Abstract
In response to Salmonella typhimurium, the intestinal epithelium generates an intense inflammatory response consisting largely of polymorphonuclear leukocytes (neutrophils, PMN) migrating toward and ultimately across the epithelial monolayer into the intestinal lumen. It has been shown that bacterial-epithelial cell interactions elicit the production of inflammatory regulators that promote transepithelial PMN migration. Although S. typhimurium can enter intestinal epithelial cells, bacterial internalization is not required for the signaling mechanisms that induce PMN movement. Here, we sought to determine which S. typhimurium factors and intestinal epithelial signaling pathways elicit the production of PMN chemoattractants by enterocytes. Our results suggest that S. typhimurium activates a protein kinase C-dependent signal transduction pathway that orchestrates transepithelial PMN movement. We show that the type III effector protein, SipA, is not only necessary but is sufficient to induce this proinflammatory response in epithelial cells. Our results force us to reconsider the long-held view that Salmonella effector proteins must be directly delivered into host cells from bacterial cells.
Salmonella can cause a variety of diseases in humans and animals, ranging from gastroenteritis to systemic infections. Comparisons of bacterial genomic sequences indicate that Salmonella evolved as pathogens by acquiring large clusters of virulence genes, called pathogenicity islands (1, 2). More than 25 virulence genes are encoded on a 40-kb segment of the bacterial chromosome designated Salmonella Pathogenicity Island 1 (SPI1) (3). SPI1 encodes structural components and secreted substrates of a type III protein-secretion system that mediates the secretion and translocation of effector proteins from Salmonella into mammalian cells (reviewed in ref. 4). Two SPI1-secreted proteins, SipB and SipD, facilitate the translocation of SPI1-secreted effector proteins into the cytosol of mammalian cells (5, 6). The Sip proteins are thought to form secretion channels connecting the bacterial and host cells or, alternatively, form translocation-competent complexes with effector proteins that are capable of penetrating the mammalian cell membrane.
Salmonella effector proteins have been shown to interact with and directly affect proteins in mammalian cells (7, 8). For example, SopE is translocated into the cytosol of epithelial cells via the Salmonella typhimurium SPI1 secretion system and acts as a guanyl-nucleotide exchange factor for Rho GTPase proteins such as Rac and Cdc42. This results in actin polymerization, membrane ruffling, and invasion of S. typhimurium into the epithelial cells (9, 10). The bacterial effector protein, SptP, preferentially binds the GTP-bound form of Rac and enhances its GTPase activity, switching off the SopE-induced signal transduction pathways and restoring the morphology of the epithelial cells after bacterial invasion (11). In vivo, the SPI1 secretion system is required for Salmonella to enter intestinal epithelial cells (12–14). Recent studies indicate that the SPI1 secretion system also plays a key role in the mechanism by which Salmonella cause nontyphoidal disease (15–18).
The pathophysiology of localized enteritis caused by Salmonella is characterized by movement of electrolytes and water as well as polymorphonuclear leukocytes (neutrophils) (PMN) into the intestinal mucosa and lumen from the underlying microvasculature (19–21). The recruitment of inflammatory cells is considered to be a key virulence determinant underlying the development of Salmonella-elicited enteritis (22). In vitro experiments have shown that interaction between Salmonella and polarized intestinal epithelial cells can induce the epithelial cells to release potent chemoattractants that direct the transepithelial migration of PMN (23–26). For example, IL-8 is secreted basolaterally and may function to attract PMN through the lamina propria to the infected mucosa (24). Pathogen-elicited epithelial chemoattractant (PEEC) is secreted apically by the epithelial cells and directs PMN migration across the epithelial monolayer (26). The concerted and polarized secretion of IL-8 and PEEC by epithelial cells is thought to be responsible for directing PMN movement into the intestinal lumen in response to nontyphoidal Salmonella infections in humans.
The SPI1 secretion system, which is required for bacterial invasion, also is required for S. typhimurium to induce IL-8 and PEEC production in vitro as well as intestinal inflammatory responses in vivo. Interestingly, although the ability of Salmonella serotypes to elicit diffuse enteritis in humans correlates well with their ability to elicit PMN transmigration, it does not correlate well with their ability to invade epithelial cells (22). It has been proposed that extracellular Salmonella can translocate effector proteins into the host cell cytosol that stimulate proinflammatory host cell signaling pathways (15, 27). These observations led us to hypothesize that a secreted effector of the SPI1 type III secretion system, one that may not be involved in invasion, induces the cellular events leading to Salmonella-elicited gastroenteritis. Consistent with our hypothesis, the results presented herein reveal that the S. typhimurium SipA protein plays an essential and direct role in activating the epithelial signaling pathways that promote PMN transepithelial migration.
Experimental Procedures
Cell Culture.
T84 intestinal epithelial cells (passages 45–65) were grown in a 1:1 mixture of Dulbecco-Vogt modified Eagles medium and Ham's F-12 medium supplemented with 15 mM Hepes buffer (pH 7.5), 14 mM NaHCO3, 40 mg/liter penicillin, 8 mg/liter ampicillin, 90 mg/liter streptomycin, and 5% newborn calf serum. Polarized monolayers of T84 cells were formed and maintained on 0.33-cm2 ring-supported collagen-coated polycarbonate filters (Costar) as described (28) with recently detailed modifications (29). T84 cell monolayers reached a steady-state resistance 4–6 days after plating with some variability largely related to cell passage number. Cell culture inserts of inverted monolayers, used to study transmigration of PMN in the physiological basolateral-to-apical direction, were constructed as described (29–31).
Bacterial Strains.
S. typhimurium EE633 (sipA∷lacZY4) and VV341 (hilA∷kan-339) are derivatives of SL1344, an invasive mouse-virulent strain (6, 32). Plasmid pAK68C, which encodes a sipA-hemagglutinin (HA) gene fusion, was constructed by using the pBH vector (Boehringer Mannheim). Briefly, a HindIII–EcoRI DNA fragment containing the entire sipA ORF was prepared by PCR amplification using the HindIISipA primer (5′-GCAAGCTTAGTTACAAGTGTAAGGACGC-3′) and the SipAEcoRI primer (5′-GCGAATTCCGCTGCATGTGCAAGC-3′) with exTaq polymerase (Intergen, Purchase, NY). The sipA fragment was first cloned into pBluescript SK (Stratagene) to generate pAK62A and was sequenced by using the Prism ready reaction dideoxy terminator sequencing kit and the model 373A DNA sequencing system using the M13 forward and reverse primers (Applied Biosystems). The sipA fragment then was subcloned into pBH to generate pAK68C. Plasmids were passaged through the r−m+ S. typhimurium strain LB5000 (33) before being transformed into SL1344 or EE633 to generate AJK51 (SL1344/pAK68C), AJK63 (EE633/pAK68C), EE817 (SL1344/pBH), and EE818 (EE633/pBH). Although expression of the sipA-HA fusion in pAK68C is under the control of the inducible tac promoter, cultures of strains containing pAK68C were grown in the absence of isopropyl β-d-thiogalactoside because significant levels of SipA-HA were produced even under the noninducing conditions.
Growth of Bacteria for Assays Using Cell Culture Inserts.
Nonagitated microaerophilic bacterial cultures were prepared by inoculating 10 ml of LB broth (34) with 0.01 ml of a stationary-phase culture, followed by overnight incubation at 37°C, as described (22, 25). Ampicillin (50 μg/ml) was added to bacterial culture media when necessary.
Invasion Assays.
Infection of T84 monolayers was performed by the method described previously (22, 25).
PMN Transmigration Assays.
The physiologically directed (basolateral-to-apical) PMN transepithelial migration assay using cell culture inserts of inverted T84 monolayers has been described (25, 31). Human PMN were isolated from normal volunteers as described (31, 35). In a subset of experiments, S. typhimurium induction of PMN transepithelial migration was performed in the presence of α-SipA mAbs (a kind gift from the Institute for Animal Health). Specifically wild-type (WT) S. typhimurium was incubated in the presence of α-SipA mAb (0–60 μg/ml) 30 min before and during infection at the apical surface of model intestinal epithelia. S. typhimurium incubation with the irrelevant control mAb KJ126 was treated in the same manner.
Isolation of S. typhimurium Secreted Proteins.
S. typhimurium strains were grown in LB medium overnight as described above for invasion assays. The culture supernatants were collected and filtered through a 0.45 μm filter (Corning). The proteins from the supernatants were precipitated with 10% (vol/vol) trichloroacetic acid as described for Yersinia Yop proteins (36).
Purification of SipA-HA Fusion Protein.
Crude lysate from Escherichia coli DH5α expressing the SipA-HA recombinant fusion protein was precleared by passing the material through a 1-ml Sepharose column to remove any proteins that bind nonspecifically to Sepharose. The HA-affinity matrix (HA.11 affinity matrix; Convance, Berkeley CA) was equilibrated at 4°C with buffer C [200 mM Mes 2-(N- morpholine) ethanosulfonic acid, pH 6.2/0.1 mM MgCl2/0.1 mM EDTA, containing 0.05% Tween 20 and 0.5 M NaCl] before the addition of the precleared E. coli lysate. The HA affinity matrix was mixed with the E. coli lysate for 18 h at 4°C with constant end-over-end shaking. The unbound lysate material was washed free from the column by washing with 40 ml of buffer C. To elute the bound protein, the column resin was mixed with 1 mg of HA peptide dissolved in 2.5 ml of column buffer, and the resin/peptide mix was warmed to 30°C for 20 min. This step was carried out twice, and the peak fractions were pooled, buffer was exchanged with Hanks' balanced salt solution(+) and analyzed via SDS/PAGE and Western blotting to verify protein purity.
Preparation and Characterization of α-SipA mAb.
A hybridoma clone and α-SipA mAb were generated as described (37). As demonstrated by Western blot analysis of secreted and whole-cell proteins from different Salmonella strains, the α-SipA mAb is specific for SipA and does not cross-react with Salmonella proteins. The specificity of the antibody was demonstrated by analyzing samples of secreted proteins and whole-cell lysates (by Western blot) from different WT Salmonella strains and an Salmonella dublin sipA mutant. Secreted proteins from polar and nonpolar mutants also were analyzed to verify that the α-SipA mAb is SipA-specific and does not cross-react with other Salmonella proteins (data not shown).
Western Blotting.
Proteins were separated by SDS/PAGE (12%), transferred to nitrocellulose (Bio-Rad; 0.45 μ membrane), and subsequently reacted with anti-HA mAb 16B12 (Babco, Richmond, CA). Reactive bands were visualized by enhanced chemiluminescence using a kit from Pierce according to the manufacturer's instructions.
Generation of Imposed Gradients of SipA.
In these experiments, the PMN transmigration assays were performed by the addition of PMN to the upper chamber of the transwell after purifed SipA (20 μg/ml) was added to the opposing (lower) chamber. Migration was directed in the apical-to-basolateral (nonphysiologic) direction to avoid direct contact of SipA with the apical surface. Transmigration in the apical-to-basolateral direction is qualitatively similar to PMN transmigration in the basolateral-to-apical direction, but it is 5-fold less efficient (38, 39). Therefore, 5-fold more PMN were added when transmigration proceeded in the apical-to-basolateral direction so that baseline transmigration signals would be equivalent in both directions. A positive control for this experiment was the evaluation of PMN transmigration in the apical-to-basolateral direction induced by imposed gradients of n-formylmethionylleucylphenylalanine (fMLP) (1 μM).
Chelerytherine Chloride Treatment.
Hanks' balanced salt solution(+)-washed T84 cell monolayers were incubated in the presence of 5 μM chelerytherine chloride (stock concentration at 5 mM in water) (Biomol, Plymouth Meeting, PA) for 30 min at 37°C. Subsequently, S. typhimurium was added to the apical surface of the T84 monolayers in the continued presence of chelerytherine chloride. After an incubation of 1 h at 37°C, the cells were washed free of both nonadherent bacteria and chelerytherine chloride and then processed for either the invasion assay or PMN transmigration assay.
Data Presentation.
Because variations exist in both transepithelial resistance between groups of monolayers (baseline resistance range 650 to 1,500 ohm⋅cm2) and in PMN obtained from different donors, individual experiments were performed by using large numbers of monolayers and PMN from single blood donors on individual days. PMN isolation was restricted to 10 different donors (repetitive donations) over the course of these studies. S. typhimurium invasion and myeloperoxidase assay data were compared by Student's t test. PMN transmigration results are represented as PMN cell equivalents (CEs) derived from a daily standard PMN dilution curve. PMN that completely traversed the monolayer are represented as the number of PMN (CE/ml in a total volume of 1 ml). Values are expressed as the mean and SD of an individual experiment done in triplicate repeated at least three times.
Results
Expression of S. typhimurium sipA Is Critical for Eliciting PMN Transepithelial Migration.
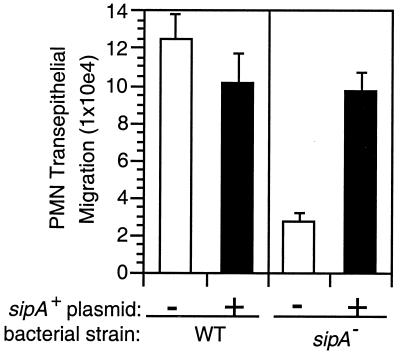
We screened S. typhimurium type III effector mutants to see whether any are altered in their ability to promote PMN transepithelial migration, but not altered in their ability to invade T84 cell monolayers. In our assay, the apical surface of T84 epithelial cell monolayers was exposed for 1 h to S. typhimurium and subsequently was rinsed to remove nonadherent bacteria. The ability of infected epithelial monolayers to elicit PMN transepithelial migration was assessed by adding human PMN to the basolateral compartment of the rinsed monolayers (27). As shown in Fig. 1, we found that a S. typhimurium sipA mutant (6) is dramatically reduced (>80%) in its capacity to induce PMN transepithelial migration as compared with the WT S. typhimurium strain. It has been shown that a S. typhimurium sipA mutant is impaired in its ability to enter epithelial cells only after very short infection times, but exhibits WT levels of invasion at times beyond 30 min (40). Similar to these previous studies, our S. typhimurium sipA mutant (EE633) entered the apical surface of T84 cell monolayers to the same extent as WT bacteria, during our 1-h assay (0.65 ± 0.10% vs. 0.64 ± 0.12% for WT vs. sipA S. typhimurium, respectively).
Figure 1.
The sipA gene is required for S. typhimurium to induce PMN transmigration. WT (SL1344) or sipA mutant (EE633) S. typhimurium strains that contain no plasmid (empty bars) or a plasmid expressing sipA (pAK68C) (filled bars) were assessed for their ability to promote PMN transmigration. Data are presented as the mean ± SD of assays performed in triplicate. WT or sipA mutant strains that contain the pBH vector plasmid behaved identically to the same strains containing no plasmid (data not shown).
SDS/PAGE and Western analyses showed that, as expected, the sipA mutant fails to express and secrete SipA (data not shown and ref. 6). The specificity of the sipA mutant defects were verified by showing that a plasmid that expresses the sipA gene restores the ability of the sipA mutant to secrete SipA and to induce PMN transepithelial migration, basically to the levels elicited by either the WT strain or the WT strain containing the sipA plasmid (data not shown; Fig. 1).
α-SipA mAb Antagonizes the Ability of S. typhimurium to Induce PMN Transepithelial Migration.
SipA is secreted via the SPI1 type III secretion system and is translocated into the cytosol of mammalian cells where it interacts with components of the actin cytoskeleton, enhancing the cytoskeletal changes that mediate the early stages of S. typhimurium entry into nonphagocytic cells (40, 41). Studies of many bacterial systems indicate that type III effector proteins are incapable of entering mammalian cells on their own and, instead, are delivered directly from adherent bacteria via type III secretion systems (4). Consistent with this idea, antiserum to YopE, a type III effector of Yersinia pestis, has no effect on the YopE-dependent cytotoxicity of Y. pestis (42).
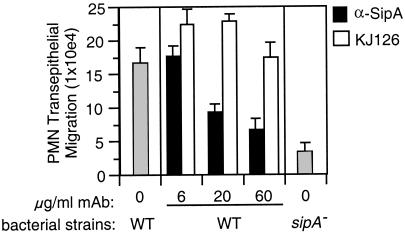
To test whether direct translocation of SipA into T84 cells induces transepithelial signaling to PMN, we incubated WT S. typhimurium in the presence of a neutralizing α-SipA mAb before and during infection of the epithelial monolayer. Surprisingly, the presence of α-SipA mAb significantly reduced the ability of S. typhimurium to induce PMN transepithelial migration (Fig. 2). We addressed whether addition of α-SipA mAb might indirectly affect PMN migration by interfering with the ability of the bacteria to interact with the epithelial cells or by directly inhibiting PMN migration. We found that the addition of α-SipA mAb neither prevented the association of WT S. typhimurium with T84 cell monolayers [0.27 ± 0.05% vs. 0.33 ± 0.06% WT S. typhimurium internalized in the absence vs. presence of α-SipA mAb (60 μg/ml), respectively] nor inhibited the transmigration of PMN to imposed gradients of a potent PMN chemoattractant, fMLP [29.21 ± 8.36 × 104 vs. 35.63 ± 9.27 × 104 PMN cell equivalents transmigrated in the absence vs. presence of α-SipA mAb (60 μg/ml), respectively]. Further substantiating the idea that α-SipA mAb specifically inhibits the ability of S. typhimurium to induce PMN transepithelial migration, an irrelevant mAb, KJ126 (a kind gift from Cathryn Nagler-Anderson, Massachusetts General Hospital), failed to inhibit the induction of PMN transepithelial signaling in response to WT S. typhimurium (Fig. 2).
Figure 2.
α-SipA mAb attenuates S. typhimurium-induced PMN migration. WT S. typhimurium was incubated in the presence of a α-SipA mAb (black bars) or an irrelevant control mAb KJ126 (white bars) before and during the infection of T84 monolayers. PMN transmigration is compared with that induced by WT and sipA mutant S. typhimurium strains in the absence of mAb (gray bars). Data represent the mean ± SD of triplicate samples.
SipA Is Sufficient to Induce PMN Transepithelial Migration.
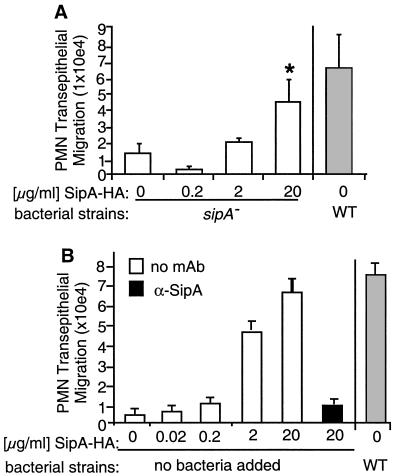
Our results indicate that the secretion of SipA and the ability of SipA to induce epithelial cell responses are not coupled to its direct delivery into epithelial cells from bacterial cells. Instead, the data suggest that an extracellular form of SipA is an important intermediate that mediates the epithelial cell responses triggered by S. typhimurium. If our hypothesis was correct, purified SipA should rescue the PMN defect of the EE633 sipA mutant strain. As shown, addition of recombinant SipA-HA epitope-tagged protein obtained from E. coli (Fig. 3) allows EE633 to induce PMN transepithelial migration (Fig. 4A). Further supporting this idea, addition of culture supernatants from WT S. typhimurium rescues the PMN transmigration defect of the sipA mutant strain (data not shown). The specificity of this induction is evidenced by the finding that mAb against SipA blocks the response (data not shown). Taken together, these results confirm the notion that an extracellular form of SipA can facilitate S. typhimurium-induced PMN transepithelial migration.
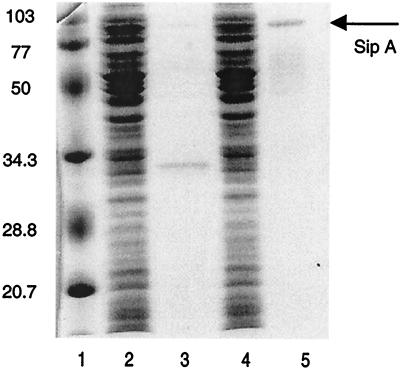
Figure 3.
Coomassie-stained SDS/PAGE gel showing SipA-HA purification. Lanes show molecular mass standards in kDa (lane 1), precleared crude lysate from E. coli DH5α/pAK68C expressing the SipA-HA recombinant fusion protein (lane 2), lysate material that did not bind to the affinity matrix (lane 4), the eluted SipA-HA (lane 5), and the uneluted proteins that remained bound to the affinity matrix (lane 3).
Figure 4.
(A) The addition of purified SipA-HA permits S. typhimurium strain EE633 to induce PMN transepithelial migration. The purified recombinant SipA-HA fusion protein was added to 1.25 × 108 sipA mutant S. typhimurium (EE633) at a final concentration of (0.2–20 μg/ml) just before and during a 1-h bacterial infection at the apical surface of T84 cell monolayers. Elicitation of PMN transepithelial migration subsequently was measured. Data represent the mean ± SD of triplicate samples; (*, not significantly different from levels induced by WT). (B) Purified SipA-HA is sufficient to induce PMN transepithelial migration. The apical surface of T84 cell monolayers was exposed to varying amounts of SipA-HA (empty bars). After 1 h at 37°C, excess SipA-HA was removed, PMN were added to the basolateral membrane interface, and PMN migration was assessed. Alternatively, 60 μg/ml of α-SipA mAb was added to purified SipA (20 μg/ml) 30 min before and during the 1 h incubation with the T84 cells (filled bar). PMN transmigration is compared with that induced by WT S. typhimurium (gray bar). Data represent the mean ± SD of triplicate samples.
To examine whether SipA alone can induce PMN transepithelial signaling, without the assistance of other Salmonella or type III factors, SipA-HA, purified from E. coli, was added to buffer (Hanks' balanced salt solution with Ca2+ and Mg2+) overlying washed T84 cell monolayers. As shown in Fig. 4B, exposure of the apical surface of epithelial cells to 20 μg/ml SipA-HA elicits a PMN transmigration response that is equivalent to that induced by WT S. typhimurium. Induction of the PMN transmigration response by purified SipA-HA was dose-dependent and completely abrogated by α-SipA mAb (Fig. 4B). This effect was not attributed to potential trace amounts of lipopolysaccharide (LPS) because this polysaccharide does not induce PMN transepithelial migration in this model system [17.57 ± 3.5 × 104, 0.17 ± 0.02 × 104, and 0.18 ± 0.03 × 104 cell equivalents of PMN transmigrated for WT S. typhimurium, negative control (Hanks' balanced salt solution(+) buffer), and 10 μg/ml LPS, respectively]. The ability of purified Sip-HA to induce the PMN transmigration response and rescue the sipA mutant defect was also not attributed to the carboxyl-terminal epitope tag because a peptide containing the HA-epitope sequence had no effect in these assays (data not shown).
These results indicate that interaction of SipA protein with epithelial cells is sufficient to induce epithelial cell responses that elicit basolateral-to-apical PMN transmigration. However, although excess SipA-HA was removed from the apical surface of T84 cells by washing, before the addition of PMN to the basolateral interface, it remained possible that PMN movement was in direct response to a transepithelial gradient of the recombinant SipA-HA protein itself. To address this possibility, we performed PMN transmigration assays in which 20 μg/ml SipA-HA was added to the basolateral compartment and PMN were added to the opposing apical compartment. Although the nonphysiologic apical-to-basolateral migration of PMN could be stimulated by fMLP, we were unable to observe PMN migration in response to a basolateral-to-apical gradient of SipA-HA protein (data not shown). Thus, the recombinant SipA-HA fusion protein does not appear to be a direct PMN chemotaxin.
An Inhibitor of Protein Kinase C (PKC) Blocks SipA-Induced PMN Migration.
Because S. typhimurium secretes and translocates a number of type III effector proteins via the SPI1 secretion system, we wondered whether purified SipA-HA induces PMN transepithelial migration via the same mechanism as WT S. typhimurium. We took a pharmacological-based approach to characterize the specific features of the signaling pathways required for SipA-induced vs. S. typhimurium-induced PMN transepithelial migration. First, we screened different drugs that inhibit a variety of signal transduction pathways for their ability to inhibit S. typhimurium-induced PMN transepithelial migration vs. fMLP-induced PMN transepithelial migration. Because S. typhimurium-induced signaling to PMN across intestinal epithelial cell monolayers occurs by a mechanism that is independent from fMLP-induced signaling (25), we predicted that drugs that inhibit the specific signal transduction components targeted by S. typhimurium should have no effect on fMLP-induced PMN migration.
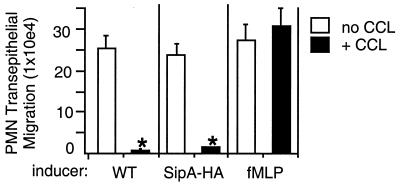
As shown in Fig. 5, treatment of T84 cell monolayers with chelerythrine chloride dramatically decreases the ability of S. typhimurium to induce PMN transepithelial migration (>95% reduction), whereas this drug has no effect on the ability of fMLP to induce PMN transepithelial migration. Chelerythrine chloride is a potent and selective inhibitor of PKC (43). Treatment with chelerythrine chloride did not alter the ability of S. typhimurium to become internalized by T84 cell monolayers (data not shown), which is consistent with other studies that have shown that PKC inhibitors do not affect S. typhimurium invasion (44). Also shown in Fig. 5, treatment of T84 cell monolayers with chelerythrine chloride completely abrogated the ability of purified S. typhimurium SipA-HA to induce PMN transepithelial migration. These data indicate that SipA that is secreted by S. typhimurium plays a direct role in activating a PKC-dependent epithelial signaling pathway that promotes PMN transepithelial migration.
Figure 5.
Chelerythrine chloride (CCL) specifically inhibits the ability of WT S. typhimurium and purified SipA-HA to induce PMN transepithelial migration. WT S. typhimurium, purified SipA-HA (20 μg/ml), or fMLP (10 nM) were incubated with T84 cells to induce PMN migration (empty bars). The effect of CCL (filled bars) was assessed by incubating T84 cell monolayers in the presence of 5 μM CCL for 30 min at 37°C. Inducers were added to the apical surface of T84 cell monolayers in the continued presence of the drug, and after 1 h (at 37°C), the monolayers were processed for the PMN transmigration assay. Data represent the mean ± SD of triplicate samples: (*, P < 0.05).
Discussion
Recruitment of inflammatory cells is considered to be a key virulence mechanism underlying Salmonella-elicited enteritis (15, 22). In vitro studies have shown that the SPI1 type III secretion system is required for S. typhimurium to activate epithelial signaling pathways that regulate PMN transmigration (22). We have determined the molecular basis of this requirement by showing that SipA, a secreted substrate of the S. typhimurium SPI1 type III secretion system, is not only necessary but is sufficient to activate epithelial signaling pathways and promote PMN transepithelial migration. We have used genetic, biochemical, and cell biological methods to establish the pivotal role that secreted SipA plays in inducing the epithelial cell responses that lead to PMN transepithelial migration.
We have found that exogenously added α-SipA mAb inhibits the bacterial-induced epithelial signal transduction events that lead to PMN transepithelial migration. We also have found that purified S. typhimurium SipA protein can trigger the PMN migration response in the absence of the type III secretion and translocation factors, such as SipB and SipD. Our results are surprising and contrast with previous ideas about how type III secreted proteins are translocated into the cytosol of host cells (4, 5, 45). Our results suggest that interaction of SipA at the apical surface of intestinal epithelial cells is sufficient to initiate the cellular events that lead to PMN transepithelial migration. SipA may not need to enter the epithelial cell cytosol to stimulate proinflammatory signal transduction pathways and, instead, may function extracellularly at the epithelial cell surface where it engages a specific receptor. In this case, SipA also would function intracellularly where it would interact with the actin cytoskeleton to enhance bacterial invasion (40, 41). Alternatively, SipA may be able to bind and cross the epithelial cell membrane, entering the host cell cytosol by a process that does not require the type III translocation system. In the cytosol, SipA may interact with one or more host cell components to alter the actin cytoskeleton as well as stimulate the production of PMN chemoattractants. Regardless, our results demonstrate that SipA has other activities that have not been previously recognized. There are precedents for bifunctional type III effectors. The SPI1-secreted protein, SipB, plays a role in protein translocation but also enters the host cell cytosol and directly activates capase-1 (5, 46). The Pseudomonas aeruginosa type III secreted protein, exoenzyme S, has two independent effector domains, and, when delivered into the cytosol of host cells, interferes with two different signal transduction pathways (47).
We have shown that purified SipA as well as culture supernatants from WT S. typhimurium can rescue the PMN defect of a sipA mutant. Interestingly, although purified SipA can induce PMN migration in the absence of bacteria, culture supernatants are not sufficient to induce transmigration (ref. 25 and data not shown). The level of soluble SipA may be critical for its productive interaction with epithelial cells. Alternatively, bacterial culture supernatants might contain factors that inhibit the interaction and activity of SipA on epithelial cells. In either case, our results suggest that the presence of S. typhimurium bacteria in the transmigration assay enhances the activity of SipA in culture supernatants. The sipA mutant bacteria may stimulate signaling pathways that when combined with the SipA-induced signals will trigger the PMN response. However, we favor the idea that S. typhimurium cells help to localize and stabilize the interactions of SipA at the epithelial cell surface. Further experiments will be required to distinguish these hypotheses and investigate exactly how SipA is presented to epithelial cells in the context of WT S. typhimurium.
We previously have shown that the apical secretion of the PMN chemoattractant, PEEC, by T84 cells directs the transepithelial migration of PMN in response to S. typhimurium infection (26). Our current results suggest that SipA affects a PKC-dependent signal transduction pathway that leads to the polarized secretion of PEEC. Because S. typhimurium-induced PMN transmigration has been shown to require epithelial cell protein synthesis (25), we speculate that a SipA-induced, PKC-dependent signaling response leads to the production and secretion of PEEC. At this point, it is not clear how SipA might induce PEEC production via activation of host PKC. PKC can initiate a phosphorylation cascade that results in the phosphorylation of a pivotal protein kinase, mitogen-activated protein (MAP) kinase, and, ultimately, activation of the AP-1 transcription factor (48, 49). PKC activation also can lead to the phosphorylation and degradation of Iκ-B, which allows NF-κB to migrate into the nucleus and act as a transcriptional regulator (50, 51). Because inhibitors of the NF-κB pathway have no effect on S. typhimurium-induced PMN migration (27), we speculate that a PKC-activated, MAP kinase-dependent signal transduction pathway governs PEEC expression and PMN transepithelial signaling in polarized epithelial cells.
Interestingly, the MAP kinase and NF-κB signal transduction pathways have been implicated in the mechanism by which S. typhimurium induces IL-8 expression and secretion. The translocation of the type III effector, SopE, into the host cell cytosol stimulates Cdc42 and Rac, which induce membrane ruffling and bacterial invasion (7). The activation of Cdc42 by SopE also appears to enhance the phosphorylation of MAP kinase and Iκ-B, which may contribute to the expression and secretion of IL-8. S. typhimurium type III effectors also may induce a Ca2+-mediated signaling pathway that leads to activation of NF-κB and IL-8 secretion (52). The overlap between signal transduction pathways affected by S. typhimurium type III effectors suggests that more than one type III effector is able to induce PEEC and IL-8 secretion. However, the bacterial factors and host cell signal events required for PEEC secretion appear to be different from those involved in IL-8 secretion. For example, disruption of the sipA gene, which abolishes PMN transepithelial migration, has no effect on S. typhimurium-induced IL-8 secretion (unpublished data). In addition, epithelial secretion of IL-8 induced by S. typhimurium is sensitive to inhibitors of the NF-κB pathway, but the induction of PMN transepithelial migration is not (27). However, it remains to be determined whether the induction of PEEC by SipA involves epithelial signaling pathways that are completely separate from those involved in IL-8 production, or if some signal transduction component, such as MAP kinase, contribute to the induction of both PEEC and IL-8.
In summary, using a physiologically relevant epithelial model, we have established that the S. typhimurium-secreted protein, SipA, is not only necessary but is sufficient to activate epithelial signaling pathways that promote PMN transepithelial migration. This work identifies SipA as an important mediator of the immune inflammatory response that results in PMN influx. The fact that purified SipA directly activates this response has prompted us to reconsider the long-held view that S. typhimurium effector proteins must be delivered into host cells by a type III secretion and translocation system. Rather, our results suggest that SipA engages a host surface receptor, or translocates into the host cell by a process independent of the type III system. The mechanism of activation of epithelial proinflammatory signaling cascades by SipA represents a potential target for therapeutic intervention. A better understanding of how SipA activates PMN transmigration also may lead to a better understanding of the pathways by which chronic states of intestinal inflammation (i.e., Crohn's disease and ulcerative colitis) are aberrantly activated.
Acknowledgments
This work was supported by National Institutes of Health Grant DK50989 (to B.A.M.) and the Giovanni Armenise-Harvard Foundation for Advanced Scientific Research (to C.A.L.). A.J.K. was a Howard Hughes Medical Institute Predoctoral Fellow.
Abbreviations
- PMN
polymorphonuclear leukocytes (neutrophils)
- PKC
protein kinase C
- SPI1
Salmonella Pathogenicity Island 1
- PEEC
pathogen-elicited epithelial chemoattractant
- WT
wild type
- HA
hemagglutinin
- fMLP
n-formylmethionylleucylphenylalanine
- MAP
mitogen-activated protein
References
- 1.Baumler A, Tsolis R M, Ficht T A, Adams L G. Infect Immun. 1998;66:4579–4587. doi: 10.1128/iai.66.10.4579-4587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Groisman E A, Ochman H. Trends Microbiol. 1997;5:343–349. doi: 10.1016/S0966-842X(97)01099-8. [DOI] [PubMed] [Google Scholar]
- 3.Darwin K H, Miller V L. Clin Microbiol Rev. 1999;12:405–428. doi: 10.1128/cmr.12.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hueck C J. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collazo C, Galan J E. Mol Microbiol. 1997;24:747–753. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- 6.Hueck C J, Hantman M J, Bajaj V, Johnson C, Lee C A, Miller S I. Mol Microbiol. 1995;18:479–490. doi: 10.1111/j.1365-2958.1995.mmi_18030479.x. [DOI] [PubMed] [Google Scholar]
- 7.Hardt W-D, Chen L-M, Schuebel K E, Bustelo X R, Galan J E. Cell. 1998;93:815–826. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- 8.Norris A F, Wilson M P, Wallis T S, Galyov E E, Majerus P W. Proc Natl Acad Sci USA. 1998;95:14057–14059. doi: 10.1073/pnas.95.24.14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L, Hobbie S, Galan J. Science. 1996;274:2115–2118. doi: 10.1126/science.274.5295.2115. [DOI] [PubMed] [Google Scholar]
- 10.Hardt W-D, Urlaub H, Galan J E. Proc Natl Acad Sci USA. 1998;95:2574–2579. doi: 10.1073/pnas.95.5.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu Y, Galan J E. Nature (London) 1999;401:293–297. doi: 10.1038/45829. [DOI] [PubMed] [Google Scholar]
- 12.Clark M A, Hirst B H, Jepson M A. Infect Immun. 1998;66:724–731. doi: 10.1128/iai.66.2.724-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones B D, Ghori N, Falkow S. J Exp Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Penheiter K L, Mathur N, Giles D, Fahlen T, Jones B D. Mol Microbiol. 1997;24:697–709. doi: 10.1046/j.1365-2958.1997.3741745.x. [DOI] [PubMed] [Google Scholar]
- 15.Galyov E E, Wood M W, Rosqvist R, Mullan P B, Watson P R, Hedges S, Wallis T S. Mol Microbiol. 1997;25:903–912. doi: 10.1111/j.1365-2958.1997.mmi525.x. [DOI] [PubMed] [Google Scholar]
- 16.Jones M A, Wood M W, Mullan P B, Watson P R, Wallis T S, Galyov E E. Infect Immun. 1998;66:5799–5804. doi: 10.1128/iai.66.12.5799-5804.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsolis R M, Adams L G, Ficht T A, Baumler A J. Infect Immun. 1999;67:4879–4885. doi: 10.1128/iai.67.9.4879-4885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson P R, Galyov E E, Paulin S M, Jones P W, Wallis T S. Infect Immun. 1998;66:1432–1438. doi: 10.1128/iai.66.4.1432-1438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Day D W, Mandal B K, Morrson B C. Histopathology. 1978;2:117–131. doi: 10.1111/j.1365-2559.1978.tb01700.x. [DOI] [PubMed] [Google Scholar]
- 20.McGovern V J, Slavutin L J. Am J Surg Pathol. 1979;3:483–490. doi: 10.1097/00000478-197912000-00001. [DOI] [PubMed] [Google Scholar]
- 21.Rout W R, Formal S B, Dammin G J, Giannella R A. Gastroenterology. 1974;67:59–70. [PubMed] [Google Scholar]
- 22.McCormick B A, Miller S I, Carnes D K, Madara J L. Infect Immun. 1995;63:2302–2309. doi: 10.1128/iai.63.6.2302-2309.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eckmann L, Kagnoff M, Fierer J. Infect Immun. 1993;61:4569–4574. doi: 10.1128/iai.61.11.4569-4574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCormick B A, Hofman P M, Kim J, Carnes D K, Miller S I, Madara J L. J Cell Biol. 1995;131:1599–1608. doi: 10.1083/jcb.131.6.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCormick B A, Colgan S P, Archer C D, Miller S I, Madara J L. J Cell Biol. 1993;123:895–907. doi: 10.1083/jcb.123.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCormick B A, Parkos C A, Colgan S P, Carnes D K, Madara J L. J Immunol. 1998;160:455–466. [PubMed] [Google Scholar]
- 27.Gewirtz A T, Siber A M, Madara J L, McCormick B A. Infect Immun. 1999;67:608–617. doi: 10.1128/iai.67.2.608-617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dharmsathaphorn K, Madara J L. Methods Enzymol. 1990;192:354–389. doi: 10.1016/0076-6879(90)92082-o. [DOI] [PubMed] [Google Scholar]
- 29.Madara J L, Parkos C A, Colgan S P, MacLeod R J, Nash S, Matthews J, Delp C, Lencer W S. J Clin Invest. 1992;89:1938–1944. doi: 10.1172/JCI115800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nash S, Parkos C A, Nusrat A, Delp C, Madara J L. J Clin Invest. 1991;87:1474–1477. doi: 10.1172/JCI115156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parkos C A, Delp C, Arnaout M A, Madara J L. J Clin Invest. 1991;88:1605–1612. doi: 10.1172/JCI115473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wray C, Sojka W J. Res Vet Sci. 1978;25:139–143. [PubMed] [Google Scholar]
- 33.Bullas L R, Ryu J I. J Bacteriol. 1983;156:471–474. doi: 10.1128/jb.156.1.471-474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Revel H R. Virology. 1966;31:688–701. doi: 10.1016/0042-6822(67)90197-3. [DOI] [PubMed] [Google Scholar]
- 35.Henson P, Oades Z G. J Clin Invest. 1975;56:1053–1061. doi: 10.1172/JCI108152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forsberg A, Bolin I, Norlander L, Wolf-Watz H. Microb Pathog. 1987;2:123–137. doi: 10.1016/0882-4010(87)90104-5. [DOI] [PubMed] [Google Scholar]
- 37.Wood M W, Rosqvist R, Mullan P B, Edwards M H, Galyov E E. Mol Microbiol. 1996;22:327–338. doi: 10.1046/j.1365-2958.1996.00116.x. [DOI] [PubMed] [Google Scholar]
- 38.Colgan S P, Parkos C A, Delp C, Arnaout M A, Madara J L. J Cell Biol. 1993;120:785–798. doi: 10.1083/jcb.120.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parkos C A, Colgan S P, Delp C, Arnaout M A, Madara J L. J Cell Biol. 1992;117:757–764. doi: 10.1083/jcb.117.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou D, Mooseker M S, Galan J E. Science. 1999;283:2092–2096. doi: 10.1126/science.283.5410.2092. [DOI] [PubMed] [Google Scholar]
- 41.Zhou D, Mooseker M S, Galan J E. Proc Natl Acad Sci USA. 1999;96:10176–10181. doi: 10.1073/pnas.96.18.10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petersson J, Holmstrom A, Hill J, Leary S, Frithz-Lindsten E, von Euler-Mateli A, Carlsson E, Titball R, Forsberg A, Wolf-Watz H. Mol Microbiol. 1999;32:961–976. doi: 10.1046/j.1365-2958.1999.01408.x. [DOI] [PubMed] [Google Scholar]
- 43.Herbert J, Augerau J, Gleye J, Maffrand J. Biochem Biophys Res Commun. 1990;171:189–195. doi: 10.1016/0006-291x(90)91375-3. [DOI] [PubMed] [Google Scholar]
- 44.Rosenshine I, Duronio V, Finlay B B. Infect Immun. 1992;60:2211–2217. doi: 10.1128/iai.60.6.2211-2217.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galan J E. Mol Microbiol. 1996;20:263–271. doi: 10.1111/j.1365-2958.1996.tb02615.x. [DOI] [PubMed] [Google Scholar]
- 46.Hersh D, Monack D M, Smith M R, Ghori N, Falkow S, Zychlinsky A. Proc Natl Acad Sci USA. 1999;96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gehring U M, Schmidt G, Peterson K J, Aktories K, Barbieri J T. J Biol Chem. 1999;274:36369–36372. doi: 10.1074/jbc.274.51.36369. [DOI] [PubMed] [Google Scholar]
- 48.Karin M. J Biol Chem. 1995;270:16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- 49.Whitmarsh A J, Davis R J. J Mol Med. 1996;74:589–607. doi: 10.1007/s001090050063. [DOI] [PubMed] [Google Scholar]
- 50.Siebenlist U, Franzozo G, Brown K. Ann Rev Cell Biol. 1994;10:405–455. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- 51.Thanos D, Maniatis T. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 52.Gewirtz A T, Roa A S, Simon P O, Merlin D, Madara J L, Neish A S. J Clin Invest. 2000;105:79–92. doi: 10.1172/JCI8066. [DOI] [PMC free article] [PubMed] [Google Scholar]