Abstract
An assay has been developed that allows the identification of molecules that function as type I IFN antagonists. Using this assay, we have identified an Ebola virus-encoded inhibitor of the type I IFN response, the Ebola virus VP35 protein. The assay relies on the properties of an influenza virus mutant, influenza delNS1 virus, which lacks the NS1 ORF and, therefore, does not produce the NS1 protein. When cells are infected with influenza delNS1 virus, large amounts of type I IFN are produced. As a consequence, influenza delNS1 virus replicates poorly. However, high-efficiency transient transfection of a plasmid encoding a protein that interferes with type I IFN-induced antiviral functions, such as the influenza A virus NS1 protein or the herpes simplex virus protein ICP34.5, rescues growth of influenza delNS1 virus. When plasmids expressing individual Ebola virus proteins were transfected into Madin Darby canine kidney cells, the Ebola virus VP35 protein enhanced influenza delNS1 virus growth more than 100-fold. VP35 subsequently was shown to block double-stranded RNA- and virus-mediated induction of an IFN-stimulated response element reporter gene and to block double-stranded RNA- and virus-mediated induction of the IFN-β promoter. The Ebola virus VP35 therefore is likely to inhibit induction of type I IFN in Ebola virus-infected cells and may be an important determinant of Ebola virus virulence in vivo.
Ebola viruses are enveloped, negative-strand RNA viruses belonging to the family Filoviridae. These viruses possess genomes of approximately 19 kb and are known to encode eight proteins, the nucleoprotein (NP), VP35, VP40, glycoprotein (GP), soluble GP, VP30, VP24, and L (polymerase) proteins (1). Ebola virus infections frequently result in severe hemorrhagic fever, and epidemics of the Ebola virus, Zaire subtype have resulted in mortality rates of greater than 80% (1, 2). The pathologic features and the immune responses characteristic of fatal and nonfatal human Ebola virus infections have begun to be characterized (3–5). Additionally, the mechanisms by which Ebola viruses induce hemorrhage and shock are beginning to be explored. Recent reports have suggested roles for both immune-mediated pathology (3) as well as pathology mediated by specific viral proteins. Membrane-bound GP has been proposed to mediate cytotoxicity in endothelial cells (4), whereas soluble GP has been proposed to inhibit early neutrophil activation (5). However, the latter mechanism is controversial (6). To fully understand the pathogenesis of Ebola virus infections, it will be important to study further the mechanisms by which the virus interacts with its host, including the ways in which the virus subverts the host antiviral response.
One important component of the host antiviral response is the type I IFN system. Type I IFN is synthesized in response to viral infection; double-stranded RNA (dsRNA) or viral infection activates latent transcription factors, including IRF-3 and NF-κB, resulting in the transcriptional up-regulation of type I IFN, IFN-α, and IFN-β, genes. Secreted type I IFNs signal through a common receptor, activating the JAK/STAT signaling pathway. This signaling stimulates transcription of IFN-sensitive genes, including a number that encode antiviral proteins, and leads to the induction of an antiviral state. Among the antiviral proteins induced in response to type I IFN are dsRNA-dependent protein kinase R (PKR), 2′,5′-oligoadenylate synthetase (OAS), and the Mx proteins (7–10).
Many viruses have evolved mechanisms to subvert the host IFN response. For example, the herpes simplex virus (HSV-1) protein ICP34.5 counteracts the PKR-mediated phosphorylation of translation initiation factor eIF-2α, preventing the establishment of an IFN-induced block in protein synthesis (11). In the negative-strand RNA viruses, several different anti-IFN mechanisms have been identified (12, 13). First, the influenza A virus NS1 protein was shown to block IFN responses in virus-infected cells (12). Subsequently, the V protein of SV5 was shown to target STAT1 for proteasome-mediated degradation, preventing signaling from both type I and type II IFN receptors (13, 14). Also, the Sendai virus C proteins were found to block types I and II IFN signaling and to counteract the establishment of an antiviral state (15–17). Recently, measles virus infection has been shown to block induction of type I IFN production (18). Also, the bovine respiratory syncytial virus NS1 and NS2 proteins have been shown to function together to antagonize the type I IFN response (51).
The best-studied example of an IFN antagonist encoded by a negative-strand RNA virus is the influenza A virus NS1 protein. A mutant influenza virus, influenza delNS1 virus, which lacks the NS1 ORF and, therefore, produces no NS1 protein, grows poorly on substrates in which type I IFN-induced antiviral pathways are intact (12). Such substrates include Madin Darby canine kidney (MDCK) cells, 10-day-old embryonated chicken eggs, and mice. It is clear that the growth of influenza delNS1 virus is impaired because of its inability to counteract IFN-mediated antiviral response(s). The virus grows similarly to wild-type virus [influenza A/PR/8/34 (H1N1) (PR8) virus] on substrates such as 6-day-old embryonated chicken eggs, Vero cells, and STAT1−/− mice, which do not mount an effective type I IFN response (12, 19). The failure of influenza delNS1 virus to grow on IFN-producing substrates correlates with its ability to induce IFN. Thus, infection of cells with this mutant virus induces large amounts of type I IFN, a circumstance associated with a potent antiviral state (ref. 19; M. Salvatore, H. Zheng, T. Muster, P.P., and A.G.-S., unpublished results). Such IFN synthesis does not occur when the same substrates are infected with the NS1 protein-producing wild-type PR8 virus (12). The ability of the NS1 protein to prevent IFN production and to facilitate growth of influenza viruses on IFN-producing substrates may be related to its ability to bind single-stranded and/or dsRNA (20–22). NS1 has been shown to inhibit activation of PKR and OAS (21). Additionally, the NS1 protein is able to prevent viral activation of IRF-3 (23) and NF-κB (52), central components in promoting type I IFN synthesis.
Ebola virus-infected endothelial cells have impaired IFN responses, suggesting that Ebola virus also may encode an IFN antagonist. After treatment with dsRNA, IFN-α, or IFN-γ, little induction of IFN-stimulated genes was seen in infected cells compared with uninfected cells (24, 25). However, this does not reflect a general inhibition of signal transduction; IL-1β-induced signaling was not inhibited in Ebola virus-infected cells (24, 25).
In this report, we describe an assay that permits identification of proteins that inhibit the type I IFN-induced antiviral response. This assay was used further to screen for an Ebola virus-encoded IFN antagonist. The assay uses transient transfection of plasmids encoding type I IFN antagonists to enhance growth on MDCK cells of the IFN-sensitive influenza delNS1 virus. Thus, expression of the influenza A virus NS1 protein or the HSV-1 ICP34.5 efficiently complemented growth of influenza delNS1 virus. Using this assay, we screened individual Ebola virus proteins for their ability to rescue growth of influenza delNS1 virus on MDCK cells. One Ebola virus protein, the VP35 protein, was found to complement influenza delNS1 virus growth. VP35 subsequently was found to block dsRNA- and virus-mediated induction of an IFN-responsive promoter and the IFN-β promoter. Therefore, the VP35 protein is likely to function as an inhibitor of the type I IFN response in Ebola virus-infected cells and may be an important determinant of Ebola virus virulence.
Materials and Methods
Cells and Viruses.
MDCK and 293 cells were maintained in DMEM/10% FBS. Virus-infected cells were maintained in DMEM/0.3% BSA (ICN). Influenza delNS1 virus was propagated at 33°C in 7-day-old embryonated chicken eggs (19). Sendai virus, strain Cantell, was propagated at 37°C in 10-day-old embryonated chicken eggs.
Plasmids.
The NS1 expression plasmid pCAGGS-PR8 NS1 SAM has been described (23). The NS1 expression plasmid pCMV-PR8 NS1 SAM was generated by subcloning the NS1 ORF from pCAGGS-PR8 NS1 SAM into pcDNA3 (Invitrogen). The HSV-1 γ134.5 gene was excised with NcoI and BamHI from the plasmid pBR4789 (a generous gift from B. Roizman, University of Chicago), which contains the HSV-1 BamHI S1 fragment, and subcloned into pCAGGS (26).
The Ebola virus strain Mayinga, subtype Zaire, was used for cloning (nucleotide numbers refer to the Ebola virus Zaire genome, GenBank accession number AF086833). The ORFs and small parts of the nontranslated regions of the NP, VP35, and VP30 genes were amplified by reverse transcription—PCR, with specific primers containing appropriate restriction sites. The products were inserted into the expression vector pcDNA3. NP was inserted by using BamHI. VP35 (nucleotides 3,126–4,175) was inserted by using BamHI and NotI. The VP30 gene (nucleotides 8,506–9,399) was inserted by using EcoRI and XhoI. The GP and soluble GP inserts were excised from the plasmids pGEM-mGP7 and pGEM-mGP8 (27) with BamHI and HindIII. The HindIII sites were blunted, and the fragments were inserted between the BamHI–EcoRV restriction sites of pcDNA3.1(+) (Invitrogen). RNA-cDNA hybrids from a cDNA library of the Ebola virus subtype Zaire, strain Mayinga, (28) were used to amplify the VP40 and VP24 genes. PCR fragments containing ORFs of VP40 (nucleotides 4,410–5,779) and VP24 (nucleotides 10,290–11,312) genes were inserted at the EcoRV restriction site of pcDNA3.1(+). The clones were confirmed by DNA sequencing.
Influenza delNS1 Virus Complementation Assays.
High-efficiency transient transfection of MDCK cells was performed by using Lipofectamine2000 (LF2000) (GIBCO/BRL). Four micrograms of the indicated expression plasmid was adjusted to 50 μl by using Optimem I medium (GIBCO/BRL). Per transfection, 10 μl of LF2000 was adjusted to 0.25 ml with Optimem I medium and incubated in a 5-ml polystyrene snap-cap tube at room temperature for 5 min. Each 50-μl DNA sample was added to the 0.25-ml LF2000/Optimem I mix, agitated gently, and incubated 20 min at room temperature. A confluent 80-cm2 flask of MDCK cells was detached with trypsin. The cells were brought to 12 ml with DMEM/10% FBS (no antibiotics), pelleted at 1,000 rpm for 5 min in a table-top centrifuge, and, after aspiration of the supernatant, resuspended in DMEM/10% FBS (no antibiotics) to a concentration of 4 × 106 cells/ml. A portion (0.25 ml) of the cell suspension was aliquoted into 35-mm tissue culture dishes. After the 20-min incubation period, 1 ml of DMEM/10% FBS (no antibiotics) was added to each DNA/LF2000 mix, and the DNA/LF2000/medium mixture was added to the dishes containing the MDCK cells. After mixing, the cells were maintained at 37°C overnight. Sixteen to 20 h posttransfection, the cells were infected with 103 plaque-forming units (pfu) of influenza delNS1 virus [multiplicity of infection (moi) = 0.001] in a volume of 0.1 ml. After removal of the inoculum, the cells were maintained in 1.5 ml of DMEM/0.3% bovine albumin/3 μg/ml trypsin (trypsin, 1:250; Difco).
Reporter Gene Assays.
293 cells were transfected by using the calcium phosphate method (29). Each transfection of 1 × 106 cells contained 0.3 μg of the IFN-stimulated response element (ISRE)-driven chloramphenicol acetyltransferase (CAT) reporter plasmid pHISG-54-CAT (30) or a mouse IFN-β promoter-driven CAT reporter pIFN-CAT (52), 0.3 μg of the simian virus 40 promoter luciferase reporter plasmid pGL2-Control (Promega), and 4 μg of the indicated expression plasmid.
Twenty-four hours posttransfection, the cells were mock-treated, transfected with 40 μg polyI:polyC (Amersham Pharmacia) by using LF2000 according to the manufacturer's recommendations, or infected with influenza delNS1 virus or Sendai virus at an moi of 1.0 for each virus. Cells treated by all methods were maintained in DMEM/0.3% bovine albumin. Twenty-four hours posttreatment, the cells were harvested and lysed in Reporter Lysis Buffer (Promega). CAT assays were performed as described (31). Luciferase assays were performed by using the Promega luciferase assay system according to the manufacturer's directions.
Western Blot Analysis.
293 cells were transfected with 4 μg of the indicated plasmids by using LF2000, as described above. Forty-eight hours posttransfection, the cells were lysed in SDS/PAGE sample buffer, and Western blots were performed by following standard procedures. The influenza A virus NS1 protein was detected by using anti-PR8 NS1 rabbit polyclonal antiserum 5091 at a 1:1,000 dilution. The Ebola virus NP and VP35 proteins were detected by using a goat polyclonal anti-Ebola virus antiserum (purchased from Vector Laboratories) at a 1:20,000 dilution. Secondary antibodies were anti-rabbit or anti-goat antibodies conjugated to horseradish peroxidase. Detection was performed by using the NEN Renaissance Western blot chemiluminescence reagent.
Northern Blot Analysis.
293 cells were transfected with 5 μg of either empty vector or the VP35 expression plasmid by using LF2000. Twenty-four hours posttransfection, the cells were infected with influenza delNS1 virus or Sendai virus at an moi of 1. Twenty-four hours later, total RNA was extracted by using TRI reagent (Molecular Research Center, Cincinnati). Ten micrograms of total RNA was used for Northern blot analysis by using Quickhyb hybridization solution (Stratagene). IFN-β or β-actin mRNAs were detected by hybridization to specific [32P]ATP-labeled probes.
Results
Expression of Known IFN Antagonists Complements Growth of Influenza delNS1 Virus.
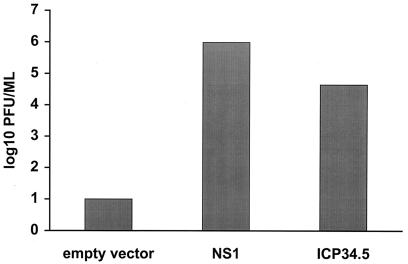
The influenza delNS1 virus grows poorly on IFN-producing cells such as MDCK cells. However, wild-type PR8 virus, isogenic with influenza delNS1 virus except that it produces the NS1 protein, grows to high titer on MDCK cells (12). It therefore was determined whether high-efficiency transfection of MDCK cells with an NS1-expression plasmid would complement growth of influenza delNS1 virus (Fig. 1). When MDCK cells were transfected with an influenza A virus NS1 plasmid and infected 24 h later with influenza delNS1 virus at a low moi, the mutant virus grew to approximately 1 × 106 pfu/ml. However, infection of cells transfected with an empty plasmid yielded 102 pfu/ml or less (Fig. 1).
Figure 1.
Growth of the influenza delNS1 virus is complemented by transient transfection of an influenza A NS1 protein or an HSV ICP34.5 expression plasmid. MDCK cells were transfected with 4 μg of empty expression plasmid (pCAGGS), pCAGGS-PR8 NS1 SAM (23), or pCAGGS-γ134.5. Twenty-four hours later, the cells were infected with influenza delNS1 virus (moi = 0.001). Forty-eight hours posttransfection, viral titers were determined by plaque assay. The results are the average of two independent experiments.
It then was determined whether expression of another known inhibitor of the type I IFN-induced antiviral response, HSV-1 ICP34.5, would complement growth of influenza delNS1 virus. Expression of the HSV-1-encoded PKR antagonist ICP34.5 (11) clearly complemented growth of the influenza delNS1 virus (Fig. 1). This result suggests that complementation of influenza delNS1 virus growth reflects an anti-IFN function. It also indicates that this complementation assay can be used to identify other proteins that inhibit the IFN-induced antiviral response.
The Ebola Virus VP35 Protein Complements Growth of Influenza delNS1 Protein.
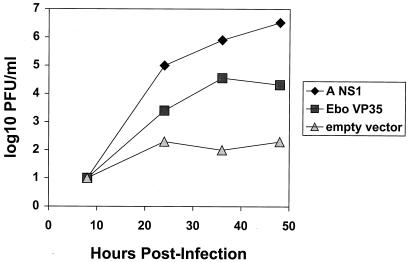
The influenza delNS1 virus complementation assay then was used to screen for an Ebola virus-encoded IFN antagonist. An empty vector, the NS1-expression plasmid, or plasmids encoding individual Ebola virus proteins were transfected into MDCK cells. Twenty-four hours posttransfection, the cells were infected with influenza delNS1 virus. Forty-eight hours postinfection, the supernatants were harvested and viral titers were determined by plaque assay (Table 1). The only Ebola virus protein that enhanced influenza delNS1 virus growth was the VP35 protein (Table 1). Time-course analysis clearly demonstrated the enhancement of influenza delNS1 virus growth by VP35 (Fig. 2).
Table 1.
Ability of Ebola virus proteins to complement growth of influenza deINS1 virus
| Expressed protein | pfu/ml |
|---|---|
| Empty vector | 10 |
| NS1 | 1.2 × 106 |
| NP | 10 |
| VP35 | 1.9 × 104 |
| VP40 | <10 |
| GP | <10 |
| sGP | 20 |
| VP30 | <10 |
| VP24 | <10 |
Figure 2.
Growth of the influenza delNS1 virus is complemented by the Ebola virus VP35 protein. MDCK cells were transfected with 4 μg of empty expression plasmid (pcDNA3), NS1 expression plasmid, or Ebola virus VP35 expression plasmid. Twenty-four hours later, the cells were infected with influenza delNS1 virus (moi = 0.001). Viral titers were determined by plaque assay at the indicated times.
Expression of the Ebola Virus VP35 Protein Blocks Induction of an ISRE Promoter.
To determine whether VP35 inhibits the dsRNA- and virus-mediated activation of IFN-sensitive gene expression, cells were transfected with an ISRE-driven CAT-reporter plasmid and a constitutively expressed, simian virus 40 promoter-driven luciferase reporter plasmid. Additionally, the cells were transfected with empty vector, NS1 expression plasmid, VP35 expression plasmid, or, as an additional control, an Ebola virus NP expression plasmid. One day later, the cells were mock-treated, transfected with dsRNA, or infected with either influenza delNS1 virus or with Sendai virus, strain Cantell (an attenuated strain known to induce large amounts of IFN). After an additional 24 h, cell lysates were prepared and assayed for CAT activity and luciferase activity (Fig. 3A). Transfection of cells with dsRNA or infection with either influenza delNS1 virus or Sendai virus gave a strong induction of the IFN-sensitive promoter. When either NS1 or VP35 was present, expression from the IFN-responsive promoter was almost completely blocked. Levels of ISRE induction, normalized to levels of luciferase activity, are shown in Fig. 3A. Expression of the control luciferase reporter plasmid was not inhibited by expression of either NS1 or VP35 (data not shown). Expression of the Ebola virus NP, which did not complement growth of influenza delNS1 virus, did not inhibit activation of the ISRE promoter (data not shown). Expression of the NS1, VP35, and NP proteins was confirmed by Western blotting (Fig. 3B). These results show that both NS1 and VP35 can block type I IFN production and/or signaling in response to either dsRNA treatment or to viral infection.
Figure 3.
Expression of Ebola virus VP35 protein inhibits dsRNA- or virus-mediated induction of an ISRE. (A) Fold induction of an ISRE promoter–CAT reporter gene in the presence of empty vector, NS1 expression plasmid, or VP35 expression plasmid. 293 cells were transfected with 4 μg of the indicated expression plasmid plus 0.3 μg each of the reporter plasmids pHISG-54-CAT and pGL2-Control. Twenty-four hours posttransfection, the cells were mock-treated or treated with the indicated IFN inducer. The CAT activities were normalized to the corresponding luciferase activities to determine fold induction. (B) Western blot showing NS1, VP35, and Ebola virus NP expression. 293 cells were transfected with 4 μg of the indicated plasmids. Forty-eight hours later, cell lysates were prepared and Western blots were performed by using the indicated antiserum.
Expression of the Ebola Virus VP35 Protein Blocks Activation of the INF-β Promoter.
In wild-type influenza A virus-infected cells, the NS1 protein blocks induction of type I IFN. This block is due, in large part, to the ability of NS1 to prevent activation of IRF-3 (23) and NF-κB (52), two transcription factors that play a critical role in stimulating the synthesis of IFN-β. Synthesis of IFN-β, in turn, plays an important role in the initiation of the type I IFN cascade (32). The Ebola virus VP35, therefore, was tested for its ability to block activation of the IFN-β promoter.
Empty vector, NS1 expression plasmid, or VP35 expression plasmid was cotransfected with a mouse IFN-β promoter-driven CAT reporter and a simian virus 40 promoter-driven luciferase reporter. When cells subsequently were transfected with dsRNA, a strong induction of the IFN-β promoter was observed in empty vector-transfected cells, but this induction was blocked when either NS1 or VP35 was expressed (Fig. 4A).
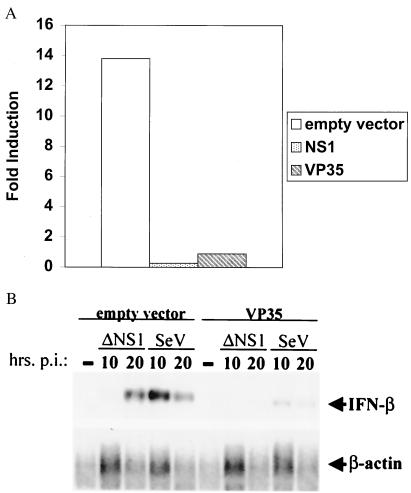
Figure 4.
The VP35 protein of Ebola virus inhibits induction of the IFN-β promoter. (A) Inhibition of induction of the mouse IFN-β promoter. 293 cells were transfected with 4 μg of the indicated expression plasmid plus 0.3 μg each of the reporter plasmids pIFN-β-CAT and pGL2-Control. Twenty-four hours posttransfection, the cells were mock-transfected or transfected with 40 μg of polyI:polyC. (B) Northern blot showing VP35-mediated inhibition of endogenous IFN-β induction. 293 cells were transfected with either empty vector or VP35 expression plasmid. Twenty-four hours later, the cells were mock-infected (−) or infected with influenza delNS1 virus (delNS1) or Sendai virus (SeV) (moi = 1). Total RNA was prepared from cells at 10 h or 20 h posttransfection. Mock-transfected cell RNA was prepared at the same time as the 20-h postinfection samples. Northern blots were performed to detect IFN-β or β-actin mRNAs. Note that less total RNA was obtained when cells, including the mock-infected cells, were lysed at the 20-h postinfection time point.
It also was determined whether VP35 could block activation of the endogenous human IFN-β promoter. Cells were transfected with empty vector or VP35 expression plasmid and, 24 h later, mock-infected or infected with influenza delNS1 virus or with Sendai virus. Ten or 20 h postinfection, total cellular RNA was isolated, and a Northern blot was performed to detect IFN-β mRNA (Fig. 4B). Expression of VP35 clearly blocked induction of the endogenous IFN-β promoter. Before infection with either virus, IFN-β mRNA was undetectable. After infection, when the IFN-β mRNA levels were normalized to β-actin mRNA levels, it was found that, in influenza delNS1 virus-infected cells, the presence of VP35 reduced IFN-β induction 8-fold at 10 h postinfection and 8.4-fold at 20 h posttransfection. In Sendai virus-infected cells, the presence of VP35 reduced IFN-β induction 6.1-fold at 10 h posttransfection and 5.9-fold at 20 h posttransfection.
The Ebola Virus VP35 Blocks INF Induction When Coexpressed with the Ebola Virus NP.
The VP35 protein is an essential component of the Ebola virus RNA synthesis complex and likely associates with the viral NP (33, 34). Therefore, it was determined whether Ebola virus VP35 retained its IFN-antagonizing properties when it was coexpressed with the Ebola virus NP. An ISRE-reporter assay was performed in which cells received either empty vector, VP35 alone, NP alone, or a combination of VP35 and NP. Twenty-four hours posttransfection, the cells were transfected with dsRNA or infected with Sendai virus. As seen previously, transfection with empty plasmid or with NP expression plasmid did not block activation of the ISRE promoter, but expression of VP35 did block its activation (Fig. 5). Further, coexpression of VP35 and NP was able to block ISRE activation to the same extent as expression of VP35 alone (Fig. 5). These data indicate that VP35, even when coexpressed with the Ebola virus NP, can act as an IFN antagonist.
Figure 5.
The Ebola virus VP35 protein inhibits type I IFN induction when coexpressed with Ebola virus NP. Fold induction of the IFN-inducible ISRE-driven reporter in the presence of empty vector, VP35, NP, or VP35 plus NP. 293 cells were transfected with a total of 4 μg of expression plasmid, including 2 μg of a plasmid encoding an individual protein and 2 μg of a second plasmid (either empty vector or a second expression plasmid) plus 0.3 μg each of the reporter plasmids pHISG-54-CAT and pGL2-Control. Twenty-four hours posttransfection, the cells were mock-treated or treated with the indicated IFN inducer. Twenty-four hours postinduction, CAT and luciferase assays were performed. The CAT activities were normalized to the corresponding luciferase activities to determine fold induction.
Discussion
In this report, the Ebola virus VP35 protein was identified as an IFN antagonist based on its ability to complement growth of the influenza delNS1 virus. VP35 shares this complementing ability with the influenza A virus NS1 protein, the HSV-1 ICP34.5 (Fig. 1), and the vaccinia virus E3L protein (unpublished observation). Each of these proteins has been shown to interfere with one or more components of the IFN-induced antiviral response. The NS1 protein of influenza A virus blocks both the production of type I IFN (12) and the activation of the IFN-induced antiviral proteins PKR (21, 35, 36) and OAS (unpublished observation). The inhibition of type I IFN production occurs, at least in part, because NS1 blocks activation of the latent transcription factors IRF-3 and NF-κB (23, 52), which are involved in the dsRNA- and virus-mediated activation of the IFN-β promoter. These inhibitory functions may be mediated by the binding of dsRNA by NS1 (21, 23, 52). The vaccinia virus E3L protein also blocks the activation of both PKR (37–40) and OAS (41). Further, E3L expression promotes growth of vaccinia virus in the presence of IFN (40, 42). The HSV-1 ICP34.5 antagonizes the type I IFN antiviral response by targeting phosphorylated eIF-2α, the product of activated PKR. Specifically, ICP34.5 binds the cellular protein phosphatase 1α and retargets it to eIF-2α (11). The resulting dephosphorylation of eIF-2α thus counteracts PKR function (11). As a result, virus mutants that fail to make ICP34.5 are attenuated and show restricted tissue tropism in mice (43, 44). However, as with the influenza delNS1 virus, virulence of these mutants is restored in mice lacking genes critical to type I IFN signaling or in mice lacking PKR (45, 46). Because the HSV-1 ICP34.5 rescues influenza delNS1 virus growth, a VP35-mediated block of PKR function likely would be sufficient to enhance influenza delNS1 virus growth. However, any number of mechanisms might accomplish this task, including a direct effect on PKR function, a block in type I IFN-induced signaling, such that IFN-responsive genes (including PKR) are not transcriptionally activated, or a block in IFN synthesis. We also demonstrated that VP35, like the influenza A virus NS1 protein, prevents dsRNA- and virus-mediated activation of ISRE-containing promoters (Fig. 3) and the IFN-β promoter (Fig. 4). In blocking virus-induced production of IFN, VP35 would prevent the establishment of an antiviral state in both uninfected cells and Ebola virus-infected cells and, thus, facilitate viral replication.
Previously, VP35 has been shown to play an essential role in Ebola virus RNA synthesis (33). VP35 appears to be an ortholog of the paramyxovirus and rhabodvirus P proteins (phosphoproteins) (33), although the filovirus VP35 proteins are only weakly phosphorylated (47). The IFN antagonists of two other nonsegmented negative-strand viruses, the SV5 V protein and Sendai virus C proteins, also are encoded within P genes. However, these paramyxovirus proteins are not equivalent to P proteins. The Sendai virus C proteins are encoded by overlapping ORFs that initiate at alternative start codons and are read from an alternate reading frame (48). The SV5 V protein shares a common amino terminus with the P protein; however, because of the insertion by the viral polymerase of a nontemplate encoded G residue, the V protein possesses a carboxyl terminus distinct from that of the P protein. There is no evidence for production of either C or V orthologs from the Ebola virus VP35 gene. There are no ORFs of significant length near the 5′ end of the VP35 gene that would encode a C protein. Additionally, although mRNA editing by the Ebola virus polymerase occurs within the GP gene (27, 49), there is no evidence for the presence of RNA-editing signals within the VP35 gene, nor is there evidence for production of an edited VP35 gene product. Given the results of our study, the P proteins of other nonsegmented negative-strand viruses also should be examined for IFN-antagonizing capability.
The production of an IFN antagonist may contribute to the virulence of Ebola viruses. In humans, it appears that an appropriate cytokine response is related to the development of asymptomatic or nonfatal Ebola virus infection (50). Thus, a viral factor that influences type I IFN production might influence viral pathology. It is clear from the study of other viruses that the presence of an IFN antagonist is required for full virulence. For example, wild-type influenza A/PR/8/34 virus is pathogenic in wild-type mice (LD50 = 102–103 pfu), but the influenza delNS1 virus is severely attenuated (LD50 > 106 PFU). However, when the IFN-induced antiviral response is absent, as in STAT1−/− mice, influenza delNS1 virus is nearly as virulent as wild-type virus (12). Additionally, influenza viruses with truncated NS1 proteins display diminished capacity to replicate in wild-type mice (19). Likewise, HSV-1 mutants that do not produce ICP34.5 are attenuated in wild-type mice but not in IFN receptor−/− mice or PKR−/− mice (45, 46). To experimentally assess the significance of the VP35 IFN-antagonizing function on Ebola virus virulence, it ultimately will be necessary to generate viral mutants that retain the ability to replicate their genomes but that lack the VP35-encoded IFN-inhibiting function. The development of an Ebola virus reverse genetics system, toward which significant progress has been made (33), will facilitate such analyses. It also will be of interest to assess the relative IFN-inhibiting potency of VP35 proteins from Ebola virus strains that have displayed different levels of pathogenicity in humans.
The influenza delNS1 complementation assay described in this manuscript provides a straightforward way in which to screen proteins for IFN-antagonizing function. It should be possible, using this system, to identify the proteins of other viruses that, like the Ebola virus VP35 protein, function as IFN antagonists. The identification of novel, viral-encoded IFN antagonists will further our understanding of how pathogens evade the innate immune response. Further, mutation of viral-encoded IFN antagonists may be an ideal way in which to generate stable, attenuated live vaccines for a variety of different viral pathogens. Such an approach already is being investigated for influenza viruses (19). Viral-encoded IFN antagonists also may become targets for new antiviral drugs against important human pathogens. Inhibition of the activity of viral IFN antagonists should result in an induction of antiviral pathways in the infected cell and a concomitant inhibition of virus replication.
Acknowledgments
We thank Louis Nguyenvu for excellent technical assistance. This work was supported by a National Institutes of Health National Research Service Award postdoctoral fellowship to C.F.B., by research grants from the National Institutes of Health to A.-G.S. and P.P. and from the Deutsche Forschungsgemeinschaft to E.M. and H.-D.K.
Abbreviations
- NP
nucleoprotein
- GP
glycoprotein
- dsRNA
double-stranded RNA
- OAS
2′,5′-oligoadenylate synthetase
- ISRE
IFN-stimulated response element
- GP
glycoprotein
- pfu
plaque-forming units
- moi
multiplicity of infection
- MDCK
Madin Darby canine kidney cells
- PKR
dsRNA-dependent protein kinase R
- CAT
chloramphenicol acetyltransferase
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220398297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220398297
References
- 1.Klenk H-D, Slenczka W, Feldmann H. In: Encyclopedia of Virology. Webster R G, Granoff A, editors. Vol. 2. New York: Academic; 1994. pp. 827–831. [Google Scholar]
- 2.Peters C J, Khan A S. Curr Top Microbiol Immunol. 1999;235:85–95. doi: 10.1007/978-3-642-59949-1_6. [DOI] [PubMed] [Google Scholar]
- 3.Villinger F, Rollin P E, Brar S S, Chikkala N F, Winter J, Sundstrom J B, Zaki S R, Swanepoel R, Ansari A A, Peters C J. J Infect Dis. 1999;179, Suppl. 1:S188–S191. doi: 10.1086/514283. [DOI] [PubMed] [Google Scholar]
- 4.Yang Z, Duckers H J, Sullivan N J, Sanchez A, Nabel E G, Nabel G J. Nat Med. 2000;6:886–889. doi: 10.1038/78645. [DOI] [PubMed] [Google Scholar]
- 5.Yang Z, Delgado R, Xu L, Todd R F, Nabel E G, Sanchez A, Nabel G J. Science. 1998;279:1034–1037. doi: 10.1126/science.279.5353.1034. [DOI] [PubMed] [Google Scholar]
- 6.Maruyama T, Buchmeier M J, Parren P W H I, Burton D R. Science. 1998;282:843a. [Google Scholar]
- 7.Clemens M J, Elia A. J Interferon Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- 8.Floyd-Smith G, Slattery E, Lengyel P. Science. 1981;212:1030–1032. doi: 10.1126/science.6165080. [DOI] [PubMed] [Google Scholar]
- 9.Haller O, Frese M, Kochs G. Rev Sci Tech. 1998;17:220–230. doi: 10.20506/rst.17.1.1084. [DOI] [PubMed] [Google Scholar]
- 10.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 11.He B, Gross M, Roizman B. Proc Natl Acad Sci USA. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy D E, Durbin J E, Palese P, Muster T. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 13.Young D F, Didcock L, Goodbourn S, Randall R E. Virology. 2000;269:383–390. doi: 10.1006/viro.2000.0240. [DOI] [PubMed] [Google Scholar]
- 14.Didcock L, Young D F, Goodbourn S, Randall R E. J Virol. 1999;73:9928–9933. doi: 10.1128/jvi.73.12.9928-9933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcin D, Latorre P, Kolakofsky D. J Virol. 1999;73:6559–6565. doi: 10.1128/jvi.73.8.6559-6565.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gotoh B, Takeuchi K, Komatsu T, Yokoo J, Kimura Y, Kurotani A, Kato A, Nagai Y. FEBS Lett. 1999;459:205–210. doi: 10.1016/s0014-5793(99)01241-7. [DOI] [PubMed] [Google Scholar]
- 17.Komatsu T, Takeuchi K, Yokoo J, Tanaka Y, Gotoh B. J Virol. 2000;74:2477–2480. doi: 10.1128/jvi.74.5.2477-2480.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naniche D, Yeh A, Eto D, Manchester M, Friedman R M, Oldstone M B. J Virol. 2000;74:7478–7484. doi: 10.1128/jvi.74.16.7478-7484.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talon J, Salvatore M, O'Neill R E, Nakaya Y, Zheng H, Muster T, Garcia-Sastre A, Palese P. Proc Natl Acad Sci USA. 2000;97:4309–4314. doi: 10.1073/pnas.070525997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida T, Shaw M W, Young J F, Compans R W. Virology. 1981;110:87–97. doi: 10.1016/0042-6822(81)90010-6. [DOI] [PubMed] [Google Scholar]
- 21.Lu Y, Wambach M, Katze M G, Krug R M. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 22.Hatada E, Fukuda R. J Gen Virol. 1992;73:3325–3329. doi: 10.1099/0022-1317-73-12-3325. [DOI] [PubMed] [Google Scholar]
- 23.Talon J, Horvath C M, Polley R, Basler C F, Muster T, Palese P, Garcia-Sastre A. J Virol. 2000;74:7989–7996. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harcourt B H, Sanchez A, Offermann M K. Virology. 1998;252:179–188. doi: 10.1006/viro.1998.9446. [DOI] [PubMed] [Google Scholar]
- 25.Harcourt B H, Sanchez A, Offermann M K. J Virol. 1999;73:3491–3496. doi: 10.1128/jvi.73.4.3491-3496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niwa H, Yamamura K, Miyazaki J. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 27.Volchkov V E, Becker S, Volchkova V A, Ternovoj V A, Kotov A N, Netesov S V, Klenk H D. Virology. 1995;214:421–430. doi: 10.1006/viro.1995.0052. [DOI] [PubMed] [Google Scholar]
- 28.Volchkov V E, Volchkova V A, Chepurnov A A, Blinov V M, Dolnik O, Netesov S V, Feldmann H. J Gen Virol. 1999;80:355–362. doi: 10.1099/0022-1317-80-2-355. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 30.Bluyssen H A, Vlietstra R J, van der Made A, Trapman J. Eur J Biochem. 1994;220:395–402. doi: 10.1111/j.1432-1033.1994.tb18636.x. [DOI] [PubMed] [Google Scholar]
- 31.Percy N, Barclay W S, Garcia-Sastre A, Palese P. J Virol. 1994;68:4486–4492. doi: 10.1128/jvi.68.7.4486-4492.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marie I, Durbin J E, Levy D E. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muhlberger E, Weik M, Volchkov V E, Klenk H D, Becker S. J Virol. 1999;73:2333–2342. doi: 10.1128/jvi.73.3.2333-2342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becker S, Rinne C, Hofsass U, Klenk H D, Muhlberger E. Virology. 1998;249:406–417. doi: 10.1006/viro.1998.9328. [DOI] [PubMed] [Google Scholar]
- 35.Tan S L, Katze M G. J Interferon Cytokine Res. 1998;18:757–766. doi: 10.1089/jir.1998.18.757. [DOI] [PubMed] [Google Scholar]
- 36.Hatada E, Saito S, Fukuda R. J Virol. 1999;73:2425–2433. doi: 10.1128/jvi.73.3.2425-2433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang H W, Watson J C, Jacobs B L. Proc Natl Acad Sci USA. 1992;89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davies M V, Chang H W, Jacobs B L, Kaufman R J. J Virol. 1993;67:1688–1692. doi: 10.1128/jvi.67.3.1688-1692.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharp T V, Moonan F, Romashko A, Joshi B, Barber G N, Jagus R. Virology. 1998;250:302–315. doi: 10.1006/viro.1998.9365. [DOI] [PubMed] [Google Scholar]
- 40.Shors T, Kibler K V, Perkins K B, Seidler-Wulff R, Banaszak M P, Jacobs B L. Virology. 1997;239:269–276. doi: 10.1006/viro.1997.8881. [DOI] [PubMed] [Google Scholar]
- 41.Romano P R, Zhang F, Tan S L, Garcia-Barrio M T, Katze M G, Dever T E, Hinnebusch A G. Mol Cell Biol. 1998;18:7304–7316. doi: 10.1128/mcb.18.12.7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beattie E, Denzler K L, Tartaglia J, Perkus M E, Paoletti E, Jacobs B L. J Virol. 1995;69:499–505. doi: 10.1128/jvi.69.1.499-505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chou J, Kern E R, Whitley R J, Roizman B. Science. 1990;250:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 44.Markovitz N S, Baunoch D, Roizman B. J Virol. 1997;71:5560–5569. doi: 10.1128/jvi.71.7.5560-5569.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leib D A, Harrison T E, Laslo K M, Machalek M A, Moorman N J, Virgin H W. J Exp Med. 1999;189:663–672. doi: 10.1084/jem.189.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leib D A, Machalek M A, Williams B R, Silverman R H, Virgin H W. Proc Natl Acad Sci USA. 2000;97:6097–6101. doi: 10.1073/pnas.100415697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Becker S, Muhlberger E. Curr Top Microbiol Immunol. 1999;235:23–34. doi: 10.1007/978-3-642-59949-1_2. [DOI] [PubMed] [Google Scholar]
- 48.Lamb R A, Kolakofsky D. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott; 1996. pp. 1177–1204. [Google Scholar]
- 49.Sanchez A, Trappier S G, Mahy B W, Peters C J, Nichol S T. Proc Natl Acad Sci USA. 1996;93:3602–3607. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baxter A G. Lancet. 2000;355:2178–2179. doi: 10.1016/S0140-6736(00)02394-1. [DOI] [PubMed] [Google Scholar]
- 51.Schlender J, Bossert B, Buchholz U, Conzelmann K-K. J Virol. 2000;74:8234–8242. doi: 10.1128/jvi.74.18.8234-8242.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, X., Li, M., Zheng, H., Muster, T., Palese, P. & García-Sastre, A. (2000) J. Virol., in press. [DOI] [PMC free article] [PubMed]