Abstract
Despite its central role in virus infection, little is known about the mechanisms of intracellular trafficking of virus components within infected cells. In this study, we followed the dynamics of tobacco mosaic virus movement protein (MP) distribution in living protoplasts after disruption of microtubules (MTs) by cold treatment and subsequent rewarming to 29°C. At early stages of infection, cold treatment (4°C) caused the accumulation of MP fused to green fluorescent protein (GFP) in large virus replication bodies that localized in perinuclear positions, whereas at midstages of infection, the association of MP:GFP with MTs was disrupted. Rewarming the protoplasts to 29°C reestablished the association of MTs with the replication bodies that subsequently spread throughout the cytoplasm and to the periphery of the cell. The role of MTs in the intracellular distribution of the MP also was analyzed by examining the distribution pattern of a nonfunctional mutant of MP (TAD5). Like MP:GFP, TAD5:GFP interacted with the endoplasmic reticulum membranes and colocalized with its viral RNA but did not colocalize with MTs. The involvement of MTs in the intracellular distribution of tobacco mosaic virus MP is discussed.
Successful plant virus infection requires introduction and replication in a single cell followed by local and systemic spread in the host. Genetic, biochemical, and cellular studies have shown that interactions between viral and host components determine the capability of viruses to infect the plant (1–3). However, despite extensive studies, little is known about these interactions or their roles in the intra- and intercellular distribution of viruses.
Cell-to-cell virus movement is an active process that requires specific virus-encoded proteins referred to as movement proteins (MPs) (4). Tobacco mosaic virus (TMV) encodes a 30-kDa MP (5, 6) that is required for local spread of viral RNA (vRNA) through transiently modified plasmodesmata (7). Recently, two groups identified a cell wall-associated enzyme, pectin methylesterase, that interacts in vitro with the MP (8, 9). The role of this enzyme in virus movement still needs to be determined.
Fluorescence microscopy of BY-2 tobacco cells infected with TMV that encodes MP:green fluorescent protein (GFP) fusion protein and in situ hybridization showed that the MP (10, 11) and vRNA (12) colocalized in endoplasmic reticulum (ER)-derived, irregularly shaped structures. These structures also contained viral replicase and are apparently sites of virus replication. Deletions of three amino acids at regular intervals throughout the MP sequence allowed the identification of important functional domains that are involved in the subcellular accumulation of the MP (13). Previous studies also have shown that a polyubiquitinated TMV MP is degraded by the 26S proteasome (14).
Colocalization of the MP with microtubules (MTs) (15) and microfilaments (16) led to the suggestion that interactions between the MP and elements of the cytoskeleton are involved in the cell-to-cell movement of complexes that contain vRNA and MP (17, 18). This hypothesis was supported by recent studies showing the colocalization of TMV RNA with MTs (12). These studies also showed the dramatic changes in distribution of vRNA after disruption of the cytoskeleton with pharmacological agents.
MTs are highly dynamic cytoskeletal components (19) that play an important role in a wide variety of biological activities including cell shape and polarity, cell division, and cellular movement (20). These cellular functions appear to be regulated by the ratio between polymerized and depolymerized forms of tubulin (21). In recent years, the use of different drugs to suppress MT dynamics has made it possible to examine how MTs regulate a wide spectrum of cellular activities (21).
In this study, we followed the dynamics of MP:GFP accumulation in single, living protoplasts after disrupting MTs by cold treatment and subsequent rewarming to 29°C. The role of MTs in intracellular distribution of the MP also was examined in protoplasts infected with a mutant of TMV encoding TADS, a nonfunctional mutant of MP that does not coalign with MTs. Immunostaining with anti-ER luminal-binding protein (BiP) antibody and in situ hybridization showed that TAD5 colocalized with ER and its vRNA. These experiments give insights regarding the central role of MTs in the intracellular distribution of TMV MP and RNA.
Materials and Methods
Plasmids.
The plasmid pTMV-M:GfusBr (7) was used to generate infectious transcripts of TMV that encoded the MP fused to S65T GFP (22); infection produces MP:GFP fusion protein. TMV mutant TAD5 contains a deletion of 3 aa in the MP in positions 49–51 and change of G52E (13). The TAD5 ORF was fused to GFP (producing TAD5:GFP) as described (13). The clone pTR447 containing a fragment of the replicase gene was described (12) and used to synthesize the nonradioactive (digoxigenin or fluorescein)-labeled RNA probes used in Northern and in situ hybridization experiments, respectively.
Transfection, Agarose-Embedment, and Cold Treatment of Protoplasts.
Protoplasts were isolated from tobacco cell line BY-2 and transfected as described (23). Approximately one million protoplasts were inoculated by electroporation with 0.2 μg of infectious RNAs. Living protoplasts were embedded in agarose as described (24). Cold treatment experiments were performed by chilling the agarose-embedded protoplasts in an ice bath. In recovery experiments, cold-treated protoplasts were rewarmed to 29°C for different periods of time before experimental analysis.
RNA Extraction and Northern Hybridization.
Protoplasts were collected by gentle centrifugation at 8, 16, and 24 h postinfection (hpi) and total RNA was isolated from 5 × 104 protoplasts by using TRIzol reagent according to the manufacturer's recommendations (GIBCO/BRL). Accumulation of plus-strand vRNA was analyzed by using glyoxal-denatured RNAs processed as described (25). RNAs extracted from equal numbers of protoplasts were loaded onto the gel and electrophoresed in 20 mM Hepes, pH 7.00/1 mM EDTA. Nucleic acids were transferred to nitrocellulose membranes according to standard procedures (26). Before hybridization, the membranes were stained with methylene blue (26). Prehybridization, hybridization, and colorimetric detection were performed as described (27). Single-stranded digoxigenin-RNA probes that recognized the plus-strand TMV RNA were obtained by in vitro transcription of pTR447 previously linearized with XbaI and using T7 RNA polymerase.
Immunofluorescent Labeling.
Protoplasts were fixed and spun onto polylysine-coated slides (Sigma) (13) and immunostained as described (12). To detect α-tubulin, the samples were incubated for 2 h at room temperature with a 1:100 dilution of a monoclonal mouse anti-α-tubulin antibody (Amersham Pharmacia). The rabbit antibody against the ER luminal protein BiP (R. S. Boston, North Carolina State University, Raleigh) was used at a 1:100 dilution. Samples were incubated for 2 h at room temperature with a 1:100 dilution of the secondary antibody, tetramethylrhodamine B isothiocyanate (TRITC)-conjugated goat anti-mouse IgG (Jackson ImmunoResearch), or TRITC-conjugated goat anti-rabbit IgG (Pierce). The samples were mounted in Mowiol containing 2.5% 1,4-diazobicyclo-[2.2.2.]-octane.
In Situ Hybridization.
In situ hybridization was performed as described (12). The samples were hybridized with 10 ng/μl fluorescein-RNA probe that was obtained by in vitro transcription of pTR447 by using the T7 RNA polymerase. The probes were labeled with fluorescein-12-UTP (Boehringer Mannheim). Because of bleaching of GFP fluorescence during the in situ hybridization procedure, the examination of both the MP and vRNA in the same cell was performed as described (12). Basically, cells showing the characteristic fluorescence pattern of TAD5:GFP accumulation were scanned by using the 488-nm argon laser of the confocal microscope and the samples then were processed for in situ hybridization. Before the hybridization reaction, the cells were imaged to verify the absence of GFP fluorescence.
Confocal Microscopy.
Confocal imaging was performed essentially as described (24) by using an Olympus Confocal Laser Scanning Microscope (IX70). Projections of serial optical sections were obtained by using the software provided by the manufacturer. Optical planes were scanned with the 488-nm argon laser by using a 550-nm (FVX-BA550RIF) barrier filter to detect GFP fluorescence. The TRITC signal was examined with the 568-nm krypton laser (FVX-BA585IF). In experiments of dual imaging (with GFP and TRITC signals), both fluorophores were scanned independently or after attenuating the intensity of the 488-nm laser line by 25% to exclude the possibility of crosstalk between the channels. Single detection controls verified the absence of fluorescence crosstalk.
Results
Effects of Cold Treatment on MP:GFP Distribution.
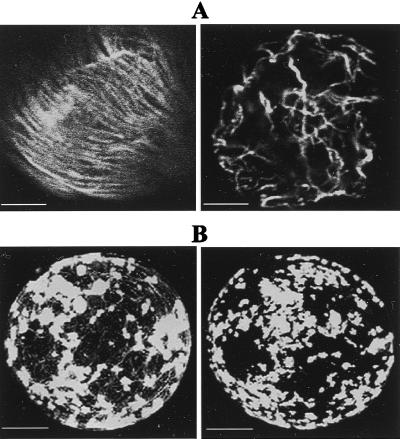
Previous studies in agarose-embedded protoplasts revealed the dynamics of MP:GFP distribution in single, living protoplasts (24). In those studies, we showed that MP:GFP accumulated in perinuclear, irregularly shaped bodies that over time, were redistributed throughout the cytoplasm and at the periphery of the cell. The irregular bodies were identified as sites of virus replication because they contain replicase, vRNA, and MP (10–12). In the present study, agarose-embedded, infected protoplasts were exposed to low-temperature treatments to analyze the effects of cold treatment on MP distribution. Low-temperature treatments of noninoculated protoplasts clearly disrupted MT organization as observed after immunostaining of fixed cells with antitubulin antibody [compare MTs in cells incubated at 29°C (Fig. 1A Left) or 4°C (Fig. 1A Right)].
Figure 1.
Effects of cold treatment on MT organization in BY-2 protoplasts. (A) Noninoculated protoplasts processed for immunofluorescence by using antitubulin antibody and a secondary antibody labeled with TRITC. (Left) Cytoplasmic filaments of tubulin in cells incubated at 29°C. (Right) Disruption of MT organization in cells maintained at 4°C for 3 h. (Scale bar: 2.5 μm.) (B) Protoplasts infected with TMV MP:GFP. At 18 hpi, MP:GFP is accumulated in replication bodies associated with MTs. (Left) A single protoplast maintained at 29°C for 18 h. (Right) Same protoplast after treatment at 4°C for 2 h. Note the absence of fluorescent filaments after cold treatment. (Scale bar: 5 μm.)
Living protoplasts infected with TMV MP:GFP and maintained at 29°C showed, at midstage of infection (18 hpi), accumulation of MP:GFP in small and large replication bodies that were associated with fluorescent filaments (MTs) (Fig. 1B and ref. 24). Treatment of these protoplasts at 4°C for 2 h was sufficient to disrupt MT organization and abolished its association with MP:GFP (Fig. 1B Right).
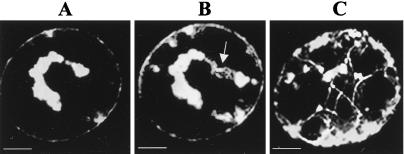
At an earlier stage of infection (12 hpi), treatment of protoplasts at 4°C for 6 h caused the virus replication bodies to localize to the region around the nucleus (Fig. 2A). In contrast, in nontreated protoplasts, MP:GFP was localized with MTs and in virus replication sites dispersed throughout the cytoplasm and at the periphery of the cell (Fig. 1B Left). When the protoplasts were rewarmed to 29°C for 1 h, MP:GFP localized with the new strands of polymerized MTs that associated with the replication bodies (arrow in Fig. 2B). Note that some of the bodies associated with MTs are localized near the periphery of the cell (Fig. 2B). After 3 h at 29°C, MP:GFP was closely associated with MTs and most of the fluorescent bodies were distributed throughout the cell. In contrast, treatment of protoplasts during late stages of infection (i.e., 30 hpi) had no apparent effect on distribution of MP:GFP (not shown).
Figure 2.
Role of MTs in MP:GFP distribution. Accumulation of MP:GFP in a single-infected protoplast examined after cold treatment at 4°C for 6 h (A) and subsequent rewarming to 29°C for 1 h (B) and 3 h (C). Note that cold treatment induced the localization of replication bodies around the nucleus (A). After rewarming the protoplasts, MP:GFP is closely associated with MTs (B and C). Most of the fluorescent bodies were localized at the periphery of the cell (C). (Scale bar: 2.5 μm.)
Based on these results, we conclude that the absence of MTs changed, either directly or indirectly, the subcellular distribution of the sites of virus replication and accumulation during early and midstages of TMV infection.
TAD5:GFP Remains Associated with Perinuclear Replication Bodies Throughout Infection.
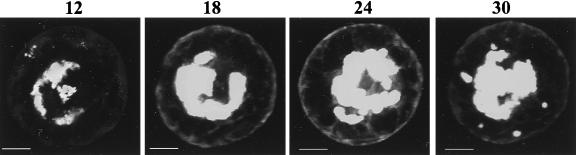
The results described above suggest but do not definitively demonstrate that MTs are involved in distribution of MP:GFP during TMV infection. To verify the relationship between MP:GFP and MTs, we investigated the distribution pattern of a mutant of the MP, TAD5 from which amino acids 49–51 were deleted (13). It was reported (13) that TAD5:GFP does not colocalize with MTs. We examined single-living protoplasts at different stages after infection with TMV that produced TAD5:GFP. At 12 hpi, TAD5:GFP was observed in large bodies near the nucleus (Fig. 3, 12 hpi). At 18–24 hpi, the bodies were exclusively localized around the nucleus (Fig. 3, 18 hpi) and grew or coalesced to form larger structures by 24 and 30 hpi (Fig. 3, 24 and 30 hpi). This pattern was very similar to that observed in cold-treated protoplasts infected with TMV that produced MP:GFP (compare with Fig. 2A).
Figure 3.
TAD5:GFP remains associated with perinuclear replication bodies throughout infection. Accumulation of TAD5:GFP in a single protoplast at 12, 18, 24, and 30 hpi. The images were obtained after overlaying multiple optical confocal images. TAD5:GFP accumulated in replication bodies that localized around the nucleus throughout infection. (Scale bar: 2.5 μm.)
The results indicated that the TAD5 mutation dramatically affects the subcellular distribution of TAD5:GFP, preventing its association with MTs and resulting in accumulation of the virus replication bodies around the nucleus.
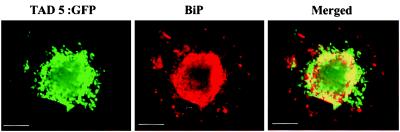
To determine whether TAD5:GFP colocalized with ER as does MP:GFP (12), protoplasts infected with TMV TAD5:GFP were stained at early stages of infection with anti-BiP antibody (BiP is a luminal ER protein; ref. 28) followed by a secondary antibody labeled with TRITC. The samples were independently scanned to minimize crosstalk between the two channels. Representative perinuclear accumulation of both TAD5:GFP (green) and BiP (red) are shown in Fig. 4. Merging the images revealed that most of TAD5:GFP colocalized with ER (yellow in the merged image).
Figure 4.
TAD5:GFP colocalizes with BiP (ER marker). BY-2 protoplasts infected with TAD5:GFP were fixed and processed for immunofluorescence by using anti-BiP antibody followed by a secondary antibody labeled with TRITC. Most of the BiP (red) was associated with sites that contain TAD5:GFP (green). Merging the two images demonstrates colocalization of the signals (yellow in merged image). (Scale bar: 2.5 μm.)
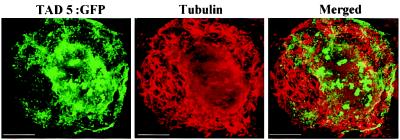
TAD5:GFP Does Not Colocalize with MTs.
A previous report showed that at midstages of infection, TAD5:GFP does not coalign with MTs (13). In our study, at early stages of infection, 20% of the infected protoplasts showed localization of TAD5:GFP in virus replication sites as well as in short strands of fluorescent filaments distributed throughout the cytoplasm. To clarify if TAD5:GFP colocalized with MTs as does MP:GFP (15), protoplasts infected with TAD5:GFP were immunostained with a mAb to α-tubulin and MTs were visualized with a TRITC-conjugated secondary antibody. A representative optical section of an infected protoplast showing accumulation in perinuclear small replication bodies and short cytoplasmic strands of fluorescent filaments is shown in Fig. 5. Immunofluorescent detection of tubulin clearly revealed the distribution of MTs throughout the cytoplasm (Fig. 5). After superimposing both images, we concluded that the TAD5:GFP signal did not significantly coalign with MTs although we cannot exclude the possibility that a minor amount of coalignment can occur.
Figure 5.
TAD5:GFP does not colocalize with MTs. Protoplasts infected with TAD5:GFP were fixed and processed for immunofluorescence by using antitubulin antibody followed by a secondary antibody labeled with TRITC. The cytoplasmic filaments of tubulin (red) did not colocalize with TAD5:GFP (green). Merging the images demonstrates absence of colocalization between the signals (merged image). (Scale bar: 2.5 μm.)
TAD5:MP Colocalizes with Its vRNA.
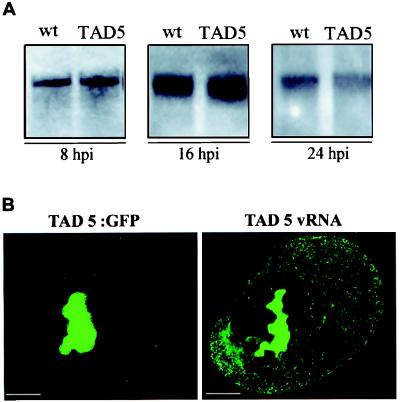
Previous studies showed that the loss of function of TAD5 mutant was not caused by instability of the MP (13). To examine possible differences in replication of vRNA as a result of the TAD5 mutation, equal numbers of protoplasts were extracted at different times after infection with TMV TAD5 or with wild-type TMV. RNAs were analyzed by Northern blot hybridization by using a probe labeled with digoxigenin. The membranes were stained with methylene blue before hybridization to confirm equal loading of samples (not shown). Time-course analysis revealed a clear, detectable hybridization signal at 8 hpi in both infections (Fig. 6A, 8 hpi). The amount of vRNA increased by 16 hpi (Fig. 6A, 16 hpi) and considerably decreased at 24 hpi (Fig. 6A, 24 hpi). These results were in agreement with those described (12) for wild-type TMV infection and indicated that the TAD5 mutation in the MP gene did not result in an increase or decrease in virus replication.
Figure 6.
(A) Accumulation of genome-length TMV plus-strand RNA in tobacco protoplasts inoculated with wild-type TMV MP:GFP or TAD5:GFP. Total RNA was extracted at 8, 16, and 24 hpi, glyoxylated, and subjected by electrophoresis on a 1% denaturing agarose gel. Northern blots were probed with a digoxigenin-labeled RNA probe that recognized the plus-strand of TMV RNA. (B) vRNA colocalizes with TAD5:GFP. Protoplasts infected with TAD5:GFP virus were fixed at midstages of infection, and the pattern of TAD5:GFP accumulation was observed by confocal microscopy. The samples were treated to eliminate fluorescence by GFP and hybridized with the fluorescein-RNA probe to detect TAD5 vRNA. A cell showing the TAD5:GFP distribution was identified by its position and shape and visualized to detect the products of hybridization (TAD5 vRNA). Comparison of both images reflects colocalization of TAD5:GFP and vRNA. (Scale bar: 2.5 μm.)
To determine whether RNA of TAD5:GFP colocalized with TAD5:MP as does MP:GFP (12), protoplasts infected with TAD5:GFP were fixed and processed for in situ hybridization. As shown in Fig. 6B, TAD5:GFP and vRNA colocalized in the large replication bodies in the region around the nucleus. The vRNA accumulated in small fluorescent cytoplasmic strands that apparently lacked TAD5:GFP (Fig. 6B, TAD5 vRNA).
The results presented here indicate that in BY-2 protoplasts, TAD5 MP colocalizes with vRNA in perinuclear bodies; it was demonstrated (12) that MP:GFP also colocalized with vRNA in infected protoplasts.
Discussion
Previous studies have shown that TMV MP (15, 16) and vRNA (12) colocalized with MTs and cytoplasmic and perinuclear ER. It was hypothesized that TMV uses the motility functions of MTs for transport of virus replication sites throughout the cytosol. However, no direct previous evidence has supported the hypothesis. In the present study, we followed the dynamics of MP:GFP distribution in living protoplasts immobilized in agarose in combination with low-temperature treatments to examine the role of MTs in the intracellular distribution of MP:GFP. It is well known that MTs are disrupted by low temperature (Fig. 1; ref. 29). When protoplasts infected with TMV MP:GFP were maintained at 4°C, MTs were disrupted as was the association with MP:GFP. When protoplasts were treated at early stages of infection, the replication bodies remained localized around the nucleus. After rewarming of the protoplasts, the MTs were reformed and some were attached to, or were or in close proximity to, the replication bodies. Concurrently, the replication bodies were relocated throughout the cytoplasm and near to the periphery of the cell (Fig. 2).
Several types of evidence corroborated the importance of MTs in TMV infection. First, immunostaining with antitubulin antibodies concurrent with in situ hybridization experiments revealed that vRNA is coaligned with MTs. Second, treatment with oryzalin, a plant MT-depolymerizing agent, prevented the dispersion of vRNA to the periphery of the cell (12) and the replication bodies coalesced to form larger bodies (10). Third, a recent study suggests that the increased efficiency of intercellular movement of TMV RNA at elevated temperatures is correlated with an increased association of MP with MTs (30).
We used the TAD5 mutant of MP to study the role of interactions with MTs in the spread of the virus to peripheral positions of the cells. Infection with TMV that produced TAD5 protein is unable to cause local or systemic infection of tobacco plants (13). Here we show that TAD5:GFP did not colocalize with MTs (Fig. 5) and accumulated in replication bodies that remained near the nucleus throughout infection (Fig. 3). Interestingly, the distribution of TAD5:GFP was similar to that of MP:GFP in infected protoplasts after cold treatment (Fig. 2A). These results indicated that the absence of association of MP with MTs results in the perinuclear accumulation of the replication bodies and suggest a role of MTs in the intracellular distribution of MP to peripheral sites within the cell.
The role of the cellular cytoskeleton in protein/RNA trafficking has been described in a variety of biological systems (31) including infection by some viruses. It is known that certain animal viruses exploit the cytoskeleton to successfully enter the cell (29), replicate (32), and spread within the cell (33). In the case of local, cell-cell spread of plant virus infection, protein–RNA complexes spread from sites of replication to the plasmodesmata, through which they pass to initiate virus infection in neighboring cells (34). Interactions between MPs produced by several types of plant viruses and the cytoskeleton have been reported (3, 35). However, the precise mechanisms by which virus proteins or virions exploit the cytoskeleton remain to be determined.
Host membranes play a key role in virus infection (36–39). In the case of TMV, previous studies showed that MP:GFP (10, 11) and TMV RNA (12) are colocalized with ER membranes. In the current study, we show that TAD5:GFP and its vRNA colocalized with ER (Figs. 4 and 6B). These results are in agreement with a previous report (12) suggesting that during its synthesis, MP remains associated with vRNA, acting as an anchoring protein and trapping vRNA on ER-derived structures or replication bodies. The differences in distribution of TAD5:GFP compared with MP:GFP were apparently not caused by the inability of TAD5:GFP to interact with ER membranes (Fig. 4), to differences in protein stability (13), or subcellular colocalization with vRNA (Fig. 6). We cannot definitively exclude the possibility that other processes such as ubiquitination (14) or phosphorylation (40) of the MP are affected by infection with TAD5:GFP. If this is the case, these processes must be also altered by cold treatment because MP:GFP distribution after cold treatment was similar to that observed for TAD5:GFP infection.
We conclude that the primary difference between wild-type virus, which is capable of causing local and systemic infection, and virus containing TAD5, which does not, resides in the lack of interaction between TAD5 MP and MTs. Our study indicates that TAD5 MP remains associated with vRNA in the replication bodies and its inability to associate with MTs impedes the spread of the virus to peripheral positions in the cell. The study also supports the conclusion that association of MP with MTs is essential for intracellular spread of TMV.
Acknowledgments
We thank Dr. C. Reichel (GPC-AG, Genome Pharmaceuticals Corporation, Munich, Germany) for critical reading of the manuscript; the Olympus Company; Drs. S. Kay and S. Halpain (Department of Cell Biology, Scripps Research Institute) for use of the confocal laser scanning microscope; and Dr. Rebecca Boston (North Carolina State University, Raleigh) for the BiP antibody. This research was supported by National Science Foundation Grant MCB 9631124 and the Scripps Family Chair. P.M. was supported by a fellowship from the Ministerio de Educación y Cultura, Spain.
Abbreviations
- TMV
tobacco mosaic virus
- MP
movement protein
- MT
microtubule
- vRNA
viral RNA
- GFP
green fluorescent protein
- TAD5
nonfunctional mutation of MP
- ER
endoplasmic reticulum
- BiP
binding protein
- hpi
h postinfection
- TRITC
tetramethylrhodamine B isothiocyanate
References
- 1.Buck K W. Adv Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lazarowitz S G, Beachy R N. Plant Cell. 1999;11:535–548. doi: 10.1105/tpc.11.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reichel C, Más P, Beachy R N. Trends Plant Sci. 1999;4:458–462. doi: 10.1016/s1360-1385(99)01490-9. [DOI] [PubMed] [Google Scholar]
- 4.Atabekov J G, Taliansky M E. Adv Virus Res. 1990;38:201–248. doi: 10.1016/s0065-3527(08)60863-5. [DOI] [PubMed] [Google Scholar]
- 5.Deom C M, Oliver M J, Beachy R N. Science. 1987;237:389–394. doi: 10.1126/science.237.4813.389. [DOI] [PubMed] [Google Scholar]
- 6.Meshi T, Watanabe Y, Saito T, Sugimoto A, Maeda T, Okada Y. EMBO J. 1987;6:2557–2563. doi: 10.1002/j.1460-2075.1987.tb02544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oparka K J, Prior D A M, Santa Cruz S, Padgett H S, Beachy R N. Plant J. 1997;12:781–789. doi: 10.1046/j.1365-313x.1997.12040781.x. [DOI] [PubMed] [Google Scholar]
- 8.Dorokhov Y L, Mäkinen K, Frolova O Y, Merits A, Saarinen J, Kalkkinen N, Atabekov J G, Saarma M. FEBS Lett. 1999;461:223–228. doi: 10.1016/s0014-5793(99)01447-7. [DOI] [PubMed] [Google Scholar]
- 9.Chen M-H, Sheng J, Hind G, Handa A K, Citovsky V. EMBO J. 2000;19:913–920. doi: 10.1093/emboj/19.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinlein M, Padgett H S, Gens J S, Pickard B G, Casper S J, Epel B L, Beachy R N. Plant Cell. 1998;10:1107–1120. doi: 10.1105/tpc.10.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reichel C, Beachy R N. Proc Natl Acad Sci USA. 1998;95:11169–11174. doi: 10.1073/pnas.95.19.11169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Más P, Beachy R N. J Cell Biol. 1999;147:945–958. doi: 10.1083/jcb.147.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kahn T W, Lapidot M, Heinlein M, Reichel C, Cooper B, Gafny R, Beachy R N. Plant J. 1998;15:15–25. doi: 10.1046/j.1365-313x.1998.00172.x. [DOI] [PubMed] [Google Scholar]
- 14.Reichel C, Beachy R N. J Virol. 2000;74:3330–3337. doi: 10.1128/jvi.74.7.3330-3337.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinlein M, Epel B L, Padgett H S, Beachy R N. Science. 1995;270:1983–1985. doi: 10.1126/science.270.5244.1983. [DOI] [PubMed] [Google Scholar]
- 16.McLean B G, Zupan J, Zambryski P. Plant Cell. 1995;7:2101–2114. doi: 10.1105/tpc.7.12.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zambryski P. Science. 1995;270:1943–1944. doi: 10.1126/science.270.5244.1943. [DOI] [PubMed] [Google Scholar]
- 18.Carrington J C, Kasschau K D, Mahajan S K, Schaad M C. Plant Cell. 1996;8:1669–1681. doi: 10.1105/tpc.8.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang D, Wadsworth P, Heplar P K. Proc Natl Acad Sci USA. 1990;87:8820–8824. doi: 10.1073/pnas.87.22.8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shibaoka H. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:527–544. [Google Scholar]
- 21.Jordan M A, Wilson L. Methods Enzymol. 1998;298:252–276. doi: 10.1016/s0076-6879(98)98024-7. [DOI] [PubMed] [Google Scholar]
- 22.Heim R, Cubitt A B, Tsien R Y. Nature (London) 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 23.Watanabe Y, Meshi T, Okada Y. FEBS Lett. 1987;219:65–69. [Google Scholar]
- 24.Más P, Beachy R N. Plant J. 1998;15:835–842. doi: 10.1046/j.1365-313X.1998.00253.x. [DOI] [PubMed] [Google Scholar]
- 25.McMaster G K, Carmichel G G. Proc Natl Acad Sci USA. 1977;74:4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T A. Molecular Cloning: A Laboratory Manual. 2nd Ed. New York: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 27.Más P, Pallás V. J Gen Virol. 1996;77:531–540. doi: 10.1099/0022-1317-77-3-531. [DOI] [PubMed] [Google Scholar]
- 28.Boston R, Viitanen P V, Vierling E. Plant Mol Biol. 1996;32:191–222. doi: 10.1007/BF00039383. [DOI] [PubMed] [Google Scholar]
- 29.Kizhatil K, Albritton L M. J Virol. 1997;71:7145–7156. doi: 10.1128/jvi.71.10.7145-7156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyko V, Ferralli J, Heilein M. Plant J. 2000;22:315–325. doi: 10.1046/j.1365-313x.2000.00740.x. [DOI] [PubMed] [Google Scholar]
- 31.Ferrandon D, Elphick L, Nusslein-Volhard C, St. Johnston D. Cell. 1994;79:1221–1232. doi: 10.1016/0092-8674(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 32.Gupta S, De B P, Drazba J A, Banerjee A K. J Virol. 1998;72:2655–2662. doi: 10.1128/jvi.72.4.2655-2662.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sodeik B, Ebersold M W, Helenius A. J Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lucas W J, Gilbertson R L. Annu Rev Phytopathol. 1994;32:387–411. doi: 10.1146/annurev.phyto.43.040204.135939. [DOI] [PubMed] [Google Scholar]
- 35.Lazarowitz S G. Curr Opin Plant Biol. 1999;2:332–338. doi: 10.1016/S1369-5266(99)80058-2. [DOI] [PubMed] [Google Scholar]
- 36.Huang M, Zhang L. Mol Plant-Microbe Interact. 1999;12:680–690. [Google Scholar]
- 37.Schaad M C, Jensen P E, Carrington J C. EMBO J. 1997;16:4049–4059. doi: 10.1093/emboj/16.13.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward B M, Medville R, Lazarowitz S G, Turgeon R. J Virol. 1997;71:3726–3733. doi: 10.1128/jvi.71.5.3726-3733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Más P, Sanchez-Pina M A, Balsalobre J M, Pallás V. Plant Sci (Limerick, Irel) 2000;153:113–124. doi: 10.1016/s0168-9452(99)00253-8. [DOI] [PubMed] [Google Scholar]
- 40.Kawakami S, Padgett H S, Hosokawa D, Okada Y, Beachy R N, Watanabe Y. J Virol. 1999;73:6831–6840. doi: 10.1128/jvi.73.8.6831-6840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]