Abstract
Some methanogenic and acetogenic microorganisms have the catalytic capability to cleave heterolytically the C O bond of methanol. To obtain insight into the elusive enzymatic mechanism of this challenging chemical reaction we have investigated the methanol-activating MtaBC complex from Methanosarcina barkeri composed of the zinc-containing MtaB and the 5-hydroxybenzimidazolylcobamide-carrying MtaC subunits. Here we report the 2.5-Å crystal structure of this complex organized as a (MtaBC)2 heterotetramer. MtaB folds as a TIM barrel and contains a novel zinc-binding motif. Zinc(II) lies at the bottom of a funnel formed at the C-terminal β-barrel end and ligates to two cysteinyl sulfurs (Cys-220 and Cys-269) and one carboxylate oxygen (Glu-164). MtaC is structurally related to the cobalamin-binding domain of methionine synthase. Its corrinoid cofactor at the top of the Rossmann domain reaches deeply into the funnel of MtaB, defining a region between zinc(II) and the corrinoid cobalt that must be the binding site for methanol. The active site geometry supports a SN2 reaction mechanism, in which the C
O bond of methanol. To obtain insight into the elusive enzymatic mechanism of this challenging chemical reaction we have investigated the methanol-activating MtaBC complex from Methanosarcina barkeri composed of the zinc-containing MtaB and the 5-hydroxybenzimidazolylcobamide-carrying MtaC subunits. Here we report the 2.5-Å crystal structure of this complex organized as a (MtaBC)2 heterotetramer. MtaB folds as a TIM barrel and contains a novel zinc-binding motif. Zinc(II) lies at the bottom of a funnel formed at the C-terminal β-barrel end and ligates to two cysteinyl sulfurs (Cys-220 and Cys-269) and one carboxylate oxygen (Glu-164). MtaC is structurally related to the cobalamin-binding domain of methionine synthase. Its corrinoid cofactor at the top of the Rossmann domain reaches deeply into the funnel of MtaB, defining a region between zinc(II) and the corrinoid cobalt that must be the binding site for methanol. The active site geometry supports a SN2 reaction mechanism, in which the C O bond in methanol is activated by the strong electrophile zinc(II) and cleaved because of an attack of the supernucleophile cob(I)amide. The environment of zinc(II) is characterized by an acidic cluster that increases the charge density on the zinc(II), polarizes methanol, and disfavors deprotonation of the methanol hydroxyl group. Implications of the MtaBC structure for the second step of the reaction, in which the methyl group is transferred to coenzyme M, are discussed.
O bond in methanol is activated by the strong electrophile zinc(II) and cleaved because of an attack of the supernucleophile cob(I)amide. The environment of zinc(II) is characterized by an acidic cluster that increases the charge density on the zinc(II), polarizes methanol, and disfavors deprotonation of the methanol hydroxyl group. Implications of the MtaBC structure for the second step of the reaction, in which the methyl group is transferred to coenzyme M, are discussed.
Keywords: conformational change, methanol metabolism, x-ray structure, zinc
Methanol is an abundant C1-compound in nature. Its major source is probably the plant cell wall component pectin from which methanol is released upon hydrolytic degradation. In oxic environments methanol is rapidly metabolized to CO2 by aerobic methylotrophic microorganisms, which thereby prevents autooxidation to the cytotoxic formaldehyde (1). Methanol also does not accumulate in anaerobic environments, where methanogenic archaea (2) and acetogenic bacteria (3) reduce the C1-compound to methane, carbonylate it to acetate, and/or oxidize it to CO2 in their energy metabolism.
Methanol metabolism is initiated in methanogenic archaea by its reaction with coenzyme M (HS-CoM) to form methyl-coenzyme M (CH3-S-CoM) (reaction 1) (4, 5). Methyl-coenzyme M is the central intermediate for reduction to methane and oxidation to CO2.
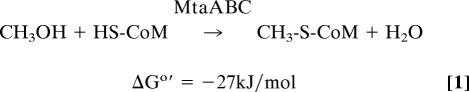 |
Reaction 1 is catalyzed by the methanol:coenzyme M methyltransferase MtaABC, which is composed of subunits MtaA, MtaB, and MtaC. MtaA and MtaB are zinc proteins, and MtaC is a corrinoid protein. MtaB catalyzes the methylation of the MtaC-bound cob(I)amide (reaction 2), and MtaA catalyzes the transfer of the methyl group from the MtaC-bound methyl-cob(III)amide to coenzyme M (reaction 3). In the two reactions MtaC can be substituted by free cob(I)alamin and free methyl-cob(III)alamin, respectively (4, 6–10).
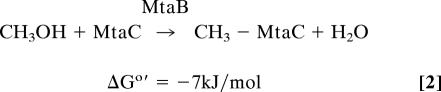 |
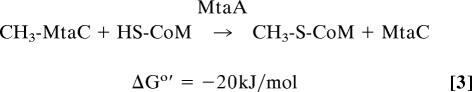 |
The two reaction steps are highly interdependent as MtaA and MtaB cooperatively interact with each other, documented by a decrease of the apparent KM of MtaB for free cob(I)alamin from >0.2 mM to 2 μM in the presence of MtaA (9). Because of the negative redox potential Eo′ = −600 mV of the cob(II)amide/cob(I)amide couple (11), cob(I)amides are even slowly oxidized by protons of water (2H+/H2; Eo′ = −414 mV) and have to be continuously rereduced by ferredoxin in an ATP-dependent enzyme-catalyzed reaction (12).
The present work is primarily focused on the methanol activation (reaction 2) catalyzed by MtaB, which forms a tight complex with MtaC (7). The 50-kDa protein MtaB does not show any sequence similarity to any protein with a studied function. It binds zinc(II) but lacks one of the known zinc-binding motifs characteristic for other zinc-containing alkyltransferases: C-Xn-G-G-C-C (e.g., cobalamin-dependent methionine synthase MetH), H-X-C-Xn-C (e.g., MtaA and cobalamin-independent methionine synthase MetE), C-X3-C-X26-C-X2-C (Ada protein), and D-X-C-X49–62-H (protein farnesyltransferase) (13–19). The corrinoid-binding protein MtaC has a molecular mass of 27 kDa and contains tightly bound 5-hydroxybenzimidazolyl cob(I)amide (factor III) as prosthetic group [vitamin B12 is cyanodimethylbenzimidazolylcob(III)amide]. MtaC shows a sequence similarity to corrinoid proteins involved in methyl transfer reactions with the highest amino acid sequence identity of 35% being to the cobalamin-binding domain of MetH from Escherichia coli. The primary structure of MtaC contains the sequence D-X-H-X2-G-X41-T-X-L-X26-G-G, which is characteristic for corrinoid proteins with the corrinoid bound in a base-off/His-on configuration (20–22). In cob(I)amide cobalt is only tetra-coordinated, in cob(II)amide it is penta-coordinated, and in methyl-cob(III)amide it is hexa-coordinated (23, 24). Therefore, during catalysis the prosthetic group of MtaC cycles through a His-off [cob(I)amide]/His-on [methyl-cob(III)amide] configuration.
The mechanism of methanol activation is not yet elucidated (25) although this question is of general chemical interest, because the hydroxyl group of methanol is a poor leaving group and CH3+ is a very unfavorable carbocation. Activation by protonation of the hydroxyl group would require very high proton concentrations because the pKa of CH3OH2+ is −1.5 (26). Alternatively, activation could be mediated by a Lewis acid, e.g., zinc(II). Methylation of cob(I)alamin in absolute methanol has been reported to depend on ZnCl2 (27).
We report here on the crystal structure of the MtaBC complex of Methanosarcina barkeri and reveal how the corrinoid protein interacts with the zinc-containing methyltransferase such that the corrinoid and zinc(II) can cooperate in the activation of methanol. Crystal structures of several other cobamide-dependent methyltransferases have been reported (28–30). Here we present the crystal structure of a corrinoid protein in complex with one of its catalytic methyltransferases.
Results and Discussion
Oligomeric Structure.
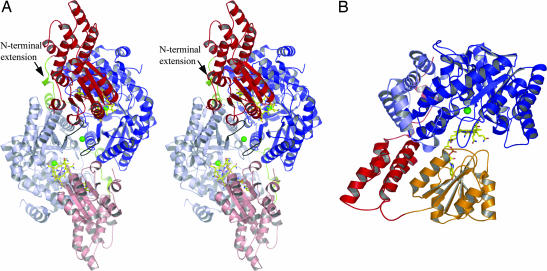
The asymmetric unit of MtaBC contains eight MtaBC units (630 kDa) related by a 422-point symmetry. In solution the enzyme is most likely present as (MtaBC)2 heterotetramers (Fig. 1A), because the buried area between the MtaBC units within the (MtaBC)2 complex is 14% of the entire surface, whereas that between two (MtaBC)2 units is only 3.5%. Native polyacrylamide gradient gel electrophoresis studies resulted in an apparent molecular mass of 130 kDa that is compatible with a (MtaBC)2 structure of 160 kDa. But although the gel electrophoresis data also match an MtaB/MtaC stoichiometry of 2:1 (6) the crystal structure clearly determines the stoichiometry to be 1:1. The interface between two MtaBC units is characterized by two prominent features (Fig. 1A). First, a noncatalytic zinc(II) ion (visible in the anomalous difference electron density map) is located between the two subunits, which was not expected because the zinc content of MtaBC was reported to be ≈1 (7). The zinc(II) ion is ligated in a tetrahedral manner by the side chain ND1 and OE2 atoms of His-316 and Glu-318 protruding from both MtaB subunits. Second, the subunits MtaC possess an extended N-terminal arm ≈50 Å in length and consisting of 25 aa. Each N-terminal extension is embedded into a groove formed at the interface between two MtaB subunits. A comparison with homologous corrinoid proteins corroborates the importance of these interactions for assembling the (MtaBC)2 complex. Corrinoid-carrying subunits from Methanosarcina species involved in methyl transfer to coenzyme M from monomethylamine, dimethylamine, and trimethylamine, respectively (31–33), lack this extension and are not purified in complex with the corresponding methyltransferases (29, 31, 34). In contrast, the corrinoid protein involved in methyl-coenzyme M formation from coenzyme M and dimethylsulfide contains an N-terminal extension and is purified in a tight complex with its methyltransferase (35).
Fig. 1.
The MtaBC structure. (A) In the (MtaBC)2 heterotetramer (in stereo) each MtaBC unit (MtaB in blue and MtaC in red) is related to its partner unit (in light blue and red) by a twofold noncrystallographic axis. The (MtaBC)2 complex has a size of 62 Å × 55 Å × 52 Å and forms two active sites separated by a distance of 38 Å. The corrinoids are shown as stick models, and the zinc(II) ions are depicted by green spheres. The contact loops between the two active sites are highlighted in black. For oligomeric assembly the interactions between the N-terminal extension of MtaC (in green) and the counter MtaB are essential. (B) In the MtaBC unit the Rossmann domain (orange) of MtaC is only loosely associated with its helical domain (red) and MtaB (blue). MtaB is subdivided into a TIM barrel core (dark blue) and a helical layer (light blue). The active site is located between the corrinoid cobalt and zinc(II).
Structure of MtaB and the Binding Mode of the Catalytic Zinc(II).
MtaB is composed of a TIM barrel and an unusual segment of 115 residues consisting of seven α-helices of different lengths that encircle the TIM barrel. In the (MtaBC)2 heterotetramer this helical layer contacts the partner subunit MtaB, the helical domain and the N-terminal arm of MtaC (Fig. 1), and perhaps also MtaA. The TIM barrel fold is reminiscent of the structures of other structurally characterized methyltransferases such as the methyl-H4F-binding domain of MetH (28), the homocysteine-binding domain of MetH (28) and of cobalamin-independent methionine synthase (36), monomethylamine methyltransferase (29), and the methyl-H4F:corrinoid iron–sulfur protein methyltransferase (MeTr) (37). The most related TIM barrel is that of the homocysteine-binding domain of MetH from Thermotoga maritima with an rmsd of 3.2 Å (70% of the Cα-trace used). However, MtaB reveals no sequence similarity with the other TIM barrel-containing methyltransferases and zinc-binding proteins, which have merely the TIM barrel scaffold in common but not the design of the active site and the zinc coordination.
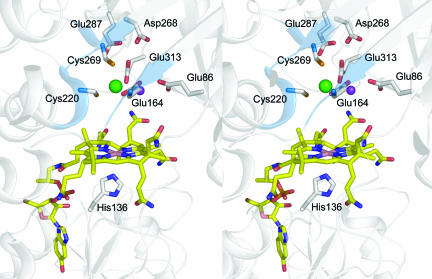
The zinc-binding site of MtaB is located at the C terminus of the TIM barrel in a deep funnel-shaped pocket. The catalytic zinc(II) binds at its bottom and is surrounded in the first coordination sphere by Cys-220, Cys-269, and Glu-164 that protrude from specifically arranged protein segments (Fig. 2). The two sulfurs and the carboxyl oxygen ligands of Cys-220, Cys-269, and Glu-164 are 2.2 Å, 2.3 Å, and 2.2 Å away from the zinc(II) and form an incomplete tetrahedral arrangement. In a second coordination sphere within a distance of 5 Å to the zinc(II) lie Asp-268, Glu-313, Asn-224, and an electron density peak that cannot be definitely assigned. The peak, which is 3.1 Å away from zinc(II), is higher than that expected for an oxygen and is ≈2.1 Å away from the carboxylate oxygens of Glu-86 and Glu-164. Therefore, we interpreted the peak as a metal ion rather than a water and fitted it tentatively as a potassium ion. The residues involved in zinc binding are strictly conserved (see Fig. 5, which is published as supporting information on the PNAS web site) and allow the definition of the MtaB recognition motif E-X2-GGK-X3-D-X55-C-X2-AN-X-A-X40-GA-X-GP-X-KDCGYE.
Fig. 2.
Active site region of the (MtaBC)2 complex. Zinc(II) (in green) is ligated to Glu-164, Cys-220, and Cys-269, forming a novel zinc coordination motif. The surrounding of the zinc(II) is dominated by acidic residues. The putative potassium ion is shown in pink. The cobalt is penta-coordinated with His-136 as axial ligand on the α-face of the 5-hydroxybenzimidazolyl cob(II)amide.
Structure of MtaC and the Binding Mode of the Cobamide.
The structure of MtaC is composed of a cobamide-binding Rossmann domain and a helical domain (Fig. 1B) in accordance with all corrinoid proteins involved in methyl transfer reactions (38). The highest structural similarity is detected with that of MetH from E. coli (2.6 Å rmsd using 58% of the Cα-trace). Whereas the fold of the individual domains is well conserved, the orientation between the two domains drastically differs, indicated by a relative rotation and translation of 51.4° and 28.0 Å, respectively, between the helical domains of MtaC and MetH. An orientation similar to that in MtaC was found in the structure of the complex between the cobalamin- and AdoMet-binding domains of MetH (39).
MtaC (and related corrinoid proteins) binds the prosthetic group at the C terminus of the parallel β-sheet in an exposed fashion (Fig. 1B) where its α-side faces the Rossmann domain and its β-side is exposed (see below). The difference between 5-hydroxybenzimidazolylcobamide of MtaC and dimethylbenzimidazolylcobamide (cobalamin) found in most corrinoid-binding proteins does not influence its binding mode but is unambiguously visible in the electron density map. The cobalt sits in the corrinoid plane and is penta-coordinated rather than hexa-coordinated, indicating that Co(III) was cryoreduced to Co(II) during x-ray exposure of the frozen crystals (40). The axial ligand is His-136 (Fig. 2), in agreement with previous site-directed mutagenesis experiments through which the base-off/His-on configuration of the corrinoid of MtaC was identified (8). His-136 is the major component of the strictly conserved and functionally essential catalytic triad consisting of His-136, Asp-134, and Thr-187 in MtaC. In the cob(III)amide oxidation state the sixth ligand is probably a water molecule.
The cobamide–protein interactions are primarily mediated by the nucleotide base whose conformation and binding pocket are well conserved among related corrinoid proteins. The tight binding of the corrinoid even in the tetra-coordinated cob(I) oxidation state was confirmed for MtaC by biochemical studies (6). Interestingly, the corrinoid rings significantly differ in their conformations because of their loose contact to the Rossmann domain and their distinctive interactions with segments of other subunits approaching the β-face. In the (MtaBC)2 complex the corrinoid interacts with MtaB and, interestingly, also with the partner MtaB in the tetramer. Whereas in MtaB only the carbonyl oxygen of Ala-294 approaches the corrinoid ring closer than 3.5 Å, the partner MtaB forms extended van der Waals interactions between Phe-321 and the substituents of the pyrrole ring A and a hydrogen bond between the acetamide substituent of pyrrole ring D and the carbonyl oxygen of Phe-321. In contrast to MtaC the β-face of the corrinoid of the isolated MetH cobalamin domain is capped by its helical domain.
The Active Site and the Binding Site of Methanol.
The methanol cleavage reaction is localized between MtaB and the Rossmann domain of MtaC. The modules are oriented in such a manner that the cobamide on the top of the Rossmann domain reaches into the funnel of MtaB (Figs. 1 and 2). Therefore, the corrinoid is mainly surrounded by residues of MtaB, which explains why methanol is also cleaved with free cobalamin in the presence of MtaB (9).
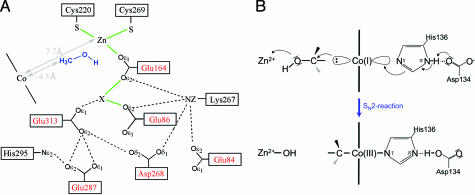
As zinc(II) is embedded at the bottom of the funnel of MtaB, the methanol binding and active sites lie most likely between the two metals that are 7.7 Å apart from each other. Methanol can access its binding site, which is partially occupied by a solvent molecule. We assume that the hydroxyl group of methanol occupies the empty fourth coordination site at zinc(II) and thereby bridges zinc(II) and the putative potassium ion from the front side (Fig. 3A). The hydroxyl group of methanol might additionally interact with the thiolate group of Cys-220 and the carboxylate group of Glu-313, both from MtaB. The methyl group of the modeled methanol points toward the cobalt; however, the distance between the carbon and the cobalt of ≈4.5 Å is too long for a van der Waals contact.
Fig. 3.
Proposed mechanism of methanol activation. (A) Scheme of a unique acidic cluster that flanks the zinc and methanol-binding site. The acidic residues (in red) might play a crucial role in polarizing zinc(II) and methanol. The unclear peak X was tentatively assigned as a potassium ion. Methanol is modeled into the protein, and its distance to the corrinoid Co is estimated based on the distance between the zinc(II) and methanol oxygen of 2 Å. (B) SN2 mechanism for the methylation of 5-hydroxybenzimidazolyl cob(I)amide with methanol. The methanol is activated by the strong electrophile zinc(II) and attacked by the supernucleophile cob(I)amide.
An accurate analysis of the interface indicates that the Rossmann domain of MtaC has only a few contacts with the helical domain and with MtaB of the same MtaBC unit. This isolated position in the crystal lattice is also reflected in the unusually high temperature factors of 74 Å2 in comparison to 22 Å2 of the adjacent MtaB. We conclude that in the (MtaBC)2 structure the active site is present in an open state (Fig. 1B) that could be transformed into a closed state by an ≈1.0-Å shift of the Rossmann domain perpendicular to the corrinoid plane toward MtaB. This shift is accompanied by only a few side chain rearrangements and might rigidify the mobile Rossmann domain. Because the lack of the axial histidine ligand in the Co(I) state might also displace the corrinoid ring further into the funnel we postulate that the reduction of Co(III) and the binding of methanol induce a conformational change from an open to a closed state. In the latter state, the methyl group of methanol and cobalt contact each other and the active site is shielded from bulk solvent. Structural analysis with the corrinoid in the potentially closed cob(I)amide state failed because the crystals were not ordered enough, perhaps because of a partial autooxidation of Co(I) to Co(II) and the concomitant change of the coordination properties.
Methanol Cleavage.
According to biochemical and chemical studies (7, 27) the heterolytic cleavage of the highly inert CH3OH is accomplished by a combined effect of the supernucleophile Co(I) and the strong electrophile zinc(II). The structural analysis confirms this finding and reveals, in addition, the geometric framework of the reaction (Fig. 3B). The zinc(II), the corrinoid cobalt, and the nitrogen τ of His-136 lie on an almost straight line, which is perpendicular to the corrinoid plane. Methanol, when positioned on this line, ligates to zinc(II) via its hydroxyl group and points with its methyl group toward the cobalt. In the closed conformation the methyl group of methanol is nucleophilically attacked by Co(I) supported by the strong Lewis acid zinc(II), which activates the OH group. The linear arrangement is optimally compatible with the proposed SN2 mechanism (Fig. 3B) and does not support an oxidative addition that requires a C O bond to be parallel to the plane of the corrinoid ring. Consistent with the proposed SN2 mechanism is the stereochemistry of the methyl group transfer from methanol to coenzyme M, which proceeds with retention (two times inversion) of stereo-configuration (41).
O bond to be parallel to the plane of the corrinoid ring. Consistent with the proposed SN2 mechanism is the stereochemistry of the methyl group transfer from methanol to coenzyme M, which proceeds with retention (two times inversion) of stereo-configuration (41).
The reaction mechanism proposed in Fig. 3 attributes a key function in Co(I) methylation to His-136 and the other components of the catalytic triad, which has been found in all corrinoid proteins catalyzing methyl transfer reactions except in the corrinoid iron–sulfur protein (42). The importance of the axial ligand His-136 for the (MtaBC)2 complex for the catalytic reaction is underscored by the finding that the methylation rate of free cob(I)inamide (which lacks the imidazole base) with methanol catalyzed by MtaB is completely dependent on the presence of imidazole, whereas the coenzyme M-dependent demethylation catalyzed by MtaA is inhibited in the presence of imidazole (9, 43).
The zinc-binding site in MtaB is unique. Zinc(II) is ligated to two thiolates and one carboxylate oxygen ligands, which constitutes a novel coordination motif (Fig. 2). Its special relevance for methanol activation, however, is so far not understood. We assume that the major catalytic strategies for cleaving methanol are directed to polarize methanol (CH3δ+-OHδ−) and perhaps to prevent deprotonation of the hydroxyl group. This strategy is realized by flanking the zinc (II) and its ligands by negatively charged residues comprising Glu-84, Glu-86, Asp-268, Glu-287 and Glu-313 of MtaB (Fig. 3A). The charged residues generate a highly dielectric environment that supports the polarization of methanol and enhances the charge density of zinc(II) by which its electrophilicity is fine tuned so as to prevent deprotonation of methanol which might happen if the zinc ion would be too electron poor. Protonated methanol might be also stabilized by a hydrogen bond to the adjacent side chain of Glu-313. The function of the electron density peak tentatively interpreted as a potassium ion is obscure. It might have a crucial role in catalysis but it might also disappear after methanol binding and in the cob(I)amide state.
The Overall MtaABC Reaction.
After the methyl group of methanol has been transferred to the cob(I)amide prosthetic group of MtaC it has to be further transferred to coenzyme M via MtaA to complete the catalytic cycle (reactions 1-3). As described, the presented (MtaBC)2 structure is in an open conformation and might represent a state of the methylated cob(III)amide after the methyl transfer from methanol before the MtaA reaction. The absence of the methyl group at the cob(III)amide indicates that the enzyme structure was determined in an inactive form.
To allow a contact between the methyl-cob(III)amide of MtaC and coenzyme M of MtaA, MtaB and the Rossmann domain of MtaC have to be separated and the corrinoid has to be positioned in front of MtaA. This process is illustrated in Fig. 4. The model for MtaA is taken from the structurally related uroporphyrinogen decarboxylase (1URO) and the position of the MtaA model onto the MtaB surface is hypothetical. The observable large-scale domain rearrangement was already reported for MetH where the cobalamin-binding domain shuttles between the tetrahydrofolate and the homocysteine-binding site (28, 44). The data of the MtaABC system are, in principle, in line with the proposed “shake, rattle and roll” model (45) which, however, requires modifications. Accordingly, the predicted large-scale motion is not accomplished by the whole MtaC subunit but only by the corrinoid-containing Rossmann domain, which is highly mobile and only weakly associated with the protein core (Figs. 1 and 4). In contrast, the helical domain and the N-terminal arm, both multiply anchored to the protein, essentially remain at their positions and prevent a dissociation of MtaC from the protein complex during the reaction cycle. Moreover, reorientation of the Rossmann relative to the helical domains is a well known process (39) owing to the flexible linker between them that serve as hinge (Figs. 1 and 4).
Fig. 4.
Proposed movement of the MtaC Rossmann domain (orange) between the actives sites of MtaB (blue), which harbors the methanol-binding site, and MtaA (green), which harbors the coenzyme M-binding site, during catalysis of methyl-coenzyme M formation from methanol and coenzyme M (reaction 1). The N-terminal arm and the helical domain of MtaC (both in red) are fixed and serve as anchor for the swinging Rossmann domain. Because the crystal structure of MtaA has not yet been determined the structurally related TIM barrel protein uroporphyrinogen decarboxylase (57) (>20% amino acid sequence identity) was modeled instead of MtaA to the surface of MtaB. The MtaA model was manually docked in a position onto MtaB that is related to the one found in the structure of MetH (45) such that the corrinoid bound to the Rossmann domain of MtaC can interact with the active site of MtaA after a conformational change involving only the Rossmann domain.
Both structural and kinetic data indicate considerable differences in the docking site between MtaA and MtaB compared with the analogous domains of MetH. In contrast to the tetrahydrofolate-binding domain of MetH, the TIM barrel of MtaB is surrounded by a helical layer and thus prevents an equivalent interface. Additionally, the limited reach of the swinging Rossmann domain and the activation of methanol cleavage in the presence of MtaA (9) makes a shorter distance between the active sites of MtaA and MtaB likely. According to the model of domain rearrangement depicted in Fig. 4 the distance between the active sites is ≈40 Å whereas that in MetH is ≈50 Å.
The two-step process catalyzed by the MtaABC complex raises the question whether the two half reactions proceed independently in two MtaABC units of the dimer or whether they are synchronized. As already described, the (MtaBC)2 heterotetramer appears to be essential for catalysis as the partner MtaB is involved in binding of the N-terminal arm and of the corrinoid of MtaC. Beyond that a coupling of the demethylation and methylation reactions within a cooperative process is conceivable as the two active sites, ≈38 Å apart, are linked via a loop and its symmetry-related partner loop (Fig. 1A). Interestingly, both loops are structurally adjacent and linked by the “interface” zinc(II). Phe-321, a residue of the mentioned loop, is involved in cobalamide binding of the one MtaBC unit and Glu-313 positioned at the end of the strand preceding this loop is involved in zinc(II) and methanol activation of the other MtaBC unit and vice versa. Therefore, methanol binding might pull the region around Glu-313 toward its binding site, which simultaneously impairs the interactions with the corrinoid of the other MtaBC unit and induces the swinging of the Rossmann domain toward MtaA. However, no biochemical data are available to support or reject an allosteric reaction mechanism between two MtaABC units.
Methods
M. barkeri strain Fusaro (DSM 804) was obtained from the Deutsche Sammlung für Mikroorganismen und Zellkulturen (Braunschweig, Germany). The organism was grown on 250 mM methanol (46). The cells were harvested anaerobically in the late exponential phase, and the MtaBC complex was purified from cell extracts as described below.
Purification and Crystallization of MtaBC.
All preparation steps were performed under aerobic conditions and at a temperature of 4°C. Ten grams of methanol-grown cells was resuspended in 50 mM Mops/KOH (pH 7.0) and disrupted via a sonicator. Cell debris was removed by centrifugation at 15,000 × g, and the cell extract was centrifuged at 150,000 × g for 90 min. The supernatant was supplemented with (NH4)2SO4 to a final concentration of 70% and incubated on ice for 30 min. Centrifugation for 20 min at 20,000 × g resulted in an orange-brown pellet that contained MtaBC. The pellet was resuspended in 50 mM Mops/KOH (pH 7.0) containing 2 M (NH4)2SO4 and applied to a phenyl-Sepharose high-performance column that was equilibrated with the same buffer. The column was developed in 80-ml steps with decreasing concentrations of (NH4)2SO4 (2.0, 0.8, 0.6, 0.4, 0.2, and 0.0 M) in 50 mM Mops/KOH (pH 7.0). MtaBC was eluted at an (NH4)2SO4 concentration of 600 mM in a volume of 32 ml, subsequently desalted via ultrafiltration, and concentrated to a volume of 0.5 ml. Finally, the protein was loaded onto a Superdex 200 gel filtration column and eluted with a solution containing 100 mM (NH4)2SO4 in 50 mM Mops/KOH (pH 7.0). The purified MtaBC was desalted and concentrated to 45 mg·ml−1 in 10 mM Mops/KOH.
MtaBC was crystallized from aerobic MtaBC preparations where the cobamide is present in the Co(III) oxidation state. Crystallization attempts were performed with the hanging drop vapor diffusion method at a temperature of 4°C by using 1 μl of the enzyme solution and 1 μl of reservoir solution [Crystal Screen kits of Hampton Research (Aliso Viejo, CA) and Jena Bioscience (Jena, Germany)] Best crystals were obtained with 100 mM sodium acetate (pH 5.6), 50 mM (NH4)2SO4, 17% (wt/vol) polyethylene glycol 8000, and 15% (wt/vol) glycerol. From a large number of well shaped crystals only a few diffracted to ≈2.5-Å resolution. Before x-ray analysis the crystals were placed into a cryoprotectant solution containing 100 mM sodium acetate, 50 mM (NH4)2SO4, 17% polyethylene glycol 8000, and 20% (wt/vol) glycerol and subsequently flash-cooled to 100 K in a nitrogen gas cold stream.
Structure Determination and Refinement.
Native data at 2.5-Å resolution were collected at the ID14-4 beamline (European Synchrotron Radiation Facility, Grenoble, France), and a single anomalous dispersion experiment at the zinc absorption edge was performed at beamline BW6 at the Deutsches Elektronen Synchrotron (Hamburg, Germany) (Table 1). The crystals adopted the space group P21 with lattice parameters of a = 101.8 Å, b = 173.2 Å, c = 190.5 Å, and β = 98.9°. Diffraction data were integrated and scaled by using XDS (47). Several crystallographic calculations were achieved with the CCP4 suite (48). All 12 zinc ions in the asymmetric unit were detected with SHELXD (49). The strong/weak correlation was 17.8/11.4 in a resolution range of 50 to 2.9 Å. The zinc sites were refined and phases were determined by using the program SHARP (50). Phases were improved by solvent flattening assuming a solvent content of 50% and by eightfold molecular averaging within DM (51). Approximately 55% of the model was built automatically by using RESOLVE (52), and the rest of the model was manually incorporated within O (53). Iterative cycles of refinement and manual model building at 2.5-Å resolution were carried out by using the programs CNS (54), REFMAC5 (55), and O. The positional parameters as well as the B values were restrained according to the eightfold noncrystallographic symmetry. Refinement of TLS parameters for the two domains of the eight copies of MtaC decreased the Rfree by 1.4% using REFMAC5 (55). The refinement statistics are given in Table 1. The quality of the model was checked with PROCHECK (56). Figs. 1, 2, and 4 were generated with PYMOL (www.pymol.org).
Table 1.
Data collection and refinement statistics
| 1D14-4 | BW6 | |
|---|---|---|
| High-resolution | Peak | |
| Data collection | ||
| Wavelength, Å | 0.9393 | 1.2824 |
| Resolution, Å | 2.5 | 2.65 |
| Multiplicity | 3.4 | 4.3 |
| Completeness, % | 96.4 | 96.7 |
| Rsym,* % | 8.5 | 18.6 |
| Cell constants, Å | 101.6, 172.5, 189.9 | 101.8, 173.2, 190.5 |
| Cell constants, ° | 98.9 | 98.9 |
| Refinement | ||
| Rcryst,† % | 18.8 | |
| Rfree,‡ % | 22.5 | |
| No. of reflections | 207,482 | |
| No. of atoms | 45,519 | |
| Bond length rms, Å | 0.011 | |
| Bond angle rms, ° | 1.3 |
*Rsym = Σ|Ii − <I>|/ΣIi, where Ii is the observed intensity and <I> is the averaged intensity obtained from multiple observations of symmetry-related reflections.
†Rcryst = Σhkl (|Fobs| − |Fcalc|)/Σhkl|Fobs|.
‡Rcrystwith 5% of the observed reflections selected randomly.
Supplementary Material
Acknowledgments
We thank Gerd Mander and the staff of the BW6 beamline (in particular Gleb Bourenkov) at the Deutsches Elektronen Synchrotron and of the 1D14-4 beamline at the European Synchrotron Radiation Facility for technical assistance and Hartmut Michel for continuous support. This work was supported by the Max-Planck-Gesellschaft and by the Fonds der Chemischen Industrie.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org. (PDB ID code 212X).
References
- 1.Vorholt JA. Arch Microbiol. 2002;178:239–249. doi: 10.1007/s00203-002-0450-2. [DOI] [PubMed] [Google Scholar]
- 2.Ferry JG, editor. Methanogenesis: Ecology, Physiology, Biochemistry & Genetics. New York: Chapman & Hall; 1993. [Google Scholar]
- 3.Drake HL. Acetogenesis. New York: Chapman & Hall; 1994. [Google Scholar]
- 4.Keltjens JT, Vogels GD. In: Methanogenesis: Ecology, Physiology, Biochemistry and Genetics. Ferry JG, editor. New York: Chapman & Hall; 1993. pp. 253–303. [Google Scholar]
- 5.Thauer RK. Microbiology. 1998;144:2377–2406. doi: 10.1099/00221287-144-9-2377. [DOI] [PubMed] [Google Scholar]
- 6.Sauer K, Harms U, Thauer RK. Eur J Biochem. 1997;243:670–677. doi: 10.1111/j.1432-1033.1997.t01-1-00670.x. [DOI] [PubMed] [Google Scholar]
- 7.Sauer K, Thauer RK. Eur J Biochem. 1997;249:280–285. doi: 10.1111/j.1432-1033.1997.t01-1-00280.x. [DOI] [PubMed] [Google Scholar]
- 8.Sauer K, Thauer RK. Eur J Biochem. 1998;253:698–705. doi: 10.1046/j.1432-1327.1998.2530698.x. [DOI] [PubMed] [Google Scholar]
- 9.Sauer K, Thauer RK. Eur J Biochem. 1999;261:674–681. doi: 10.1046/j.1432-1327.1999.00355.x. [DOI] [PubMed] [Google Scholar]
- 10.Sauer K, Thauer RK. In: Chemistry and Biochemistry of B12. Banerjee R, editor. New York: Wiley; 1999. pp. 655–679. [Google Scholar]
- 11.Lexa D, Saveant JM. Acc Chem Res. 1983;16:235–243. [Google Scholar]
- 12.Daas PJH, Hagen WR, Keltjens JT, vanderDrift C, Vogels GD. J Biol Chem. 1996;271:22346–22351. doi: 10.1074/jbc.271.37.22346. [DOI] [PubMed] [Google Scholar]
- 13.Matthews RG, Goulding CW. Curr Opin Chem Biol. 1997;1:332–339. doi: 10.1016/s1367-5931(97)80070-1. [DOI] [PubMed] [Google Scholar]
- 14.Hightower KE, Fierke CA. Curr Opin Chem Biol. 1999;3:176–181. doi: 10.1016/s1367-5931(99)80030-1. [DOI] [PubMed] [Google Scholar]
- 15.Wilker JJ, Lippard SJ. Inorg Chem. 1997;36:969–978. doi: 10.1021/ic961082o. [DOI] [PubMed] [Google Scholar]
- 16.Sauer K, Thauer RK. Eur J Biochem. 2000;267:2498–2504. doi: 10.1046/j.1432-1327.2000.01245.x. [DOI] [PubMed] [Google Scholar]
- 17.Peariso K, Zhou ZHS, Smith AE, Matthews RG, Penner-Hahn JE. Biochemistry. 2001;40:987–993. doi: 10.1021/bi001711c. [DOI] [PubMed] [Google Scholar]
- 18.Gencic S, LeClerc GM, Gorlatova N, Peariso K, Penner-Hahn JE, Grahame DA. Biochemistry. 2001;40:13068–13078. doi: 10.1021/bi0112917. [DOI] [PubMed] [Google Scholar]
- 19.Breksa AP, Garrow TA. Arch Biochem Biophys. 2002;399:73–80. doi: 10.1006/abbi.2001.2751. [DOI] [PubMed] [Google Scholar]
- 20.Ludwig ML, Matthews RG. Annu Rev Biochem. 1997;66:269–313. doi: 10.1146/annurev.biochem.66.1.269. [DOI] [PubMed] [Google Scholar]
- 21.Marsh ENG. Essays Biochem. 1999;34:139–154. doi: 10.1042/bse0340139. [DOI] [PubMed] [Google Scholar]
- 22.Banerjee R, Ragsdale SW. Annu Rev Biochem. 2003;72:209–247. doi: 10.1146/annurev.biochem.72.121801.161828. [DOI] [PubMed] [Google Scholar]
- 23.Van Doorslaer S, Jeschke G, Epel B, Goldfarb D, Eichel RA, Krautler B, Schweiger A. J Am Chem Soc. 2003;125:5915–5927. doi: 10.1021/ja021218j. [DOI] [PubMed] [Google Scholar]
- 24.Fasching M, Schmidt W, Kräutler B, Stupperich E, Schmidt A, Kratky C. Helv Chim Acta. 2000;83:2295–2316. [Google Scholar]
- 25.Matthews RG. Acc Chem Res. 2001;34:681–689. doi: 10.1021/ar0000051. [DOI] [PubMed] [Google Scholar]
- 26.Olah GA. Angew Chem Int Ed. 1993;32:767–788. [Google Scholar]
- 27.Schnyder A, Darbre T, Keese R. Angew Chem Int Ed. 1998;37:1283–1285. doi: 10.1002/(SICI)1521-3773(19980518)37:9<1283::AID-ANIE1283>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 28.Evans JC, Huddler DP, Hilgers MT, Romanchuk G, Matthews RG, Ludwig ML. Proc Natl Acad Sci USA. 2004;101:3729–3736. doi: 10.1073/pnas.0308082100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hao B, Gong WM, Ferguson TK, James CM, Krzycki JA, Chan MK. Science. 2002;296:1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 30.Doukov T, Seravalli J, Stezowski JJ, Ragsdale SV. Structure (London) 2000;8:817–830. doi: 10.1016/s0969-2126(00)00172-6. [DOI] [PubMed] [Google Scholar]
- 31.Burke SA, Krzycki JA. J Biol Chem. 1997;272:16570–16577. doi: 10.1074/jbc.272.26.16570. [DOI] [PubMed] [Google Scholar]
- 32.Ferguson DJ, Krzycki JA. J Bacteriol. 1997;179:846–852. doi: 10.1128/jb.179.3.846-852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferguson DJ, Gorlatova N, Grahame DA, Krzycki JA. J Biol Chem. 2000;275:29053–29060. doi: 10.1074/jbc.M910218199. [DOI] [PubMed] [Google Scholar]
- 34.Zhou WH, Das A, Habel JE, Liu ZJ, Chang J, Chen LR, Lee D, Nguyen D, Chang SH, Tempel W, et al. Acta Crystallogr F. 2005;61:537–540. doi: 10.1107/S1744309105010511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tallant TC, Paul L, Krzycki JA. J Biol Chem. 2001;276:4485–4493. doi: 10.1074/jbc.M007514200. [DOI] [PubMed] [Google Scholar]
- 36.Pejchal R, Ludwig ML. PloS Biol. 2005;3:254–265. doi: 10.1371/journal.pbio.0030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doukov T, Seravalli J, Stezowski JJ, Ragsdale SW. Structure (London) 2000;8:817–830. doi: 10.1016/s0969-2126(00)00172-6. [DOI] [PubMed] [Google Scholar]
- 38.Drennan CL, Huang S, Drummond JT, Matthews RG, Ludwig ML. Science. 1994;266:1669–1674. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- 39.Bandarian V, Pattridge KA, Lennon BW, Huddler DP, Matthews RG, Ludwig ML. Nat Struct Biol. 2002;9:53–56. doi: 10.1038/nsb738. [DOI] [PubMed] [Google Scholar]
- 40.Champloy F, Gruber K, Jogl G, Kratky C. J Synchrotron Radiat. 2000;7:267–273. doi: 10.1107/S0909049500006336. [DOI] [PubMed] [Google Scholar]
- 41.Zydowsky LD, Zydowsky TM, Haas ES, Brown JW, Reeve JN, Floss HG. J Am Chem Soc. 1987;109:7922–7923. [Google Scholar]
- 42.Stich TA, Seravalli J, Venkateshrao S, Spiro TG, Ragsdale SW, Brunold TC. J Am Chem Soc. 2006;128:5010–5020. doi: 10.1021/ja054690o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jensen KP, Ryde U. J Mol Struct Theochem. 2002;585:239–255. [Google Scholar]
- 44.Bandarian V, Ludwig ML, Matthews RG. Proc Natl Acad Sci USA. 2003;100:8156–8163. doi: 10.1073/pnas.1133218100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matthews R. FASEB J. 2004;18:C223–C223. [Google Scholar]
- 46.Karrasch M, Bott M, Thauer RK. Arch Microbiol. 1989;151:137–142. [Google Scholar]
- 47.Kabsch H. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 48.Bailey S. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 49.Schneider TR, Sheldrick GM. Acta Crystallogr D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 50.La Fortelle E de, Bricogne G. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 51.Cowtan KD. Joint CCP4 ESF-EACBM Newsletter Protein Crystallogr. 1994;31:83–91. [Google Scholar]
- 52.Terwilliger TC. Acta Crystallogr D. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 54.Brünger A, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve R, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 55.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 56.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 57.Whitby FG, Phillips JD, Kushner JP, Hill CP. EMBO J. 1998:03650. doi: 10.1093/emboj/17.9.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.