Abstract
Cellular transformation by the BCR/ABL oncogene depends on the ABL-encoded tyrosine kinase activity. To block BCR/ABL function, we created a unique tyrosine phosphatase by fusing the catalytic domain of SHP1 (SHP1c) to the ABL binding domain (ABD) of RIN1, an established binding partner and substrate for c-ABL and BCR/ABL. This fusion construct (ABD/SHP1c) binds to BCR/ABL in cells and functions as an active phosphatase. ABD/SHP1c effectively suppressed BCR/ABL function as judged by reductions in transformation of fibroblast cells, growth factor independence of hematopoietic cell lines, and proliferation of primary bone marrow cells. In addition, the leukemogenic properties of BCR/ABL in a murine model system were blocked by coexpression of ABD/SHP1c. Both the “escort” function provided by ABD and the inhibitor function provided by the phosphatase of SHP1c were necessary for effective BCR/ABL interference. Expression of ABD/SHP1c also reversed the transformed phenotype of K562, a human leukemia-derived cell line. These results have direct implications for leukemia therapeutics and suggest an approach to block aberrant signal transduction in other pathologies through the use of appropriately designed escort/inhibitors.
Alterations in the ABL tyrosine kinase are characteristic genetic events in multiple forms of leukemia. In humans, leukemogenic forms of ABL arise from chromosomal translocations. In the resulting fusion proteins, residues encoded by the first exon of ABL are replaced by sequence from the BCR protein, resulting in 185-kDa and 210-kDa isoforms or, less frequently, from the TEL protein (1). In chronic myelogenous leukemia, p210 BCR/ABL is found in 95% of all cases (2). These same hybrid proteins can transform cultured cells and induce leukemia in mice. ABL sequences encoding tyrosine kinase activity are essential for transformation (3–5). Also present are ABL regulatory sequences, including Src homology (SH)3, SH2, and actin-binding domains. The fusion of ABL with BCR leads to increased kinase activity and an apparent shift in subcellular localization (6), changes that alter the magnitude and characteristics of downstream signals.
BCR/ABL-induced transformation depends on its continued expression (7–9). Dominant negative-acting proteins that block BCR/ABL-mediated transformation of cultured cells have been described (10–14). These proteins typically rely on disruption of broadly used signaling components required indirectly for BCR/ABL function. Ideally, an effective BCR/ABL inhibitor should directly suppress the activity that most closely correlates with transformation. The ABL kinase-specific inhibitor STI-571, for instance, has shown promise as an effective BCR/ABL-suppressing drug (15).
We propose an alternate approach to inhibition of BCR/ABL and downstream pathways, using modular peptides to combine tyrosine phosphatase and ABL-binding functions resulting in efficient ABL-targeted tyrosine phosphatases.
Protein tyrosine phosphatases (PTPs) (16, 17) might act as BCR/ABL inhibitors through the dephosphorylation of ABL and/or its substrates. Overexpression of PTP1B inhibit fibroblast transformation by p210 BCR/ABL (18), and this effect may be caused, in part, by direct dephosphorylation of BCR/ABL (19). SHP1 (SHPTP1) also can interact with and partially inhibit the function of c-ABL (20) and BCR/ABL (21). These phosphatases may be endogenous down-regulators of ABL that can inhibit BCR/ABL. Their effectiveness as BCR/ABL suppressers is moderate, however, and may be tempered by deleterious effects from overexpression. The inhibition potency of these phosphatases might be improved markedly if they could be liberated from normal regulation and targeted to BCR/ABL.
RIN1 is both a substrate and binding partner of ABL (22). RIN1 also interacts with BCR/ABL, and this association is detected in leukemia-derived cell (23). The ABL binding domain (ABD) of RIN1 interacts with both the SH3 and SH2 domains of ABL (ref. 23; this work). We have used the RIN1-ABD as an “escort” peptide to deliver the SHP1 tyrosine phosphatase catalytic domain to BCR/ABL. The resulting escort/inhibitor (ABD/SHP1c) showed potent suppressive activity against BCR/ABL in a variety of assays including leukemogenesis. These results suggest a useful strategy for targeted therapeutic intervention in diseases resulting from unregulated signaling pathways.
Materials and Methods
Plasmid Construction.
The C-terminal portions of SHP1 and SHP2 (gift of Benjamin Neel, Harvard University, Cambridge, MA) were generated by PCR. Primers introduced EcoRI and KpnI sites at 5′ ends and EcoRI/BamHI sites at 3′ ends. The products (SHP1c and SHP2c) were sequenced, digested with KpnI–BamHI, and ligated into a QE60 (Qiagen, Chatsworth, CA) clone of ABD. ABD/SHP1cCS was created by PCR (oligo: 5′-TAG GAT CCT CAC GCA GGG CCC ATC ATC GTG CAC TCC AGC GCC GGC ATC G). Wild-type sequences in pBluescript-ABD/SHP1c (EcoNI–XhoI) were replaced with the mutated fragment. An NcoI/KpnI fragment from pSRα-ABDTM (23) was used to create ABDTM/SHP1c in a QE60 vector. Phosphatase fragments, fusion constructs, and mutants were released and cloned into retroviral vectors with EcoRI.
Retroviral Vectors and Virus Stocks.
Two retrovirus vectors, pSRαMSVtk (24) and pMSCV (25), were used. pSRα constructs (Fig. 1) were made as described (23). For other constructs, p210 BCR/ABL was ligated to the internal ribosome entry site (IRES) and then cloned into pMSCV. Fusion proteins, ABD, and green fluorescent protein (GFP) cDNAs were inserted into a unique EcoRI site. In constructs without BCR/ABL, GFP cDNA was cloned into the IRES position. Retrovirus stocks were prepared by transient cotransfection of 293T cells with retroviral vectors and Psi− ecotropic or amphotropic packaging vector (24, 26). Supernatants were collected 24–48 h posttransfection. Virus titers were characterized by immunoblot analysis 48–72 h after infection of Rat1 cells. Human leukemic cells were infected with amphotropic pseudotyped virus, and fluorescence-activated cell sorting was used to monitor transfer/expression of GFP.
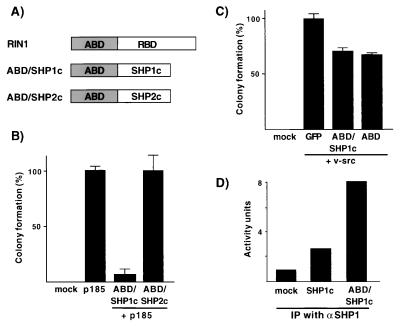
Figure 1.
Escort/inhibitor properties. (A) Depiction of fusion proteins including the ABD of RIN1 (amino acids 1–295) and the catalytic domains of SHP1 (SHP1c: amino acids 250–595) and SHP2 (SHP2c: amino acids 253–593). The sequence of RIN1 that encodes a RAS binding domain (RBD) that is eliminated in the fusion proteins. (B) ABD/SHP1c blocks BCR/ABL-induced transformation. Soft agar colony formation of Rat1 cells infected with virus expressing BCR/ABL (p185) and the indicated constructs was assayed and normalized to results from cells receiving BCR/ABL only (566 colonies = 100%). Data represent mean values and standard deviations from triplicate experiments. (C) Soft agar colony formation of Rat1 cells infected with v-src and indicated constructs. (D) PTP activity measured by using immunoprecipitated material from extracts of 293 cells transfected with the indicated constructs.
Soft Agar Colony Assays.
Cell transformation was quantified by growth in soft agar as described (24). Infected cells were grown for 48 h, and 1 × 104 cells were plated in soft agar per 6-cm dish in duplicate. Colonies were counted 3 weeks after plating (2 weeks for v-src transformation assays). For K562 cells, 1 × 103 cells were plated and incubated for 2 weeks.
Growth Factor Independence Assays.
Growth factor independence assays were performed as described (5, 27). 32D (gift of Joel Greenberger, University of Pittsburgh) and DAGM cells were grown in media containing granulocyte/macrophage colony-stimulating factor (provided by Judith Gasson, University of California, Los Angeles) for 2 days postinfection. Cells then were washed twice by using media without growth factor and plated into 96-well dishes. Two weeks after infection factor-independent cell growth was scored.
Bone Marrow Pre-B Cell Assay and Leukemogenesis Assay.
Fresh bone marrow from femurs and tibias of BALB/c mice was infected with retrovirus and plated at a density of 5 × 106 cells/6-cm dish (27). The number of nonadherent cells was counted from day 6 until the cell count reached a density greater than 2 × 106 cells per ml. Reconstitution experiments were performed as described (23). Bone marrow cells were injected into irradiated (200–275 rad) severe combined immunodeficient (SCID) mice within 6 h of infection.
Immunoprecipitation (IP) and Immunoblotting.
Cell lysates were prepared by using RIPA buffer (0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) as described (22), incubated with polyclonal anti-Abl (Oncogene Research Products, Cambridge, MA) and immunoprecipitated by using protein A-Sepharose (Amersham Pharmacia). Immunoblotting was performed as described (22) with minor modifications. Monoclonal anti-SHP1 (Transduction Laboratories, Lexington, KY) and pex-5, a monoclonal anti-Abl, were used to detect ABD/SHP1c and BCR/ABL, respectively. Polyclonal anti-RIN1 (22) and anti-phosphotyrosine (Upstate Biotechnology, Lake Placid, NY) were used to analyze fusion protein expression and cellular phosphotyrosine levels, respectively. Immunoreactive proteins were visualized by using ECL (Amersham Pharmacia).
IP-PTP Assays.
Whole-cell extracts were prepared from transfected 293T cells or infected Rat1 cells. IP was performed with anti-SHP1 antibody using normalized total cellular protein. IP material was assayed for PTP activity (New England BioLabs) by using 33P-labeled myelin basic protein as substrate. Units of activity are reported in picomole phosphate released per min.
Cell Staining and Immunofluorescence.
Benzidine staining was performed as described (28). Infected K562 cells were grown for 48 h before staining. Immunofluorescence assays were carried out as described (6) with modifications. Anti-SHP1 and polyclonal anti-Abl were used for SHP1c or ABD/SHP1c and BCR/ABL immunolocalization, respectively. Secondary antibodies were FITC-conjugated goat anti-rabbit and CY3-conjugated goat anti-mouse (Jackson Laboratories).
Results
ABD/SHP1c Is a Potent Escort/Inhibitor of BCR/ABL.
The amino terminal 295 residues of RIN1 encode an ABD that mediates interactions with c-ABL and BCR/ABL (22, 23). To test the feasibility of using the RIN1 ABD for targeted inhibition of BCR/ABL, we created fusions of ABD to the catalytic domains of SHP1 and SHP2, two tyrosine phosphatases that encode tandem SH2 domains (Fig. 1A). The resulting fusion proteins (ABD/SHP1c and ABD/SHP2c) were expressed from retroviral vectors, and each product showed the expected molecular weight (data not shown).
We next tested whether these fusion proteins could block transformation by BCR/ABL. The ABD/SHP1c fusion protein produced a 92% reduction in the number of soft agar colonies resulting from p185 BCR/ABL expression in Rat1 fibroblasts (Fig. 1B). In addition, the size of the remaining colonies was reduced. Similarly, p210 BCR/ABL was strongly inhibited by ABD/SHP1c (data not shown). Expression of the ABD/SHP1c fusion protein in wild-type cells (no BCR/ABL) had no deleterious effects; these cells were cultured extensively without loss of ABD/SHP1c expression. The SHP2 catalytic domain fusion exhibited a much weaker suppressive effect; colony size was significantly decreased but colony number was unchanged.
The transformation suppression effects observed with ABD/SHP1c were specific to BCR/ABL. When introduced with v-src, ABD/SHP1c had only a weak effect (30% reduction) on soft agar colony formation (Fig. 1C). The same result was seen for ABD, however, suggesting signaling interference that might result from low level interaction with SRC (22). ABD/SHP1c had no effect on H-RASV12-transformed cells (data not shown). These data support the model that ABD/SHP1c acts as a targeted phosphatase.
Both Escort and Inhibitor Domains Are Required for BCR/ABL Suppression.
We next tested whether the fusion protein was associated with BCR/ABL. Extracts from cells transfected with a retrovirus construct expressing both ABD/SHP1c and p210 BCR/ABL were prepared and analyzed by IP with an ABL antibody followed by Western blot probing with antibodies to ABL, SHP1, or RIN1 ABD. The results confirm that ABD/SHP1c is indeed bound to p210 BCR/ABL in cells (Fig. 2). Similar results were obtained with the p185 BCR/ABL allele (data not shown).
Figure 2.
The ABD escort domain promotes binding to BCR/ABL. Immunoblot of material immunoprecipitated from infected Rat1 cells. Lanes 1, Mock infection; lanes 2, p210 (BCR/ABL) only; lanes 3, p210 with ABD/SHP1c; lanes 4, p210 with ABDTM/SHP1c; lanes 5, p210 with ABD/SHP1cCS. Molecular mass markers are shown at left. IB, immunoblot.
The ABD of RIN1 encodes three tyrosine residues that are substrates for ABL-mediated phosphorylation. We previously have shown that, unlike wild-type ABD, an ABD construct harboring mutations in all three tyrosine residues (ABDTM) cannot rescue the transforming activity of a mutant p185 BCR/ABL (23). The ABDTM construct also shows greatly reduced binding to BCR/ABL (Y.-M.L. and J.C., unpublished data), confirming the requirement of direct binding for ABD enhancement of BCR/ABL transformation. As predicted, an ABDTM/SHP1c fusion protein showed extremely weak association with BCR/ABL (Fig. 2). These cells also gave soft agar colonies at approximately the same frequency as cells expressing BCR/ABL alone (Table 1). This result demonstrated the requirement for BCR/ABL binding by the escort domain (ABD) in transformation blockade.
Table 1.
Transformation blockade by ABD/SHP1c requires both escort and phosphatase functions
| Infection constructs | No. of soft agar colonies/104 cells | Colony size |
|---|---|---|
| Mock | 0 | |
| BCR/ABL | ||
| + GFP | 343 ± 73 (n = 8) | Med-large |
| + ABD/SHP1c | 37 ± 9 (n = 5) | Small |
| + ABD/SHP1cCS | 303 ± 76 (n = 5) | Med-large |
| + ABD™/SHP1c | 372 ± 87 (n = 6) | Med-large |
Rat1 cells were infected with retrovirus constructs expressing p210 BCR/ABL and the indicated gene then was cultured in soft agar. Colony number and average size were determined after 3 weeks (small, 0.3–0.5 mm; med, 0.5–1 mm; large, >1 mm).
We confirmed that both the ABD/SHP1c and ABD/SHP2c fusion proteins are functional tyrosine phosphatases. Extracts from 293 cells transfected with ABD/SHP1c had a significant level of tyrosine phosphatase activity that was immunoprecipitated by using antibody to the SHP1 domain (Fig. 1D). Cells transfected with SHP1c only and subjected to an IP phosphatase assay also showed activity well above what could be accounted for by endogenous SHP1 (mock-infected cells). We also examined Rat1 cells infected with either ABD/SHP1c or ABD/SHP2c viruses, by using antibody to ABD, and detected comparable levels of PTP activity (data not shown). However, ABD/SHP2c showed much weaker BCR/ABL suppression activity than ABD/SHP1c (Fig. 1B), suggesting an intrinsic difference between the tyrosine phosphatase catalytic domains in their ability to suppress BCR/ABL signaling.
We considered that the SHP1c domain might be acting through steric inhibition rather than its enzymatic properties. This possibility was examined by introducing a mutation (Cys453/Ser) that eliminates tyrosine phosphatase enzymatic activity (16). The ABD/SHP1cCS construct retained the ability to interact with BCR/ABL when both proteins were expressed in Rat1 cells (Fig. 2). However, ABD/SHP1cCS failed to inhibit BCR/ABL-dependent transformation (Table 1). This result is consistent with a requirement for tyrosine phosphatase activity in the transformation blockade.
Subcellular Localization of ABD/SHP1c.
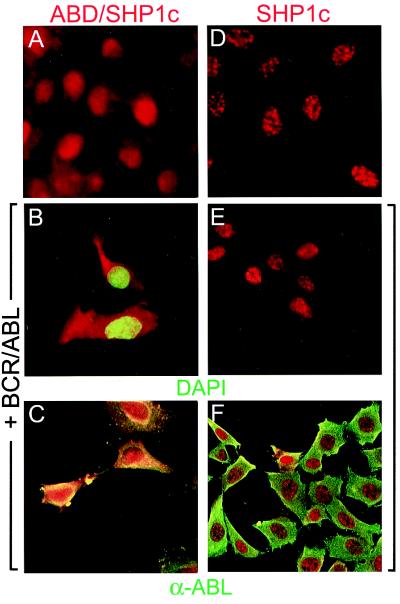
The BCR/ABL protein is sequestered primarily in the cytoplasm, and this localization is required for transformation and tumorigenesis (6). The ABD/SHP1c fusion protein, however, was found primarily in the nucleus of infected Rat1 cells stained with anti-SHP1 (Fig. 3A), or by direct fluorescence detection of an ABD/SHP1c/GFP fusion protein (data not shown). This localization appeared to be directed by the SHP1c domain; SHP1c alone showed nuclear localization in the presence or absence or BCR/ABL (Fig. 3 D and E). Wild-type SHP1, which has two SH2 domains, is known to be cytoplasmic (29), but translocates into the nucleus after cell stimulation (30). Examination of the SHP1c amino acid sequence revealed lysine- and arginine-rich regions similar to nuclear localization signals (31) near the carboxyl terminus that could mediate this sequestration. The ABD peptide itself is primarily cytoplasmic (data not shown), suggesting that targeting signals within the SHP1c domain are dominant in ABD/SHP1c localization.
Figure 3.
ABD/SHP1c is colocalized with BCR/ABL. Rat1 cells expressing ABD/SHP1c (A–C) or SHP1c (D–F) were subjected to staining with SHP1 antibody (red). Cells also expressing p210 BCR/ABL were stained with either DAPI (4′,6-diamidino-2-phenylindole) to detect nuclei (green; B and E) or ABL antibody (green; C and F). Endogenous SHP1 was barely detectable in these cells (data not shown).
Strikingly, in Rat1 cells that coexpress p210 BCR/ABL there is a dramatic shift of ABD/SHP1c into the cytoplasm (Fig. 3C) with both proteins colocalized. This result depended on the ABD sequences because no such redistribution was observed in cells expressing SHP1c only with BCR/ABL (Fig. 3F). The localization of ABD/SHP1c, therefore, is altered by the presence of its target, BCR/ABL.
Altered Tyrosine Phosphorylation in BCR/ABL Cells Expressing ABD/SHP1c.
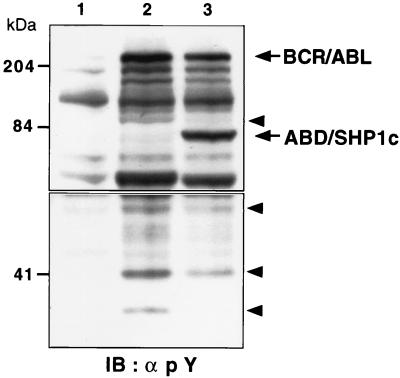
To determine the physiological substrates of the escort/phosphatase, we analyzed the profile of tyrosine-phosphorylated proteins from Rat1 cells expressing BCR/ABL with or without ABD/SHP1c. The level of tyrosine phosphorylation on BCR/ABL itself was reduced in cells that also expressed the ABD/SHP1c construct (Fig. 4), although total BCR/ABL protein levels were equivalent. An examination of other tyrosine-phosphorylated cellular proteins in cells expressing BCR/ABL revealed that ABD/SHP1c had a clear suppressive effect although some substrate signals showed greater reduction than others. In particular, bands migrating at 100 kDa, 90 kDa, 60 kDa, and 35 kDa had significantly reduced intensity in extracts from ABD/SHP1c-expressing cells (Fig. 4). The data also demonstrate that the ABD/SHP1c fusion protein itself is tyrosine-phosphorylated to a large degree. We previously have shown that the ABD of RIN1 is a substrate for both c-ABL and BCR/ABL. Under some conditions, c-ABL also is known to phosphorylate SHP1 on residues that are present in the ABD/SHP1c construct (20). However, tyrosine phosphorylation on the escort/phosphatase indicates little or no autodephosphorylation activity. This may reflect steric hindrance that blocks the reaction or substrate specificity of SHP1 that prohibits recognition of ABD.
Figure 4.
Alterations in phosphotyrosine profile correlate with transformation suppression. Extracts from Rat1 cells mock-infected (lane 1), infected with p210 BCR/ABL virus (lane 2), or infected with an ABD/SHP1c and BCR/ABL virus (lane 3) were analyzed by immunoblot (IB) by using phosphotyrosine antibody. BCR/ABL and ABD/SHP1c are indicated with arrows. Bands with the greatest reduction in signal intensity are marked with arrowheads. Upper and lower panels are derived from different exposures of the same immunoblot. Molecular mass markers are presented at left.
ABD/SHP1c Blocks BCR/ABL Action in Hematopoietic Cells in Vitro.
We next assessed the ability of ABD/SHP1c to block the effects of BCR/ABL in hematopoietic cells. DAGM (32) and 32D (33) are myeloid cell lines that normally require a growth factor supplement (e.g., granulocyte/macrophage colony-stimulating factor) to propagate in culture. Expression of BCR/ABL confers growth factor independence to these cells (34). DAGM and 32D cells were infected with retrovirus constructs then plated at multiple densities in medium without growth factor supplementation. Cells infected with ABD/SHP1c or vector showed no growth whereas cells receiving BCR/ABL gave rise to proliferating cells in 34% of low-density independent cultures (Table 2). In contrast, a virus expressing both ABD/SHP1c and BCR/ABL produced growth in only 6% of equivalent cultures. Suppression of growth factor independence by ABD/SHP1c was more pronounced in low-density cultures (Table 2) and at earlier time points (data not shown) perhaps reflecting a “conditioned medium” effect. The eventual appearance of a stable proliferation phenotype may result from genetic or epigenetic alterations that reduce expression of the escort/phosphatase relative to BCR/ABL. Such changes, although expected to be infrequent, are strongly selected for in this experimental system.
Table 2.
Repression of BCR/ABL-induced growth factor independence by ABD/SHP1c
| Retrovirus construct | % of Cultures that
proliferate
|
|||
|---|---|---|---|---|
| 32 D
|
DAGM
|
|||
| *104 | 103 | 103 | 102 | |
| Vector | 0 | 0 | 0 | 0 |
| ABD/SHP1c+GFP | 0 | 0 | 0 | 0 |
| GFP+BCR/ABL | 87 | 34 | 84 | 34 |
| ABD/SHP1c + BCR/ABL | 68 | 6 | 37 | 6 |
| GM-CSF | 100 | 100 | 100 | 100 |
32D and DAGM cells were infected with the indicated retrovirus construct (p210 BCR/ABL was used in 32D cells; p185 BCR/ABL was used in DAGM cells) and plated into 32-well dishes at 102, 103, or 104 cells/well. As a control, uninfected cells were plated into growth factor (granulocyte/macrophage colony-stimulating factor, GM-CSF) supplemented medium. The percent of wells that showed cell growth after 13 days is reported.
Cells/culture.
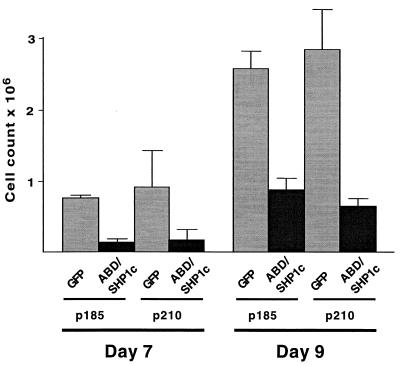
We also examined the in vitro proliferation of murine bone marrow cells in response to BCR/ABL expression (27). These cells can be induced to proliferate in culture by BCR/ABL under conditions that normally would not allow their growth. As shown in Fig. 5, the inclusion of ABD/SHP1c led to a marked suppression of BCR/ABL effects, as seen by a 2.9- to 5.5-fold decrease in outgrowth of the cultures. These results demonstrate that ABD/SHP1c can indeed block BCR/ABL action in the context of hematopoietic cells.
Figure 5.
ABD/SHP1c inhibits BCR/ABL-induced primary bone marrow cell proliferation. Murine bone marrow cells were infected with retroviral constructs expressing either the p185 or p210 form of BCR/ABL. Each construct also included either GFP or ABD/SHP1c. Cell numbers were determined at days 7 and 9 after infection. Mean cell count values from triplicate data sets are presented with standard deviations.
The Escort/Phosphatase Fusion Inhibits Leukemogenesis in Vivo.
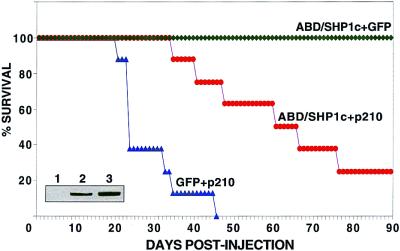
Development of cancers in a murine model system closely approximates the complexity of de novo human tumorigenesis. Some genetic alterations that result in transformation of cultured cells do not produce tumors in mice, and many transformation inhibitors that potently arrest isolated tumor cells prove ineffective in animal systems. To assay the leukemia suppression properties of the fusion protein, sublethally irradiated SCID mice were reconstituted with BALB/c-derived bone marrow cells infected with equal titers of virus constructs expressing GFP + p210 BCR/ABL, ABD/SHP1c + GFP, or ABD/SHP1c + p210 BCR/ABL. Mice receiving GFP + BCR/ABL began to develop leukemias after 21 days with 75% dead after 26 days. By 46 days all had succumbed to leukemia (Fig. 6). In contrast, animals that received ABD/SHP1c + p210 BCR/ABL did not show disease symptoms until day 35 and showed only 50% lethality at day 62. These results demonstrate that the ABD/SHP1c fusion protein can substantially block leukemogenesis in an in vivo system, a property not previously attributed to any natural or artificial protein product. The mice that received the ABD/SHP1c + GFP construct showed no signs of disease after 90 days (Fig. 6).
Figure 6.
Suppression of BCR/ABL-induced murine leukemia by ABD/SHP1c. SCID mice were reconstituted by using bone marrow cells infected with retrovirus constructs encoding GFP + p210 BCR/ABL (▴, nine mice), ABD/SHP1c + p210 BCR/ABL (●, nine mice), or ABD/SHP1c + GFP (⧫, three mice). All deaths were diagnosed as leukemias based on gross, microscopic, and FACS analysis. Survival data are presented as a Kaplan–Meyer plot. (Inset) p210 BCR/ABL expression from infected fibroblast cells demonstrating that equivalent virus titers were used for bone marrow infection (lane 1, mock; lane 2, GFP + p210; lane 3, ABD/SHP1c + p210).
Suppressed Transformation and Induced Differentiation of Human Leukemic Cells by ABD/SHP1c.
The cell culture and murine model systems described above demonstrate the ability of ABD/SHP1c to block the initiation and/or establishment of transformation by BCR/ABL. Chronic myelogenous leukemia-derived human cell lines, such as K562 (35), express BCR/ABL but may use some downstream effectors distinct from those recruited by ectopically expressed BCR/ABL in infected murine cells. In addition, the compound mutations typically found in human cancer cells might reduce the effectiveness of a direct BCR/ABL inhibitor. We therefore tested whether ABD/SHP1c could reverse the transformed phenotype of long-term cultured human leukemia cells.
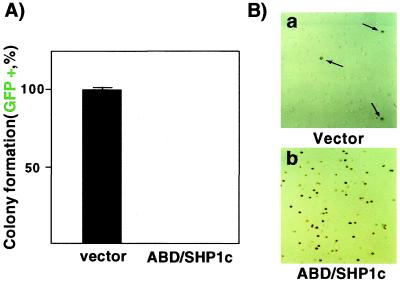
K562 cells were infected with retroviruses encoding ABD/SHP1c or no insert (vector). Each virus also included GFP to facilitate the identification of infected cells. The normalized results showed a strong reduction in colony number by ABD/SHP1c (Fig. 7A), consistent with a reversal of at least some transformation-associated phenotypes.
Figure 7.
Expression of ABD/SHP1c reverts the transformed phenotype of a chronic myelogenous leukemia line. (A) K562 cells were infected with the indicated virus (59% infection efficiency for ABD/SHP1c). Soft agar colonies that expressed GFP were counted and mean (duplicate) values were reported as a percentage (vector = 478 ± 25 colonies, all GFP positive; ABD/SHP1c = 228 ± 14 colonies, none GFP positive). (B) Globin expression was examined by benzidine staining in K562 cells infected with the indicated virus construct (95% infection efficiency for ABD/SHP1c). Vector only, 3% of cells stained positive (arrows). ABD/SHP1c, 73% of cells stained positive.
K562 cells enter an erythrocyte differentiation pathway after pharmacological inhibition of p210 BCR/ABL (36). Similarly, we found that ABD/SHP1c induced hemoglobin production in K562 cells, as judged by benzidine staining (Fig. 7B). This result indicates that these fully transformed cells, which have exited the normal differentiation pathway, can be induced to reenter a hematopoietic pathway by a targeted phosphatase. Taken together, these data demonstrate that ABD/SHP1c is able to inhibit the function of endogenous BCR/ABL in a human leukemia cell line.
Discussion
Cellular transformation by the BCR/ABL oncoprotein, both in vitro and in vivo, absolutely depends on tyrosine kinase activity. We have demonstrated that it is possible to co-opt the natural ABL binding property of RIN1 to create an escort/phosphatase that effectively suppresses transformation by BCR/ABL. This property depended on BCR/ABL binding as well as tyrosine phosphatase activity. In addition, the signal block was specific to BCR/ABL and had little or no effect on transformation by other oncoproteins.
The SHP2 catalytic domain had only a weak effect on transformation by BCR/ABL, although the ABD/SHP2c protein showed tyrosine phosphatase activity. These results suggest that the SHP1 and SHP2 catalytic domains encode determinants of substrate specificity. SHP1 expression is confined primarily to hematopoietic cells (37), where wild-type ABL is active, and SHP1 mutant mice show extensive defects in lymphocyte development (38). Some endogenous substrates of SHP1 have been identified (29). Others may include ABL-phosphorylated proteins required for cellular transformation. Indeed, overexpression of SHP1 has been reported to give a 2-fold reduction in BCR/ABL-mediated stress-activated protein kinase activation and fibroblast transformation (21). The tyrosine phosphatase PTP1B is another proposed negative regulator of BCR/ABL. PTP1B protein levels were induced after p210 BCR/ABL expression, and the two proteins were found in association (18). In addition, overexpression of PTP1B gave a 2- to 2.5-fold reduction in p210 BCR/ABL-mediated transformation (19). Our results demonstrate that an appropriate tyrosine phosphatase, when detached from its regulatory domains and appended to an ABL binding sequence, can potently suppress the transformation properties of BCR/ABL in fibroblasts and a human leukemia cell line with no apparent deleterious effects on normal cells.
Unexpectedly, the SHP1 catalytic domain construct was localized primarily to the nucleus, suggesting that the amino terminus and SH2 domains of the endogenous SHP1, known to be involved in regulation of catalytic activity (39, 40), may serve an important function in cytoplasmic retention of the protein. Nuclear localization signals in the SHP1 catalytic domain were also sufficient to retain most of the ABD/SHP1c fusion protein in the nucleus, despite the fact that RIN1 and ABD alone are normally cytoplasmic. This nuclear sequestration may be important in preventing deleterious effects from the escort/phosphatase. The shift to cytoplasmic localization in cells expressing BCR/ABL is consistent with a high affinity association between the ABD/SHP1c fusion protein and the primarily cytoplasmic oncoprotein.
There was a marked reduction in the phosphotyrosine content of BCR/ABL in cells expressing ABD/SHP1c. Several cellular proteins, normally tyrosine-phosphorylated in cells expressing BCR/ABL, also showed reduced phosphorylation when ABD/SHP1c was expressed. This could reflect a reduction in the tyrosine kinase activity of BCR/ABL. However, some signals were suppressed to a greater extent than others. This finding suggests that ABD/SHP1c may function to directly dephosphorylate BCR/ABL substrates—with a stronger activity toward some (Fig. 8). A “substrate trap” (41) mutant form of this escort/phosphatase might capture these preferred physiological substrates.
Figure 8.
Model of ABD/SHP1c-mediated inhibition of BCR/ABL signaling. The ABD of the fusion protein directs binding to BCR/ABL through complementary interactions with the SH2 and SH3 domains of ABL (23). This brings the PTP domain (SHP1c) of the fusion protein into proximity with the ABL kinase domain. The active PTP may inhibit signaling by reversing the autophosphorylation of ABL (A) and/or by releasing phosphate from newly phosphorylated ABL substrates (B).
Expression of ABD/SHP1c also suppressed the effects of BCR/ABL in hematopoietic cells, as determined by growth factor independence assays and primary bone marrow proliferation assays. In all cases, reductions and delays in transformation effects were observed. In addition, the escort/phosphatase construct strongly restrained the leukemogenic properties of BCR/ABL in a murine model system. It should be noted, however, that the level of BCR/ABL expression in the retrovirus-infected bone marrow cells likely exceeds that found in naturally occurring chronic phase leukemic cells, resulting in a highly stringent assay of functional blockade. These findings suggest that ABD/SHP1c (or a derivative construct) could be an effective inhibitor of leukemogenesis and both high-efficiency viral vectors and direct protein uptake (42) delivery systems are being pursued for this purpose. Such affinity-guided inhibitors could function to augment the therapeutic benefits of drugs such as STI-571 that block ABL kinase activity. This approach might enhance long-term treatment regimens where spontaneous drug resistance can emerge from expanding populations of BCR/ABL-expressing cells. In addition, our findings suggest that the escort/inhibitor fusion strategy may prove useful in targeting signal transduction components involved in other disease states.
Acknowledgments
We thank Drs. Benjamin Neel, Anne Marie Pendergast, Joel Greenberger, and Judith Gasson for providing critical reagents, Jami McLaughlin and Scott Thi for providing technical help, and David Rawlings and Leo Tsuda for critical discussions. Support for this research was provided by the Leukemia & Lymphoma Society (Y.-M.L.), the Margaret Early Research Foundation (J.C.), National Institutes of Health Grant CA56301 (J.C.), and National Institutes of Health Grant CA 76204 (O.N.W.). O.N.W. is a Howard Hughes Medical Institute Investigator.
Abbreviations
- ABD
ABL binding domain
- SHP1c
catalytic domain of SHP1
- SH
Src homology
- PTP
protein tyrosine phosphatase
- GFP
green fluorescent protein
- IP
immunoprecipitation
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.210253497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.210253497
References
- 1.Gotoh A, Broxmeyer H E. Curr Opin Hematol. 1997;4:3–11. doi: 10.1097/00062752-199704010-00002. [DOI] [PubMed] [Google Scholar]
- 2.Kurzrock R, Gutterman J U, Talpaz M. N Engl J Med. 1988;319:990–998. doi: 10.1056/NEJM198810133191506. [DOI] [PubMed] [Google Scholar]
- 3.Lugo T G, Pendergast A M, Muller A J, Witte O N. Science. 1990;247:1079–1082. doi: 10.1126/science.2408149. [DOI] [PubMed] [Google Scholar]
- 4.McWhirter J R, Wang J Y. Mol Cell Biol. 1991;11:1553–1565. doi: 10.1128/mcb.11.3.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pendergast A M, Gishizky M L, Havlik M H, Witte O N. Mol Cell Biol. 1993;13:1728–1736. doi: 10.1128/mcb.13.3.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Etten R A, Jackson P, Baltimore D. Cell. 1989;58:669–678. doi: 10.1016/0092-8674(89)90102-5. [DOI] [PubMed] [Google Scholar]
- 7.Huettner C S, Zhang P, Van Etten R A, Tenen D G. Nat Genet. 2000;24:57–60. doi: 10.1038/71691. [DOI] [PubMed] [Google Scholar]
- 8.Era T, Witte O N. Proc Natl Acad Sci USA. 2000;97:1737–1742. doi: 10.1073/pnas.97.4.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cobaleda C, Sánchez-García I. Blood. 2000;95:731–737. [PubMed] [Google Scholar]
- 10.Sawyers C L, Callahan W, Witte O N. Cell. 1992;70:901–910. doi: 10.1016/0092-8674(92)90241-4. [DOI] [PubMed] [Google Scholar]
- 11.Sawyers C L, McLaughlin J, Witte O N. J Exp Med. 1995;181:307–313. doi: 10.1084/jem.181.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gishizky M L, Cortez D, Pendergast A M. Proc Natl Acad Sci USA. 1995;92:10889–10893. doi: 10.1073/pnas.92.24.10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raitano A B, Halpern J R, Hambuch T M, Sawyers C L. Proc Natl Acad Sci USA. 1995;92:11746–11750. doi: 10.1073/pnas.92.25.11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reuther J Y, Reuther G W, Cortez D, Pendergast A M, Baldwin A S., Jr Genes Dev. 1998;12:968–981. doi: 10.1101/gad.12.7.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Druker B J, Tamura S, Buchdunger E, Ohno S, Segal G M, Fanning S, Zimmermann J, Lydon N B. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z Y, Dixon J E. Adv Enzymol Relat Areas Mol Biol. 1994;68:1–36. doi: 10.1002/9780470123140.ch1. [DOI] [PubMed] [Google Scholar]
- 17.Neel B G, Tonks N K. Curr Opin Cell Biol. 1997;9:193–204. doi: 10.1016/s0955-0674(97)80063-4. [DOI] [PubMed] [Google Scholar]
- 18.LaMontagne K R, Jr, Flint A J, Franza B R, Jr, Pandergast A M, Tonks N K. Mol Cell Biol. 1998;18:2965–2975. doi: 10.1128/mcb.18.5.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaMontagne K R, Jr, Hannon G, Tonks N K. Proc Natl Acad Sci USA. 1998;95:14094–14099. doi: 10.1073/pnas.95.24.14094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kharbanda S, Bharti A, Pei D, Wang J, Pandey P, Ren R, Weichselbaum R, Walsh C T, Kufe D. Proc Natl Acad Sci USA. 1996;93:6898–6901. doi: 10.1073/pnas.93.14.6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liedtke M, Pandey P, Kumar S, Kharbanda S, Kufe D. Oncogene. 1998;17:1889–1892. doi: 10.1038/sj.onc.1202117. [DOI] [PubMed] [Google Scholar]
- 22.Han L, Wong D, Dhaka A, Afar D, White M, Xie W, Herschman H, Witte O, Colicelli J. Proc Natl Acad Sci USA. 1997;94:4954–4959. doi: 10.1073/pnas.94.10.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Afar D E, Han L, McLaughlin J, Wong S, Dhaka A, Parmar K, Rosenberg N, Witte O N, Colicelli J. Immunity. 1997;6:773–782. doi: 10.1016/s1074-7613(00)80452-5. [DOI] [PubMed] [Google Scholar]
- 24.Muller A J, Young J C, Pendergast A M, Pondel M, Landau N R, Littman D R, Witte O N. Mol Cell Biol. 1991;11:1785–1792. doi: 10.1128/mcb.11.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hawley R G, Lieu F H, Fong A Z, Hawley T S. Gene Therapy. 1994;1:136–138. [PubMed] [Google Scholar]
- 26.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLaughlin J, Chianese E, Witte O N. Mol Cell Biol. 1989;9:1866–1874. doi: 10.1128/mcb.9.5.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper M C, Levy J, Cantor L N, Marks P A, Rifkind R A. Proc Natl Acad Sci USA. 1974;71:1677–1680. doi: 10.1073/pnas.71.5.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Timms J F, Carlberg K, Gu H, Chen H, Kamatkar S, Nadler M J, Rohrschneider L R, Neel B G. Mol Cell Biol. 1998;18:3838–3850. doi: 10.1128/mcb.18.7.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ram P A, Waxman D J. J Biol Chem. 1997;272:17694–17702. doi: 10.1074/jbc.272.28.17694. [DOI] [PubMed] [Google Scholar]
- 31.Dingwall C, Laskey R A. Trends Biol Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 32.Rennick D, Yang G, Gemmell L, Lee F. Blood. 1987;69:682–691. [PubMed] [Google Scholar]
- 33.Greenberger J S, Sakakeeny M A, Humphries R K, Eaves C J, Eckner R J. Proc Natl Acad Sci USA. 1983;80:2931–2935. doi: 10.1073/pnas.80.10.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goga A, McLaughlin J, Afar D E, Saffran D C, Witte O N. Cell. 1995;82:981–988. doi: 10.1016/0092-8674(95)90277-5. [DOI] [PubMed] [Google Scholar]
- 35.Lozzio C B, Lozzio B B. Blood. 1975;45:321–334. [PubMed] [Google Scholar]
- 36.Anafi M, Gazit A, Zehavi A, Ben-Neriah Y, Levitzki A. Blood. 1993;82:3524–3529. [PubMed] [Google Scholar]
- 37.Plutzky J, Neel B G, Rosenberg R D. Proc Natl Acad Sci USA. 1992;89:1123–1127. doi: 10.1073/pnas.89.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shultz L D, Rajan T V, Greiner D L. Trends Biotechnol. 1997;15:302–307. doi: 10.1016/S0167-7799(97)01060-3. [DOI] [PubMed] [Google Scholar]
- 39.Townley R, Shen S-H, Banville D, Ramachandran C. Biochemistry. 1993;32:13414–13418. doi: 10.1021/bi00212a006. [DOI] [PubMed] [Google Scholar]
- 40.Niu T, Liang X, Yang J, Zhao Z, Zhou G W. J Cell Biochem. 1999;72:145–150. doi: 10.1002/(sici)1097-4644(19990101)72:1<145::aid-jcb15>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 41.Flint A J, Tiganis T, Barford D, Tonks N K. Proc Natl Acad Sci USA. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwarze S R, Ho A, Vocero-Akbani A, Dowdy S F. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]