medical Institute (to R.Y.T.) and the National Biomedical Computational Resource (to K.K.B.).
Abbreviations
- GFP
green fluorescent protein
- HDFT
hybrid density functional theory
- TDHDFT
time-dependent HDFT
- ZINDO
Zerner's modified intermediate neglect of differential overlap
Footnotes
Terskikh, A., Fradkov, A., Savitsky, A., Siebert, P. & Matz, M., 11th International Symposium on Bioluminescence and Chemiluminescence, Pacific Grove, CA, Sept. 6–10, 2000, p. 64, abstract.
References
- 1.Baird G S, Zacharias D A, Tsien R Y. Proc Natl Acad Sci USA. 2000;97:11984–11989. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matz M V, Fradkov A F, Labas Y A, Savitsky A P, Zaraisky A G, Markelov M L, Lukyanov S A. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 3.Tsien R Y. Nat Biotechnol. 1999;17:956–957. doi: 10.1038/13648. [DOI] [PubMed] [Google Scholar]
- 4.Niwa H, Inouye S, Hirano T, Matsuno T, Kojima S, Kubota M, Ohashi M, Tsuji F I. Proc Natl Acad Sci USA. 1996;93:13617–13622. doi: 10.1073/pnas.93.24.13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frisch M J, Trucks G W, Schlegel H B, Scuseria G E, Robb M A, Cheeseman J R, Zakrzewski V G, Montgomery J A, Jr, Stratmann R E, Burant J C, et al. gaussian 98 Revision A.7. Pittsburgh: Gaussian; 1998. [Google Scholar]
- 6.Schmidt M W, Baldridge K K, Boatz J A, Elbert S T, Gordon M S, Jensen J H, Koseki S, Matsunaga N, Nguyen K A, Su S, Windus T L. J Comp Chem. 1993;14:1347–1363. [Google Scholar]
- 7.Becke A D. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- 8.Lee C, Yang W, Parr R G. Physiol Rev. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 9.Miehlich B, Savin A, Stoll H, Preuss H. Chem Phys Lett. 1989;157:200–206. [Google Scholar]
- 10.Dunning T H J, Hay P J. In: Applications of Electronic Structure Theory. Schaefer H F I, editor. New York: Plenum; 1976. pp. 1–28. [Google Scholar]
- 11.Zerner M C. In: Reviews of Computational Chemistry. Lipkowitz K B, Boyd D B, editors. New York: VCH; 1991. pp. 313–365. [Google Scholar]
- 12.Correa de Mello P, Hehenberger M, Zerner M C. Int J Quant Chem. 1982;21:251–257. [Google Scholar]
- 13.Bauernschmitt R, Ahlrichs R. Chem Phys Lett. 1996;256:454–464. [Google Scholar]
- 14.Kupfer R, Meier S, Würthwein E-U. Synthesis. 1984;1984:688–670. [Google Scholar]
- 15.Mason S F. In: The Chemistry of Synthetic Dyes. Venkataraman K, editor. Vol. 3. New York: Academic; 1970. pp. 169–221. [Google Scholar]
- 16.McCapra F, Razavi Z, Neary A P. J. Chem. Soc. Chem. Commun. 1988. 790–791. [Google Scholar]
- 17.Malassa V I, Matthies D. Chemiker-Zeitung. 1987;111:253–261. [Google Scholar]
- 18.Heim R, Prasher D C, Tsien R Y. Proc Natl Acad Sci USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsien R Y. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 20.Lukyanov K A, Fradkov A F, Gurskaya N G, Matz M V, Labas Y A, Savitsky A P, Markelov M L, Zaraisky A G, Zhao X, Fang Y, et al. J Biol Chem. 2000;275:25879–25882. doi: 10.1074/jbc.C000338200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.