Abstract
Mitochondria is believed to play a central role in p53-mediated apoptosis. However, the signal transduction pathways leading to mitochondria remain unclear. Here, we report that translocation of Bax protein from cytosol to mitochondria is required for p53-induced apoptosis. Cytosolic Bax is unable to induce apoptosis, and blocking Bax translocation inhibits cell death. Expression of Bcl-2 blocks cytochrome c release and apoptosis but has no effect on Bax translocation, suggesting that Bax translocation acts upstream of Bcl-2. We further demonstrate that Peg3/Pw1, a protein up-regulated in p53-mediated cell death process, induces Bax translocation independent of apoptosis. The results suggest that Bax translocation represents an important regulatory step in p53-mediated apoptosis, and Peg3/Pw1 functions as a modulator downstream of p53 to regulate Bax redistribution in the cells, thus favoring the cellular decision toward apoptosis over growth arrest following p53 induction.
The p53 tumor suppressor plays a pivotal role in preventing tumorigenesis in both human and mouse. Mutations of the p53 gene or inactivation of its activity by viral and cellular proteins are the most frequent events associated with human cancers (reviewed in refs. 1–3). The ability of p53 to suppress tumor growth is mainly attributed by its ability in mediating two biological processes, cell cycle arrest and apoptosis (recently reviewed in refs. 4–6). It is thought that p53-mediated growth arrest prevents the replication of damaged DNA and reduces genetic instability, whereas apoptosis induced by p53 is necessary for eliminating aberrant cells. Although the pro-apoptotic activity of p53 has been shown to play the most crucial role in suppressing tumor formation both in vitro and in vivo, the molecular events that determine whether the cell enters growth arrest or undergoes apoptosis upon p53 induction remain obscure.
The pathways responsible for p53-mediated growth arrest have mostly been defined. It has been shown that p53-mediated induction of p21 leads to inhibition of cyclin-dependent kinase activity and blocks cell entrance to DNA synthesis (7–9). Unlike p53-mediated growth arrest where p21 acts as a major effector in the process, a large body of evidence suggests that p53-mediated apoptosis is much more complex and involves multiple pathways via both transcription-dependent and independent mechanisms (see recent reviews in refs. 10–12). Thus, it is likely that multiple factors contribute to the decision making between growth arrest and apoptosis upon p53 induction. The pro-apoptotic gene Bax has been shown to be a p53 target and is up-regulated in a number of systems during p53-mediated apoptosis (13–16). However, the fact that p53-induced cell death remains intact in Bax-deficient thymocytes suggests that Bax only accounts for p53-mediated apoptosis in certain cell types (17). The finding that Bax is frequently induced to similar levels in p53-dependent growth arrest further supports the notion that Bax contributes only part of p53-induced cell death response, and other factors must be involved in p53-mediated apoptosis (18–20). A number of other genes such as PIGs, Fas/CD95, and Killer/DR5 inducible by p53 have also been implicated in p53-mediated apoptosis (21–23). However, like Bax, these genes are also induced by p53 in cells that do not undergo apoptosis (21, 24, 25), suggesting that simple induction of these genes cannot account for the cellular decision between growth arrest and apoptosis in response to p53.
The apoptotic pathway in general is carried out by the activation of a class of cysteine-dependent asparate-directed proteases called caspases, which induce a cascade of proteolytic cleavage of many essential proteins and commit cells to the suicide pathway (reviewed in ref. 26). One of the first signs of apoptosis is the loss of mitochondrial membrane potential and release of cytochrome c to cytosol. Cytochrome c interacts with apaf-1 to activate caspase-9 and initiates caspase degradation pathway. It has been shown recently that caspase-9 and apaf-1 are required for p53/c-myc-induced apoptosis (27), suggesting that p53-dependent cell death shares the common downstream apoptotic machinery. However, the upstream pathway, i.e., how p53 relays its signal to the mitochondria, remains to be elucidated.
To dissect the p53-mediated apoptosis pathway and to understand the molecular processes underlying the choice between growth arrest and cell death upon p53 induction, we developed mammalian cell lines that undergo either p53-mediated growth arrest (called VHD) or apoptosis (named VM10) (28, 29). Using this system, we previously identified a gene named Peg3/Pw1 as a potential mediator for p53-dependent cell death process as it is specifically induced during p53-mediated apoptosis but not growth arrest (30). In this report, we show that Bax is up-regulated to similar levels by p53 during either growth arrest or apoptosis in VHD and VM10 cells, respectively, confirming that induction of Bax alone is not sufficient for apoptosis. However, immunostaining of the Bax protein shows that there is a key difference in its subcellular localization; Bax is in cytosol during growth arrest and localizes to mitochondria during apoptosis. We further show that translocation of Bax from cytosol to mitochondria is required for apoptosis, and this event is mediated by Peg3/Pw1 in our system. Expression of Peg3/Pw1 induces Bax translocation. Blocking Peg3/Pw1 expression inhibits Bax translocation, cytochrome c release, and subsequent activation of caspases and apoptosis. Our data suggest that Bax translocation from cytosol to mitochondria is a critical step in p53-mediated apoptosis, and Peg3/Pw1 functions as a coactivator or modulator of apoptosis to regulate the subcellular localization of Bax protein. This regulation may play a pivotal role in determining cell death vs. survival in response to p53.
Materials and Methods
Plasmids, Cell Lines, and Antibodies.
The EGFP-Peg3 fusion protein was constructed by fusing the Peg3/Pw1 coding sequences in frame to the carboxyl-terminal of EGFP in pEGFP-C1 (CLONTECH) based on a Peg3/Pw1 cDNA fragment and the published full-length cDNA sequences (29–31). The Bcl-2 expressing plasmid and antisense Peg3/Pw1 vector were described elsewhere (29, 30). The EGFP-Bax was constructed by fusing the ORF of human Bax into the carboxyl terminus of EGFP in pEGFP-C1 (CLONTECH). The red fluorescent protein (RFP) expression DNA was obtained from CLONTECH. VHD and VM10 cells were maintained in DMEM, supplemented by 10% FBS. They were routinely grown in incubators (Forma Scientific, Marietta, OH) under 5% CO2 at 39°C and shifted to 32°C for 24–48 h for growth arrest or apoptosis assays. The DNA was transfected into cells using Effectene (Qiagen) as described by the manufacturer. In transient transfection experiments, cells were harvested 48–72 h after transfection. In some experiments, 100 μM caspase inhibitor z-VAD-fmk (benzyloxycarbonyl-Val-Ala-Asp fluoromethylketone; Promega) were added. Stable cell lines were generated by selecting the transfected cells in puromycin (2.5 μg/ml) for 2–3 weeks. Antibodies against Bcl-2, Bax, and cytochrome c and secondary antibodies were purchased from Santa Cruz Biotechnology and Sigma. Anti-Peg3/Pw1 antibody was generated in rabbit using a bacteria-produced N-terminal fragment of Peg3/Pw1 protein.
Immunofluorescent Staining.
Cells grown on glass coverslips were fixed and permeabilized in 4% paraformaldehyde and 0.1% saponin in PBS. Mitochondria was stained using MitoTracker Red CMX Ros (Molecular Probes) according to manufacturer's instruction. Immunostaining procedure was described previously (30) using monoclonal antibody against Bcl-2, a rabbit polyclonal antibody against cytochrome c, anti-Bax monoclonal antibody (all from Santa Cruz Biotechnology), and rhodamine or fluorescein-conjugated secondary antibodies (Sigma). Hochst 3342 (Molecular Probes) was used to visualize the nuclei. The coverslips were mounted and viewed by fluorescence microscopy (Zeiss Axiovert).
Subcellular Fractionation and Western Blotting.
Subcellular fraction was performed as described with minor modifications (32, 33). Cell pellets were resuspended in sucrose-supplemented cell extract buffer (300 mM sucrose/10 mM Hepes, pH 7.4/50 mM KCl/5 mM EGTA/5 mM MgCl2/1 mM DTT/protease inhibitor mixture). The cells were homogenized on ice with a Dounce homogenizer. Unbroken cells and nuclei were removed by centrifugation at 2,000 × g for 10 min at 4°C. The postnuclear supernatant was further spun at 10,000 × g for 10 min at 4°C. The crude mitochondrial pellet was purified by passing a sucrose gradient (0.1 to 0.3 M) at 9,000 × g for 8 min. The purity of mitochondria fraction was determined by the absence of cytosolic β-actin using Western blots. The supernatant was further ultracentrifuged at 14,000 × g for 10 min and then filtered by passing through a 0.22-μm ultrafilter (Millipore) to generate purified cytosolic fraction. The nuclear extracts were prepared as described (34).Western analysis was performed as described and developed using an ECL chemiluminescence kit (Amersham Pharmacia) (30).
Results
Distinct Subcellular Localization of Bax Protein in p53-Mediated Growth Arrest vs. Apoptosis.
VHD and VM10 cells are derived from a p53-deficient immortalized mouse embryo fibroblast cell line. The VHD cell expresses a temperature-sensitive mutant p53 (Ala to Val mutation at codon 135, tsp53), which expresses a nonfunctional p53 protein at 37–39°C and a fully functional p53 protein at 32°C. VM10 cells also express c-myc in addition to tsp53. At 37–39°C, both cells grow normally. However, at 32°C, VHD cells undergo G1 growth arrest in a p53-dependent manner, and VM10 cells undergo p53-mediated apoptosis (28, 29).
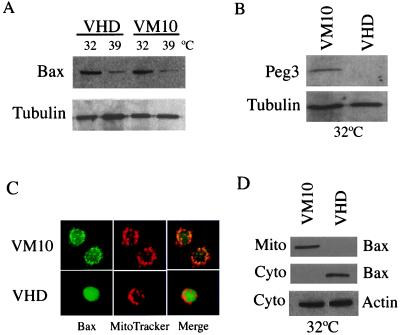
When assayed by Western analysis, a 5- to 8-fold induction of Bax was observed in both VHD and VM10 cells at 32°C where tsp53 is activated, as compared with cells at 39°C where p53 is inactive (Fig. 1A). The levels of Bax protein induced were similar in cells that are growth arrested (VHD 32°C) relative to cells that undergo apoptosis (VM10 32°C). In contrast, Peg3/Pw1 was expressed only in VM10 cells at 32°C during p53-mediated apoptosis (Fig. 1B), consistent with previous Northern results (30). The induction of Bax by p53 is consistent with the fact that Bax is a p53 target gene. However, high levels of Bax protein in cells undergoing p53-mediated growth arrest suggests that Bax induction alone is not sufficient to induce apoptosis.
Figure 1.
The VHD and VM10 cells showed similar Bax protein levels but distinct subcellular localization. (A) Expression of Bax protein in VHD and VM10 cells. Whole cell extracts were prepared from VHD and VM10 cells at 32°C (24 h after temperature shift) and 39°C, respectively. The Bax protein was detected by Western Blotting using a monoclonal antibody against Bax. The same blot was stripped and probed with an anti-α-tubulin antibody for loading control. Quantitation was done on a phosphorimager (Molecular Dynamics). (B) Induction of Peg3/Pw1 protein in VM10 cells at 32°C. Protein extracts isolated from VHD and VM10 cells at 32°C were blotted with an anti-Peg3/Pw1 antibody. Peg3/Pw1 is only expressed in VM10 cells at 32°C. (C) Immunostaining of the Bax protein in VHD and VM10 cells at 32°C. The mitochondria were stained with MitoTracker Red CMX Ros (red), and the Bax protein was stained with the monoclonal antibody and visualized with an FITC-conjugated secondary antibody (green). A representation of the staining pattern is shown. (D) Subcellular fractionation of Bax protein in VHD and VM10 cells. Cytosolic and mitochondria extracts were prepared from VHD and VM10 cells at 32°C. The protein concentration was measured using a protein assay kit (Bio-Rad). An equal amount of protein was loaded onto the gel. The Bax protein was detected by Western blots, and cytosolic β-actin was used as a control.
Previous studies in several models of apoptosis have shown that Bax translocates from cytosol to mitochondria when overexpressed or in response to certain cell death stimuli (35–40). To explore the expression pattern of Bax protein in VHD and VM10 cells, we examined its subcellular localization in the two cell lines by immunofluorescent staining and biochemical subcellular fractionation. In growth-arrested VHD cells at 32°C, the Bax protein exhibited a diffused staining pattern, indicating a predominantly cytosolic localization (Fig. 1C). The pattern of Bax staining in apoptotic VM10 cells at 32°C was dramatically different from that in VHD cells. The Bax protein showed a punctuate staining pattern and was colocalized with mitochondria marker, MitoTracker (Fig. 1C). To further confirm the immunofluorescent staining results, subcellular fractionation was performed to examine the distribution of Bax protein in both cytosol and mitochondria by Western blotting. Consistent with the immunofluorescence data, the Bax protein was detected mainly in cytosolic fraction in arrested VHD cells and redistributed to mitochondria fraction in apoptotic VM10 cells (Fig. 1D). These data indicate that although Bax is induced to the same levels by p53, its subcellular distributions are clearly distinct in these two pathways.
Expression of Bcl-2 Blocks Cytochrome c Release and Apoptosis but Has No Effect on Bax Translocation.
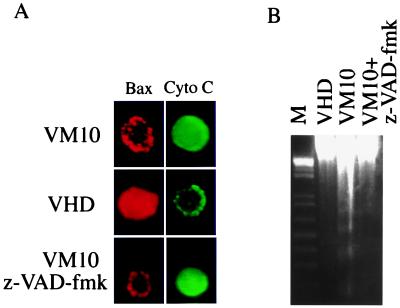
Translocation of Bax to mitochondria has been linked to cytochrome c release from mitochondria to cytosol and the activation of apoptosis in a number of systems (reviewed in refs. 41 and 42). This is also the case in VM10 cells at 32°C, as immunostaining of cytochrome c showed diffused cytosolic localization while the Bax protein displayed a mitochondrial staining pattern (Fig. 2A). In contrast, opposite staining pattern was observed in growth-arrested VHD cells as Bax was localized to cytosol and cytochrome c remained in mitochondria (Fig. 2A). All VM10 cells displaying diffused cytochrome c staining were apoptotic because their nuclei were condensed and fragmented (data not shown). Treatment of VM10 cells with a broad caspase inhibitor, z-VAD-fmk, protected the cells from apoptosis (Fig. 2B) but had not effect on redistribution of either Bax or cytochrome c (Fig. 2A). Subcellular fractionation analysis of Bax and cytochrome c confirmed the immunostaining results (data not shown). Taken together, the data are consistent with the fact that caspases act downstream of Bax-mediated cytochrome c release during apoptosis.
Figure 2.
Bax translocation in VM10 cells coincides with cytochrome c release and apoptosis. (A) Immunostaining of Bax and cytochrome c in VM10 and VHD cells at 32°C. The Bax staining was carried out using anti-Bax antibody and a rhodamine-conjugated secondary antibody (red), and cytochrome c was stained with a polyclonal antibody and an FITC-conjugated secondary antibody (green). (B) Inhibition of apoptosis in VM10 cells by z-VAD-fmk. VM10 cells were transferred to 32°C with 100 μM z-VAD-fmk added. The DNA was extracted, and DNA fragmentation was detected by agarose gel electrophoresis.
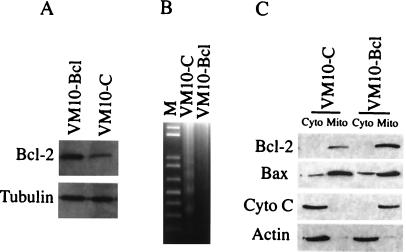
The anti-apoptotic factor Bcl-2 interacts with Bax and blocks Bax-induced apoptosis (reviewed in refs. 41 and 42). To elucidate the effect of Bcl-2 on Bax translocation, cytochrome c release, and apoptosis in VM10 cells, stable cell lines expressing Bcl-2 in VM10 cells were established (Fig. 3A). Although the VM10 cells transfected with control vector showed a typical DNA ladder characteristic of apoptosis when shifted to 32°C, VM10 cells expressing Bcl-2 were protected from apoptosis (Fig. 3B). To examine the subcellular localization of Bax and cytochrome c, mitochondria and cytosol fractions were prepared, and the protein distributions were analyzed by Western blots. Consistent with previous reports, Bcl-2 was mainly located in mitochondria, and overexpression of Bcl-2 blocked the release of cytochrome c from mitochondria to cytosol (Fig. 3C). However, Bcl-2 was incapable of blocking Bax translocation as Bax protein colocalized with mitochondria in VM10 cells overexpressing Bcl-2. Immunofluorescence data were consistent with the subcellular fractionation results (data not shown).
Figure 3.
Expression of Bcl-2 blocks cytochrome c release and apoptosis but does not affect Bax translocation. (A) VM10-bcl cells are generated by stably transfecting a Bcl-2 expressing plasmid, and VM10-C is a cell line transfected by the control vector. The Western blot was probed with a monoclonal antibody against Bcl-2. (B) Bcl-2 blocks apoptosis in VM10 cells. The cells were shifted to 32°C for 24 h, and DNA was analyzed by agarose gel electrophoresis. (C) Bcl-2 expression blocks cytochrome c release but has no effect on Bax translocation. Extracts were prepared from mitochondria and cytosolic fraction of VM10-bcl and VM10-C cells at 32°C. The protein concentration was measured using a protein assay kit (Bio-Rad). An equal amount of protein was loaded onto the gel. Bcl-2, Bax, and cytochrome c proteins were detected by Western blotting. Cytosolic actin was used as a control.
Expression of Peg3/Pw1 Induces Bax Translocation Independent of Apoptosis.
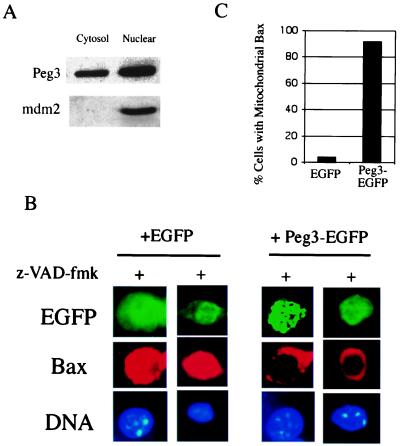
Our previous study identified Peg3/Pw1 as a gene product specifically induced during p53-mediated apoptosis in VM10 cells, and coexpression of Peg3/Pw1 with p53 leads to apoptosis (30). Although the Peg3/Pw1 protein was mainly localized to cell nucleus, cytosolic Peg3/Pw1 was detected in VM10 cells undergoing apoptosis (Fig. 4A). To test the effect of Peg3/Pw1 expression on Bax translocation, the Peg3/Pw1 expression vector was transfected into VHD cells at 32°C, where the Bax protein is localized in cytosol and endogenous Peg3/Pw1 expression is undetectable. Expression of Peg3/Pw1 in VHD cells at 32°C induced Bax translocation and apoptosis (data not shown), consistent with our previous data that coexpression of Peg3/Pw1 and p53 leads to cell death. To exclude the possibility that the translocation of Bax protein in this case was caused by apoptosis, the caspase inhibitor, z-VAD-fmk, was added to the transfected cells to prevent caspase activation and apoptosis as indicated by the intact nuclei morphology (Fig. 4B). Although expression of a control green fluorescent protein (EGFP) had no effect on Bax distribution, the Bax protein displayed a typical punctuated mitochondrial staining after transfecting a Peg3-EGFP fusion protein (Fig. 4B). Quantitation of cells displaying mitochondrial Bax staining patterns showed that Bax was translocated to mitochondria in 92% of Peg3-EGFP transfected cells, whereas only 4% of EGFP-transfected cells showed mitochondrial Bax staining (Fig. 4C). These data indicate that Peg3/Pw1 induces Bax translocation to mitochondria before apoptosis.
Figure 4.
Expression of Peg3/Pw1 induces Bax translocation. (A) Subcellular localization of Peg3/Pw1 in VM10 cells during apoptosis. Cytosolic and nuclear extracts were prepared from VM10 cells at 32°C, and Peg3/Pw1 protein was detected by a Western blot. Western blots for nuclear protein mdm2 were used as the cell fractionation control. (B) The VHD cells at 39°C were transiently transfected with either an EGFP plasmid or a plasmid expressing EGFP-Peg3/Pw1 fusion protein. The cells were shifted to 32°C for 24 h, and caspase inhibitor z-VAD-fmk was added immediately. The transfected cells were marked by GFP fluorescence (green), Bax staining is in red, and DNA was stained by Hochst 3342. Representative staining patterns are chosen from two independent experiments. The Bax staining is diffused (cytosolic) in EGFP-expressing cells and punctuated (mitochondria) in Peg3-EGFP-expressing cells. (C) Percentage of cells with mitochondrial Bax. Different fields of green cells were observed under the fluorescent microscope. A total of 30 cells were counted each time, and the data represent the average of two independent experiments.
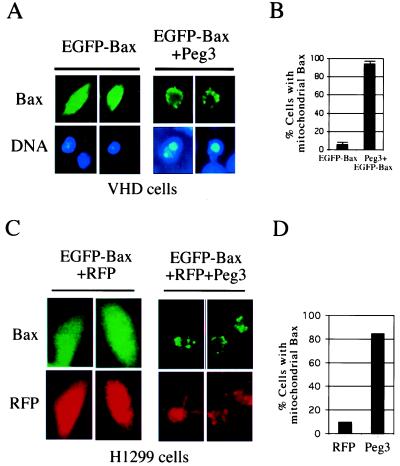
To further confirm that expression of Peg3/Pw1 induces Bax translocation, an EGFP-Bax fusion protein was generated. The EGFP was fused in frame to the amino-terminal of Bax ORF. This EGFP-Bax is fully capable of inducing apoptosis as the wild-type Bax protein (data not shown and Fig. 5). Transfecting EGFP-Bax into VHD cells resulted in most of EGFP-Bax displaying cytosolic localization patterns and normal DNA staining patterns (Fig. 5A), supporting our observation that translocation of Bax from cytosol to mitochondria is required for apoptosis. To confirm that Bax translocation induces cell death, a Peg3/Pw1 expression plasmid was cotransfected with EGFP-Bax into the cells. Instead of 90% of green cells display cytosolic Bax and remained intact when transfected with EGFP-Bax alone, cotransfection of Peg3/Pw1 and EGFP-Bax resulted in Bax translocation in 90% of transfected cells and cell death (Fig. 5 A and B). Similar results were obtained in H1299 cells, a human lung carcinoma cell line with p53 deficiency. In these experiments, a plasmid expressing an RFP was cotransfected into cell to mark the transfected cells (Fig. 5 C and D). Cotransfecting of EGFP-Bax and RFP resulted in mostly cytosolic Bax, whereas coexpression of EGFP-Bax, RFP, and Peg3/Pw1 resulted in punctuated Bax staining patterns indicative of mitochondria localization (Fig. 5 C and D). These results showed that expression of Peg3/Pw1 induces Bax translocation and apoptosis independent of p53.
Figure 5.
Cotransfection of Peg3/Pw1 and Bax resulted in Bax translocation. (A) The VHD cells at 39°C were transiently transfected with either an EGFP-Bax plasmid or EGFP-Bax plus a plasmid expressing Peg3/Pw1 at a molar ration of 1 to 3. The Bax staining was visualized by GFP fluorescence (green), and DNA was stained by Hochst 3342. Representative staining patterns are chosen from two independent experiments. The Bax staining is diffused (cytosolic) in EGFP-expressing cells and punctuated (mitochondria) in Peg3-expressing cells. (B) Percentage of cells with mitochondrial Bax. Different fields of green cells were observed under the fluorescent microscope. A total of 30 cells were counted each time, and the data represent the average of three independent experiments. Standard deviations were given. (C) The H1299 cells were transiently transfected with either an EGFP-Bax and RFP DNA or EGFP-Bax and RFP plus a plasmid expressing Peg3/Pw1. The Bax staining was visualized by GFP fluorescence (green), and transfected cells were marked by RFP fluorescence (red). Representative staining patterns are chosen from two independent experiments. (D) Percentage of cells with mitochondrial Bax. Different fields of green cells were observed under the fluorescent microscope. A total of 50 cells were counted each time, and the data represent the average of two independent experiments.
Expression of Peg3/Pw1 in VM10 Cells Is Required for Bax Translocation and Apoptosis.
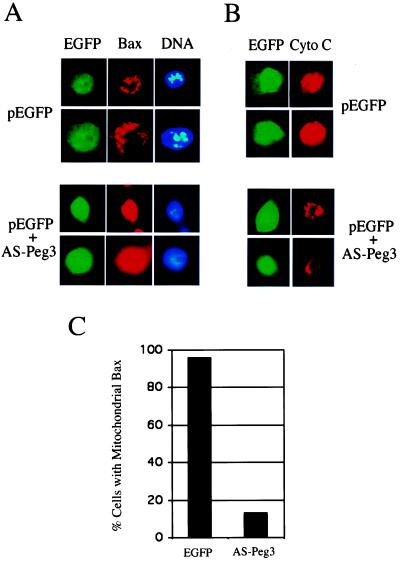
We have reported previously that transiently transfecting a plasmid expressing Peg3/Pw1 antisense mRNA is capable of inhibiting Peg3/Pw1 expression and blocking cell death in VM10 cells (30). To determine whether Peg3/Pw1 is required for Bax translocation in VM10 cells, the Peg3/Pw1 antisense vector was transiently transfected into VM10 cells together with an EGFP reporter plasmid to allow the marking of transfected cells. The VM10 cells that received the Peg3/Pw1 antisense plasmid maintained flat morphology and were well attached to the tissue culture dish, whereas control EGFP-alone transfected cells were round with blebbing membrane (data not shown). Coimmunostaining of Bax protein and cell nuclei was then performed to examine the effect of Peg3/Pw1 antisense on Bax subcellular localization. Like in VM10 cells, the Bax protein in control EGFP-alone expressing cells showed punctuated mitochondrial distribution with condensed and fragmented nuclei (Fig. 6A). Expression of Peg3/Pw1 antisense mRNA reverted the Bax localization to a diffused cytosolic distribution pattern and normal nuclear morphology (Fig. 6A). Reciprocally, cytochrome c was localized mainly to mitochondria in Peg3/Pw1 antisense-expressing cells but translocated to cytosol in control EGFP-only cells (Fig. 6B). Quantitation of cells displaying mitochondrial Bax staining patterns showed that Bax was translocated to mitochondria in 95% of EGFP transfected cells, whereas cotransfection of EGFP and Peg3/Pw1 antisense mRNA resulted in only 15% of cells with mitochondrial Bax staining patterns (Fig. 6C). These results provide further evidence supporting that Peg3/Pw1 regulates Bax translocation from cytosol to mitochondria, and this translocation is required for p53-mediated apoptosis in our cell system.
Figure 6.
Antisense Peg3/Pw1 blocks Bax translocation, cytochrome c release, and apoptosis in VM10 cells. (A) VM10 cells were transfected with either EGFP or EGFP plus a plasmid expressing antisense Peg3/Pw1 mRNA at a molar ration of 1 to 3. The cells were stained for Bax protein and DNA as described in Fig. 4. The Bax staining is punctuated (mitochondria) in EGFP-expressing cells and diffused (cytosolic) in AS-Peg3-expressing cells. (B) Duplicate of the same transfected cells were stained with anti-cytochrome c antibody. Representative staining patterns are chosen from two independent experiments. A revised staining pattern compared to Bax was observed for cytochrome c. (C) Percentage of cells with mitochondrial Bax. Different fields of green cells were observed under the fluorescent microscope. A total of 30 cells were counted each time, and the data represent the average of two independent experiments.
Discussion
One of the most intriguing questions in the p53 field is how a cell makes the decision to enter either growth arrest or undergo apoptosis upon p53 induction. In an attempt to address this question, it has been proposed that p53 may induce two sets of genes upon stress signals. One set mainly functions in cell growth control, such as p21 and GADD45. The other set acts on apoptosis, such as Bax and PIGs. The outcome of p53 induction depends on which set of signals is more dominant, i.e., a stronger p21 signal favors growth arrest and a higher level of Bax leads to cell death. This working model, however, is challenged by many previously published results and by our data presented in this paper. First, we, as well as many others, showed that induction of p21 is comparable in both p53-mediated growth arrest and apoptosis in various systems (18–20, 30). Loss of p21 does not seem to tip the balance toward apoptosis both in vitro and in vivo (43, 44). Second, we reported here that induction of Bax at the protein level is indistinguishable in arrested VHD and apoptotic VM10 cells. Similar results were also observed in a number of other systems (18–20), suggesting that expression of Bax protein alone cannot account for the distinct pathways induced by p53. The notable finding in this study is that the subcellular localization of the Bax protein plays a key role in deciding the fate of a cell; cytosolic Bax is unable to induce cell death, and translocation of Bax to mitochondria is a critical step in p53-mediated apoptosis. This finding is consistent with other studies in different cell systems showing the association of Bax translocation to mitochondria with release of cytochrome c and activation of apoptosis (41, 42). Most importantly, we demonstrate that Peg3/Pw1, a potential cell death effector of p53, can induce Bax translocation, thus linking p53 to the general apoptotic machinery mediated by mitochondria. The antisense blocking experiment strengthens the notion that Bax translocation is required for p53-mediated apoptosis, and Peg3/Pw1 plays a pivotal role in this process. Based on these results and previous published reports, we propose that one of the mechanisms underlining the decision making between growth arrest and apoptosis upon p53 induction is determined by the so called “modulators” like Peg3/Pw1. Induction of p53 results in an up-regulation of both cell growth-regulating genes as well as pro-apoptotic factors. However, the pro-apoptotic factors, such as cytosolic Bax, remain inactive and growth arrest prevails. The modulating factors serve as coactivators of pro-apoptotic proteins to stimulate their apoptotic functions and induce full-scale cell death. The presence or absence of these coactivators or modulators could account for the distinct responses to p53 induction in a wide variety of cells and under diverse conditions. This hypothesis, however, does not exclude the possibility that under certain circumstances p53 could act alone to preferentially induce either cell growth arrest or cell death. The transcription-independent cell death mediated by p53 may carry out through other mechanisms. It is also likely that modulation of survival factors, such as Bcl-2, can affect the cell death process. However, we believe that Bax translocation represents an important regulatory step during p53-mediated apoptosis, and in certain transcription-dependent p53 responses, growth arrest and apoptosis could occur in a sequential order.
Peg3/Pw1 was isolated as a gene induced during p53/c-myc-mediated apoptosis (30); however, expression of Peg3/Pw1 alone was unable to induce cell death in cells deficient of p53 such as 10.1 (30), H1299, and VHD cells (data not shown), suggesting that Peg3/Pw1-mediated apoptosis in these cells requires other factors. Because these cells express low levels of Bax protein due to p53 deficiency (Fig. 1A and data not shown) and Peg3/Pw1 causes Bax translocation, Bax seems to be one of the factors required for Peg3/Pw1-mediated apoptosis. This is consistent with our previous report showing that Peg3/Pw1 is able to cooperate with p53 to induce cell death (30), as expression of p53 induces Bax protein levels. It is also in agreement with studies reported here that expression of Bax alone leads to inefficient apoptosis, and the cell death process is greatly enhanced when Peg3/Pw1 is expressed at the same time. However, Peg3/Pw1 may induce apoptosis through other mechanisms, as Peg3/Pw1 cooperates with Siah1a to lead to cell death (30) and Peg3/Pw1 can activate NF-kb, which has been recently shown to be required for p53-mediated apoptosis (45).
The Bax translocation has been observed in a number of p53-independent apoptosis systems (reviewed in refs. 41 and 42). Our data in this report clearly show that blocking the translocation of endogenous Bax inhibits apoptosis. Although our results show that expression of Peg3/Pw1 induces Bax translocation independent of apoptosis, the molecular basis for this process remains unclear. Because Peg3/Pw1 induced Bax translocation in VHD cells at 39°C and in H1299 cells, both are p53 deficient, and it is likely that Peg3/Pw1-induced Bax translocation does not require p53. Peg3/Pw1 is a zinc finger-containing protein with largely unknown functions. Although the Peg3/Pw1 protein is localized mainly to the nucleus, a significant fraction of Peg3/Pw1 is found in cytoplasmic fraction. This is confirmed both by immunohistochemical staining (data not shown) and by subcellular fractionation (Fig. 4A). Thus, it is possible Peg3/Pw1 may either directly or indirectly act on Bax to induce either dimerization or conformational changes of Bax protein, which has been implicated in causing Bax redistribution in cells (46, 47). It is also likely that Peg3/Pw1 represents only one of the factors that affect apoptosis by modulating Bax subcellular distribution.
Acknowledgments
We thank Drs. Hui Zheng and Xuefeng Xia for critical discussion and comments on this work. We are grateful for the technical help provided by Yahong Lin and Nadia Aithmitti. We also thank Dr. Xiaodong Wang at the University of Texas Southwestern Medical Center, Dallas, for providing a Bax cDNA. This work is funded by National Cancer Institute Grant R01-CA73687 (to X.W.).
Abbreviations
- RFP
red fluorescent protein
- z-VAD-fmk
benzyloxycarbonyl-Val-Ala-Asp fluoromethylketone
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 2.Greenblatt M S, Bennett W P, Hollstein M, Harris C C. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 3.Hainaut P, Hernandez T, Robinson A, Rodriguez-Tome P, Flores T, Hollstein M, Harris C C, Montesano R. Nucleic Acids Res. 1998;26:205–213. doi: 10.1093/nar/26.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 5.May P, May E. Oncogene. 1999;18:7621–7636. doi: 10.1038/sj.onc.1203285. [DOI] [PubMed] [Google Scholar]
- 6.Oren M, Rotter V. Cell Mol Life Sci. 1999;55:9–11. doi: 10.1007/s000180050265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 8.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 9.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. Nature (London) 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 10.Bates S, Vousden K H. Cell Mol Life Sci. 1999;55:28–37. doi: 10.1007/s000180050267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sionov R V, Haupt Y. Oncogene. 1999;18:6145–6157. doi: 10.1038/sj.onc.1203130. [DOI] [PubMed] [Google Scholar]
- 12.Burns T F, El-Deiry W S. J Cell Physiol. 1999;181:231–239. doi: 10.1002/(SICI)1097-4652(199911)181:2<231::AID-JCP5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 13.Miyashita T, Reed J C. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 14.Zhan Q, Fan S, Bae I, Guillouf C, Liebermann D A, O'Connor P M, Fornace A J., Jr Oncogene. 1994;9:3743–3751. [PubMed] [Google Scholar]
- 15.Blum D, Wu Y, Nissou M F, Arnaud S, Alim Louis B, Verna J M. Brain Res. 1997;751:139–142. doi: 10.1016/s0006-8993(96)01358-3. [DOI] [PubMed] [Google Scholar]
- 16.McCurrach M E, Connor T M, Knudson C M, Korsmeyer S J, Lowe S W. Proc Natl Acad Sci USA. 1997;94:2345–2349. doi: 10.1073/pnas.94.6.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knudson C M, Tung K S, Tourtellotte W G, Brown G A, Korsmeyer S J. Science. 1995;270:96–99. doi: 10.1126/science.270.5233.96. [DOI] [PubMed] [Google Scholar]
- 18.Guillouf C, Grana X, Selvakumaran M, De Luca A, Giordano A, Hoffman B, Liebermann D A. Blood. 1995;85:2691–2698. [PubMed] [Google Scholar]
- 19.Han J, Sabbatini P, Perez D, Rao L, Modha D, White E. Genes Dev. 1996;10:461–477. doi: 10.1101/gad.10.4.461. [DOI] [PubMed] [Google Scholar]
- 20.Bouvard V, Zaitchouk T, Vacher M, Duthu A, Canivet M, Choisy-Rossi C, Nieruchalski M, May E. Oncogene. 2000;19:649–660. doi: 10.1038/sj.onc.1203366. [DOI] [PubMed] [Google Scholar]
- 21.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. Nature (London) 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 22.Owen-Schaub L B, Zhang W, Cusack J C, Angelo L S, Santee S M, Fujiwara T, Roth J A, Deisseroth A B, Zhang W W, Kruzel E, et al. Mol Cell Biol. 1995;15:3032–3040. doi: 10.1128/mcb.15.6.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu G S, Burns T F, McDonald E R, III, Jiang W, Meng R, Krantz I D, Kao G, Gan D D, Zhou J Y, Muschel R, et al. Nat Genet. 1997;17:141–143. doi: 10.1038/ng1097-141. [DOI] [PubMed] [Google Scholar]
- 24.Reinke V, Lozano G. Oncogene. 1997;15:1527–1534. doi: 10.1038/sj.onc.1201316. [DOI] [PubMed] [Google Scholar]
- 25.Sheikh M S, Burns T F, Huang Y, Wu G S, Amundson S, Brooks K S, Fornace A J, Jr, el-Deiry W S. Cancer Res. 1998;58:1593–1598. [PubMed] [Google Scholar]
- 26.Budihardjo I, Oliver H, Lutter M, Luo X, Wang X. Annu Rev Cell Dev Biol. 1999;15:269–290. doi: 10.1146/annurev.cellbio.15.1.269. [DOI] [PubMed] [Google Scholar]
- 27.Soengas M S, Alarcon R M, Yoshida H, Giaccia A J, Hakem R, Mak T W, Lowe S W. Science. 1999;284:156–159. doi: 10.1126/science.284.5411.156. [DOI] [PubMed] [Google Scholar]
- 28.Wu X, Levine A J. Proc Natl Acad Sci USA. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Wu X, Lin J, Levine A J. Mol Cell Biol. 1996;16:2445–2452. doi: 10.1128/mcb.16.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Relaix F, Wei X, Li W, Pan J, Lin Y, Bowtell D D, Sassoon D A, Wu X. Proc Natl Acad Sci USA. 2000;97:2105–2110. doi: 10.1073/pnas.040378897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L L, Szeto I Y, Cattanach B M, Ishino F, Surani M A. Genomics. 2000;63:333–340. doi: 10.1006/geno.1999.6103. [DOI] [PubMed] [Google Scholar]
- 32.Juin P, Hueber A O, Littlewood T, Evan G. Genes Dev. 1999;13:1367–1381. doi: 10.1101/gad.13.11.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jia L, Macey M G, Yin Y, Newland A C, Kelsey S M. Blood. 1999;93:2353–2359. [PubMed] [Google Scholar]
- 34.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 35.Wolter K G, Hsu Y T, Smith C L, Nechushtan A, Xi X G, Youle R J. J Cell Biol. 1997;139:1281–1292. doi: 10.1083/jcb.139.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pawlowski J, Kraft A S. Proc Natl Acad Sci USA. 2000;97:529–531. doi: 10.1073/pnas.97.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu Y T, Wolter K G, Youle R J. Proc Natl Acad Sci USA. 1997;94:3668–3672. doi: 10.1073/pnas.94.8.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosse T, Olivier R, Monney L, Rager M, Conus S, Fellay I, Jansen B, Borner C. Nature (London) 1998;391:496–499. doi: 10.1038/35160. [DOI] [PubMed] [Google Scholar]
- 39.Putcha G V, Deshmukh M, Johnson E M., Jr J Neurosci. 1999;19:7476–7485. doi: 10.1523/JNEUROSCI.19-17-07476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khaled A R, Kim K, Hofmeister R, Muegge K, Durum S K. Proc Natl Acad Sci USA. 1999;96:14476–14481. doi: 10.1073/pnas.96.25.14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsujimoto Y, Shimizu S. FEBS Lett. 2000;466:6–10. doi: 10.1016/s0014-5793(99)01761-5. [DOI] [PubMed] [Google Scholar]
- 42.Gross A, McDonnell J M, Korsmeyer S J. Genes Dev. 1999;13:1899–1911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- 43.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 44.Brugarolas J, Chandrasekaran C, Gordon J I, Beach D, Jacks T, Hannon G J. Nature (London) 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 45.Ryan K M, Ernst M K, Rice N R, Vousden K H. Nature (London) 2000;404:892–897. doi: 10.1038/35009130. [DOI] [PubMed] [Google Scholar]
- 46.Gross A, Jockel J, Wei M C, Korsmeyer S J. EMBO J. 1998;17:3878–3885. doi: 10.1093/emboj/17.14.3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nechushtan A, Smith C L, Hsu Y T, Youle R J. EMBO J. 1999;18:2330–2341. doi: 10.1093/emboj/18.9.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]