Abstract
As the leading nanodevice candidate, single-walled carbon nanotubes (SWNTs) have potential therapeutic applications in gene therapy and novel drug delivery. We found that SWNTs can inhibit DNA duplex association and selectively induce human telomeric i-motif DNA formation by binding to the 5′-end major groove under physiological conditions or even at pH 8.0. SWNT binding to telomeric DNA was studied by UV melting, NMR, S1 nuclease cleavage, CD, and competitive FRET methods. These results suggest that SWNTs might have the intriguing potential to modulate human telomeric DNA structures in vivo, like biologically relevant B-A and B-Z DNA transitions, which is of great interest for drug design and cancer therapy.
Keywords: circular dichroism, DNA conformation, i-motif DNA, ligand–DNA interactions
Since the discovery of the DNA i-motif, the formation and biological function of this unique structure have attracted much attention, and significant progress in characterizing the i-motif has been made in recent years (1, 2). The biological importance of the intramolecular i-motif is evidenced by its involvement in human telomeric and centromeric DNA structures and RNA intercalated structures, and by the discovery of several proteins that bind specifically to C-rich telomeric DNA fragments capable of forming i-motifs (2). Along with human telomeric G-quadruplex DNA, the i-motif has been an attractive drug target for cancer chemotherapy and for modulation of gene transcription (2–9). Human telomeres consist of tandem repeats of the double-stranded DNA sequence (5′-TTAGGG):(5′-CCCTAA) (3–9). The G-rich strand can form a four-stranded G-quadruplex consisting of G-quartets, whereas its complementary C-rich strand may adopt i-motif structures with intercalated C·C+ base pairs (Fig. 1).
Fig. 1.
Quadruplex “building blocks” and duplex equilibrium. (A) The G-quartet. (B) The C·C+ hemiprotonated base pair of the “building blocks” for quadruplex formation. (C) Duplex equilibrium shifted by SWNTs.
Significantly, G-quadruplex formation was shown to inhibit the activity of telomerase, an enzyme that stabilizes chromosome ends by adding tandem telomeric DNA repeats to the 3′ single-stranded overhangs (4, 5, 10). In contrast to the majority of normal human somatic cells, most tumor cells express telomerase, and telomerase expression is essential for their indefinite proliferative capacities. Therefore, the therapeutic targeting of telomerase activity has been considered a promising approach to cancer therapy (4, 5, 10). Several small molecules have been shown to efficiently inhibit telomerase activity through the stabilization of G-quadruplex DNA (4, 5). Only two molecules that can stabilize both G-quadruplex and i-motif DNA have been identified (11, 12). To our knowledge, a ligand that can selectively stabilize i-motif DNA but not G-quadruplex DNA has not been reported.
As the leading nanodevice candidate, single-walled carbon nanotubes (SWNTs) have potential applications ranging from gene therapy to novel drug delivery to membrane separation (13–17). B-Z DNA transition has been achieved on the surfaces of SWNTs (18). Recently we reported that SWNTs bind to the DNA major groove and can induce a sequence-dependent B-A DNA transition (19). In the present study we show that SWNTs can selectively stabilize human telomeric i-motif DNA, induce i-motif DNA formation by binding to the 5′-end major groove under physiological conditions or even at pH 8.0, and compete with DNA duplex association. These properties of SWNTs were investigated by UV melting, NMR, S1 nuclease cleavage, CD, and competitive FRET methods.
Results and Discussion
To increase the solubility of SWNTs in aqueous solution, we treated SWNTs (ϕ = 1.1 nm) by a routine method and functionalized the open ends of SWNTs with a carboxyl group (19). Fig. 2A and B shows the UV melting profiles of G-quadruplex and i-motif DNA in the absence or presence of carboxyl-modified SWNTs at different binding ratios. A striking difference between G-quadruplex and i-motif DNA was observed. For G-quadruplex DNA at pH 7.0, the melting temperature at which half of the DNA base pairs disassociate (Tm) was 61°C (Fig. 2B) and did not change in the presence of SWNTs. For i-motif DNA at pH 5.5, the Tm was 38°C (Fig. 2A). However, the i-motif Tm gradually increased with increasing SWNT concentration, and the Tm was salt-dependent (2) [ supporting information (SI) Fig. 7]. At 10 μg·ml−1 SWNTs and 100 mM NaCl (pH 5.5), the i-motif Tm was 60°C (ΔTm = 22°C). The increase in Tm was independent of DNA concentration at the same ratio of DNA:SWNTs, indicating a monomolecular dissociation process. These results are consistent with the formation of a stable intramolecular i-motif structure (8) and with the selective stabilization of i-motif DNA by SWNTs. DNA CD spectra (SI Fig. 7) demonstrated that SWNT binding did not disturb higher-order DNA structures because the CD intensity of i-motif DNA decreased only slightly, and hardly any change was observed for G-quadruplex DNA in the presence of SWNTs. SI Table 1 summarizes the thermodynamic parameters calculated from melting profiles (8, 20). i-motif stabilization in the presence of SWNTs is driven by a net favorable ΔΔG20°C = −9.4 ± 0.1 kJ·mol−1, with a large favorable enthalpy (ΔH20°C = −180.9 ± 1.9 kJ·mol−1) compensating for the unfavorable entropy (ΔS20°C = −544.9 ± 5.6 J·mol−1·K−1). The stock solution of SWNTs was prepared in pH 7.0 solution (see Materials and Methods). To exclude the pH effect on DNA solution with addition of SWNTs, we measured the solution pH before the experiments and found no change in pH. Even at pH 4.2 buffer solution in which half of the cytosine is normally protonated, SWNTs can still increase i-motif DNA Tm by ≈4°C. In sodium acetate buffer (10 mM sodium acetate/200 mM NaCl, pH 5.5) SWNTs can also increase i-motif DNA Tm by 10°C. In addition, under the same conditions used for the human telomeric i-motif DNA, we also investigated other C-rich DNA sequences that can form tetraplexes, such as d(CCCCAAAACCCC) (21) and d(ACCCCA) (2), and we found that SWNTs can increase their Tm by 8°C and 4°C, respectively, not as significantly as the human telomeric i-motif DNA.
Fig. 2.
DNA UV meltings and CD spectral changes at different pH. UV melting profiles of i-motif (A) and G-quadruplex (B) in the absence (black lines) or presence of SWNTs: 2 μg·ml−1 (red lines), 3 μg·ml−1 (green lines), 5 μg·ml−1 (blue lines), 8 μg·ml−1 (cyan lines), and 10 μg·ml−1 (magenta lines) in pH 5.5 or pH 7.0 cacodylic buffer for i-motif and G-quadruplex, respectively. DNA concentration was 9.2 μg/ml for i-motif and 9.5 μg/ml for G-quadruplex in base. Normalized absorption changes at 260 nm for i-motif and at 295 nm for G-quadruplex were plotted against temperature. (C) CD spectra of i-motif at pH 8.0 (solid line), pH 7.0 (open circles), and pH 5.5 (stars). (D) CD spectra of i-motif at pH 8.0 in the presence of SWNTs: 1 μg·ml−1 (red line), 2 μg·ml−1 (green line), 3 μg·ml−1 (blue line), 5 μg·ml−1 (cyan line), 8 μg·ml−1 (magenta line), and 10 μg·ml−1 (yellow line) in cacodylic buffer (0.1 mM cacodylic acid/sodium cacodylate/100 mM NaCl, pH 8.0). (E) Plot of CD intensity at 258 nm (open circles) and at 288 nm (solid circles) vs. concentration of SWNTs. The data were adopted from B. (F) UV melting profiles of i-motif at pH 5.5 (black line), pH 8.0 in the absence (green line) or presence of SWNTs at 10 μg·ml−1 (red line) in cacodylic buffer (0.1 mM cacodylic acid/sodium cacodylate/100 mM NaCl). DNA concentration was 1 μM in strand.
Fig. 2C shows that the observed DNA structure was pH-dependent, consistent with the fact that human telomeric i-motif structure involves systematic intercalated CC+ base pairs that are dependent on solution pH (8). At pH 5.5 two i-motif-characteristic CD bands appeared, a positive band near 288 nm and a negative band near 256 nm (Fig. 2C). The red shifts of CD bands with pH change from pH 8.0 to 5.5 have been attributed to the pH-dependent formation of i-motif structure (8). When the C-rich human telomeric DNA was titrated with SWNTs in buffer solution at pH 7.0 or even at pH 8.0, we found that SWNTs could induce i-motif formation. Typical CD spectra at pH 8.0 (Fig. 2D) indicate that SWNTs could not only stabilize i-motif structure but also induce its formation at pH 8.0. The structural transition was cooperative (22–25), with a midpoint at 5 μg·ml−1 SWNTs at pH 8.0 (Fig. 2E). UV melting studies showed that the C-rich DNA alone at pH 8.0 did not have a clear transition. However, in the presence of SWNTs at 10 μg·ml−1, there was an unambiguous transition at 38°C (Fig. 2F), the same Tm observed for DNA alone at pH 5.5 (Fig. 2B). This result is consistent with the similar free energy change for i-motif formation of DNA alone at pH 5.5 and of DNA in the presence of SWNTs at pH 8.0 (ΔGDNApH5.5 = −11.8 ± 0.2, ΔGSWNTpH8.0 = −11.3 ± 0.1 kJ·mol−1) (SI Table 1).
Similarly, 1H NMR spectra provided evidence that SWNTs could induce i-motif formation. Chemical shifts at 15–16 ppm for i-motif imino protons in C·C+ have been considered a hallmark of i-motif DNA (26). DNA samples were analyzed at different pH conditions in the absence or presence of SWNTs. Fig. 3A shows that in the absence of SWNTs the i-motif structure was formed at pH 5.0 but disappeared at pH 8.0, consistent with previous NMR studies (26). Interestingly, in the presence of SWNTs the i-motif structure was detectable at pH 8.0, confirming that SWNTs bind to DNA and induce i-motif formation.
Fig. 3.
i-motif NMR and native-PAGE images in the absence or presence of SWNTs. (A) One-dimensional 600-MHz 1H NMR spectra of i-motif under different conditions: line 1, pH 5.0; line 2, pH 8.0; line 3, pH 8.0 in the presence of SWNTs. Imino protons of thymines in the i-motif structure were labeled with a star. Individual DNA samples were prepared at different pH. Experimental conditions were according to ref. 25: strand concentration, 0.15 mM; temperature, 10°C; 100 mM NaCl (details as described in Materials and Methods). (B) Native PAGE images of G/C duplex in the absence or presence of the SWNT. Lane 1, dT22; lane 2, G-quadruplex; lane 3, i-motif; lane 4, G/C duplex alone; lane 5, G/C duplex with SWNTs. Twenty percent PAGE and TB running buffer were used in the experiments at room temperature.
Human telomeric G-rich and C-rich strands are complementary. A duplex competitive gel mobility shift assay showed that SWNTs could disrupt G-rich/C-rich double-strand association to induce i-motif formation. In the presence of SWNTs, new bands with mobilities even faster than that of i-motif DNA alone (8, 9) were observed (Fig. 3B), indicating formation of a more compact structure. Furthermore, competitive DNA duplex CD melting experiments (Fig. 4A) showed that there were two obvious transitions in the presence of SWNTs at pH 7.0: one at 38°C for i-motif melting and the other at 65°C for duplex melting. At pH 5.5 SWNTs increased the i-motif Tm by 15°C and even inhibited duplex formation because no duplex melting was observed (Fig. 4A). Instead, we observed both i-motif and G-quadruplex meltings that had sigmoid-shaped profiles with positive slopes.
Fig. 4.
DNA CD meltings, S1 digestion, and fluorescence changes in the presence of SWNTs. (A) CD melting profiles of the G/C duplex under different conditions: at pH 7.0 in the absence (line 1) or presence (line 2) of 10 μg·ml−1 SWNTs and at pH 5.5 in the absence (line 3) or presence (line 4) of 10 μg·ml−1 SWNTs in cacodylic buffer (0.1 mM cacodylic acid/sodium cacodylate/100 mM NaCl). DNA concentration was 9.2 μg/ml for i-motif and 9.5 μg/ml for G-quadruplex in base. Normalized CD changes at 260 nm were plotted against temperature. (B) Image of fluorescent nondenaturating PAGE after S1 digestion. Lane 1, untreated 5′-fluorescein-labeled 21-mer sequence; lane 2, S1 treated at pH 5.5; lanes 3–5, S1 treated the complexes of 5′-fluorescein-labeled DNA and SWNT-COOH (0.75, 1.75, and 5 μg·ml−1). For details see Materials and Methods. (C) Plot of fluorescence intensity at 370 nm of 2-Ap individually labeled i-motif vs. concentration of SWNTs. The DNA sequence was shown in Materials and Methods. DNA concentration was fixed at 4 μM in base titrated by SWNTs in pH 5.5 cacodylic buffer at 15°C. Open squares, T(2-Ap)A loop near 5′ end in the major groove; open circles, loop in the narrow groove; open triangles, loop near 3′ end in the major groove. Fluorescence emission spectra were recorded from 320 nm to 500 nm with an excitation wavelength at 305 nm.
Competitive duplex binding was further studied by FRET (12). The C-rich strand was labeled (12) with fluorescein and a rhodamine derivative, 3′-tetramethylrhodamine (TAMRA), at its 5′ and 3′ ends, respectively (see SI). In this system, fluorescein emission is increased upon duplex formation because of increased distance between the two dye molecules (12) in comparison with the folded i-motif structure. Therefore, it is possible to monitor duplex formation by recording the fluorescein emission at 520 nm (12). At pH 7.0 the duplex association constant was determined by fluorescence titration of i-motif DNA with the G-rich strand (SI Fig. 8). Nonlinear least-squares analysis of the titration data yielded an association constant of 2.7 ± 0.2 × 106 M−1. However, when 42 nM i-motif DNA was titrated with the G-rich strand in the presence of SWNTs at 10 μg·ml−1, fluorescence emission at 520 nm did not increase, showing that SWNTs blocked duplex association (19) and that the estimated binding affinity of SWNTs to i-motif DNA is likely >106 M−1.
The i-motif has two very wide grooves and two very narrow grooves with interstrand phosphorus–phosphorus distances as close as 5.9 Å (27). The helix twists in a right-handed manner, and the distance between nearest base pairs is only 3.1 Å. In contrast with the G-rich quadruplex, little is known about the interaction of small ligands with this structure (2, 11, 12). Only two molecules, a porphyrin derivative and an acridine dimer, have been reported to stabilize both G-quadruplex and i-motif DNA via a nonintercalative mechanism (11, 12). Our results show that SWNTs have the intriguing potential for modulating human telomeric DNA structures in vivo, like biologically relevant B-A and B-Z DNA transitions (18, 19, 22–24), which is of great importance in drug design and cancer therapy.
As for the SWNT binding site, it seems unlikely that the rigid SWNTs (ϕ = 1.1 nm) bind to i-motif DNA in the very narrow grooves or by intercalation between base pairs (11, 12). Alternatively, SWNTs may bind to the wide grooves without disturbing higher-order DNA structure (SI Fig. 7). This binding could be stabilized by various favorable interactions, including hydrophobic and van der Waals interactions and electrostatic interactions between carboxyl-modified SWNTs and CC+ base pairs, TAA loops, and the DNA backbone, as observed in our previous studies on SWNT duplex binding (19). This binding mechanism is supported by our CD results, S1 nuclease cleavage patterns (28), and fluorescence changes of 2-aminopurine (2-Ap)-labeled loops (29) of i-motif DNA in the presence of SWNTs (labeled sequences shown in Materials and Methods).
The i-motif used in our studies has three TAA loops (Fig. 1): one in the narrow groove and the other two in the wide grooves near the 5′ and 3′ ends, respectively. Fig. 4B shows the cleavage pattern of 5′-fluorescein-labeled i-motif DNA after S1 nuclease digestion at pH 5.5. Digestion by S1, a single-strand- and hairpin-loop-specific nuclease (28), resulted in three major cleavages. All three bands appeared to be due to cleavage within TAA loops, consistent with previous studies on c-myc promoter cleavage (28). The first S1 cleavage occurred near the 5′ end in the major groove with the shortest length, the second cleavage in the narrow groove with intermediate length, and the third cleavage near the 3′ end in the major groove with the longest length. In the presence of SWNTs a higher amount of S1 nuclease cleavage was observed for the major groove near the 5′ end (28). At the same time, in the presence of SWNTs cleavage in the narrow groove was decreased, and a modest decrease in S1 cleavage was also observed for the site in the major groove near the 3′ end. This cleavage pattern suggests that SWNTs bind to the i-motif at the 5′-end major groove by interacting with CC+ base pairs and the TAA loop, thereby increasing the accessibility of this loop to S1 nuclease (28). As shown in melting and CD studies (Fig. 2), SWNT binding resulted in i-motif stabilization and increased stacking of A-T Hoogsteen base pairs in the loops. Stacking in the TAA loop of the narrow groove was especially increased by SWNTs because this loop was the most labile to S1 nuclease in the absence of SWNTs but was substantially protected from cleavage after SWNT binding (28).
These results were further supported by 2-Ap fluorescence quenching experiments. As a fluorescent probe for nucleic acids (29), 2-Ap has been widely used to verify the mode of ligand binding to quadruplex DNA (29). CD and melting results (Fig. 5) showed that i-motif DNA labeled at T(2-Ap)A loops was formed, and the substitution of 2-Ap for adenine decreased the Tm by 4°C. At a low binding ratio SWNTs decreased 2-Ap fluorescence (29) in the order 2-Ap in 5′-end major groove → narrow groove → 3′-end major groove (Fig. 4C), consistent with the S1 nuclease cleavage pattern. These results indicate that SWNTs preferentially bind to the i-motif at the 5′-end major groove, making this loop the most sensitive to S1 cleavage and decreasing 2-Ap fluorescence to the greatest extent (28, 29). The decreased 2-Ap fluorescence observed for the other grooves is likely due to increased stacking of loop structures resulting from SWNT-induced i-motif stabilization.
Fig. 5.
CD spectra and UV meltings of i-motif and 2-Ap-modified i-motif. (A) CD spectra of i-motif (1 μM in strand) not labeled (black line) or labeled with 2-Ap in the T(2-Ap)A loop positioned near the 5′ end in the major groove (open squares), in the narrow groove (open circles), and near the 3′ end in the major groove (open triangles) in aqueous cacodylic buffer (0.1 mM cacodylic acid/sodium cacodylate/100 mM NaCl, pH 5.5). (B) UV melting profiles of i-motif (9.2 μg/ml for i-motif and 9.5 μg/ml for G-quadruplex in base) not labeled (black line) or labeled with 2-Ap in the T(2-Ap)A loop positioned near the 5′ end in the major groove (open squares), in the narrow groove (open circles), and near the 3′ end in the major groove (open triangles) in aqueous cacodylic buffer (0.1 mM cacodylic acid/sodium cacodylate/100 mM NaCl, pH 5.5). DNA concentration was 9.2 μg/ml for i-motif and 9.5 μg/ml for G-quadruplex in base. Normalized absorption changes at 260 nm were plotted against temperature.
There are two obvious mechanisms for stabilization of the i-motif by SWNTs. The first mechanism is charge stabilization due to interactions between the positively charged CC+ base pairs and SWNTs, lowering the pKa of the CC+ base pairs and inducing i-motif formation. Previous studies have shown that SWNTs can promote protonation of conjugated polymers by lowering their pKa values (30). The second possibility is that SWNTs may simply serve as condensation nuclei to increase the propensity for DNA aggregation. Aggregation would indirectly facilitate CC+ protonation by increasing DNA/DNA strand interactions, resulting in charge stabilization. However, the faster migration of i-motif DNA bands in the presence of SWNTs argues against the aggregation model (Fig. 3B).
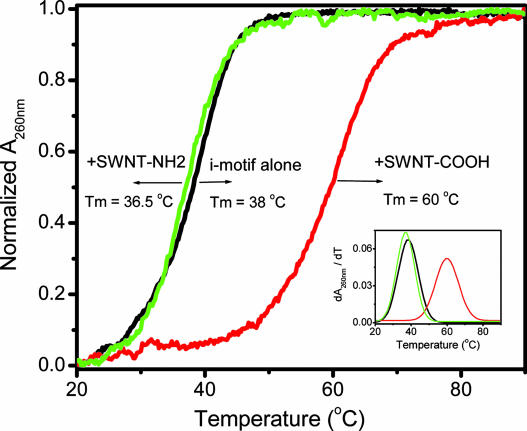
Notably, the SWNTs we used were modified with carboxyl groups (SWNT-COO−) on the open end of the side wall (19). However, when SWNT-COOH was covalently modified with ethylenediamine by a routine method (31), the positively charged amino group-modified SWNTs (SWNT-CONH-CH2CH2-NH2+) decreased the i-motif Tm by 2°C (Fig. 6). This result indicates that a negatively charged carboxyl group on the open end of the side wall not only improves SWNT solubility but also enhances interactions with i-motif CC+ base pairs by providing favorable electrostatic attractions (19). The carboxyl group was crucial for SWNT interactions with i-motif CC+ base pairs (Fig. 1).
Fig. 6.
UV melting profiles of i-motif alone (black line), in the presence of 10 μg·ml−1 SWNT-COOH (red line), or in the presence of 10 μg·ml−1 SWNT-CONHCH2CH2-NH2 (green line) in aqueous cacodylic buffer (0.1 mM cacodylic acid/sodium cacodylate/100 mM NaCl, pH 5.5). DNA concentration was 9.2 μg/ml for i-motif and 9.5 μg/ml for G-quadruplex in base. Normalized absorption changes at 260 nm were plotted against temperature. (Inset) The plot of their individual derivative of dA260 nm/dT vs. temperature.
In summary, we showed by several methods that SWNTs selectively stabilize i-motif DNA. Furthermore, SWNTs inhibit DNA duplex formation, as shown by competitive gel mobility shift assay, CD melting experiments, and FRET. S1 nuclease cleavage patterns and 2-Ap fluorescence quenching results indicated that SWNTs bind to the 5′-end major groove of the i-motif. Our results suggest that the SWNT/i-motif interaction substantially decreases the pKa of i-motif CC+ base pairs. Thus, we conclude that SWNTs directly stabilize the charged CC+ base pairs in i-motif DNA by binding at the 5′-end major groove. We are aware that the natural occurrence of i-motifs in cells has not been demonstrated and that the biological effects of induction of these structures need to be clarified.
Materials and Methods
Materials.
SWNTs (ϕ = 1.1 nm, purity >90%) were purchased from Aldrich (St. Louis, MO), purified as described previously by sonicating SWNTs in a 3:1 vol/vol solution of concentrated sulfuric acid (98%) and concentrated nitric acid (70%) for 24 h at 35–40°C, and washed with water, leaving an open hole in the tube side and functionalizing the open end of SWNTs with carboxyl group to increase their solubility in aqueous solution (19). The stock solution of SWNTs (0.15 mg·ml−1) was obtained by sonicating the SWNTs for 8 h in pH 7.0 aqueous solution (19). S1 nuclease was purchased from Takara (Tokyo, Japan).
DNA oligomers (9, 12, 23, 25, 32) 5′-CCCTAACCCTAACCCTAACCCT-3′ (i-motif), its corresponding complementary strand 5′-AGGGTTAGGGTTAGGGTTAGGG-3′ (G-quadruplex), and fluorescent analogs (see sequences below): 2-Ap labeled individually on TAA loop in three different positions, 5′-fluorescein-labeled, and 5′-fluorescein and TAMRA-labeled i-motif DNA were purchased from Sangon (Shanghai, China) and used without further purification. Concentrations of these oligomers were determined by measuring the absorbance at 260 nm after melting. Extinction coefficients were estimated by the nearest-neighbor method by using mononucleotide and dinucleotide values (9, 12, 23, 25, 32). All experiments were carried out in aqueous cacodylic buffer (33) (0.1 mM cacodylic acid/sodium cacodylate/100 mM NaCl) unless stated otherwise. The sequences of the fluorescent analogs (12, 28, 29) were 5′-Fluoro-CCCTAACCCTAACCCTAACCC-3′, 5′-Fluoro-TACCCTAACCCTAACCCTAACCC-TAMRA-3′, 5′-CCCT(2-Ap)ACCCTAACCCTAACCCT-3′, 5′-CCCTAACCCT(2-Ap)ACCCTAACCCT-3′, and 5′-CCCTAACCCTAACCCT(2-Ap)ACCCT-3′.
Bioassay.
Absorbance measurements and melting experiments were carried out on a Cary 300 UV/Vis spectrophotometer equipped with a Peltier temperature control accessory (9, 12, 19, 23). All UV/Vis spectra were measured in 1.0-cm-path-length cell with the same concentration of SWNT aqueous solution accordingly as the reference solution. Absorbance changes at either 260 nm or 295 nm vs. temperature were collected (19, 23, 25) at a heating rate of 0.5°C·min−1. Primary data were transferred to the graphics program Origin for plotting and analysis.
The thermodynamic parameters of oligomer DNA molecules were calculated by using their melting profiles according to a well established method (8, 20). The enthalpy change, ΔH, was determined from the temperature dependence of equilibrium association constant where ΔH was the slope of lnKa vs. 1/T plot according to the equation lnKa = −(ΔH/RT) + ΔS/R, where ΔS was the entropy change that was calculated according to the y axis intercept. The free energy change (ΔG) at 20°C was calculated from the standard Gibbs's equation, ΔG = ΔH − TΔS.
CD spectra and CD melting experiments were measured on a JASCO J-810 spectropolarimeter equipped with a temperature-controlled water bath (19, 22, 23, 25). The optical chamber of CD spectrometer was deoxygenated with dry purified nitrogen (99.99%) for 45 min before use and kept the nitrogen atmosphere during experiments. Three scans were accumulated and automatically averaged. SWNTs alone did not contribute to the CD signal between 200 nm and 380 nm in our experimental conditions (19). 1H NMR spectra (25, 34) were carried out on a Bruker Avance 600 MHz NMR Spectrometer at 10°C. Individual DNA samples were used at different pH conditions (26) in the absence or presence of SWNTs in 100 mM NaCl, 0.1 mM EDTA, and 0.01 mM DSS solution. Experimental conditions were according to ref. 25. DNA concentration was 0.15 mM in strand.
Fluorescence measurements were carried out on a JASCO FP-6500 spectrofluorometer at 20°C (19, 23). Duplex competitive bindings were studied by the FRET method (12). The C-rich strand used here was labeled (12) with fluorescein and a rhodamine derivative, TAMRA, at its 5′ and 3′ ends, respectively. Fluorescein emission would enhance when the duplex formed because the distance between the two dye molecules, in comparison with the folded i-motif structure, was increased (12). Therefore, it is possible to monitor duplex formation by recording the emission at 520 nm. At pH 7.0 the duplex association constant was determined by fluorescence titration (SI Fig. 8). Fixed labeled i-motif (42 nM) concentrations were titrated by its complementary G-rich strand in the absence or presence of SWNTs in pH 7.0 cacodylic buffer. An excitation wavelength of 480 nm was used, and the fluorescence emission was monitored from 500 to 700 nm.
Fluorescence titration data (22, 23, 25) were fit directly to get binding constants by using a fitting function incorporated into the program FitAll (MTR Software, Toronto, ON, Canada). The observed fluorescence is assumed to be a sum of the weighted concentrations of free and bound labeled i-motif DNA: F = F0 (Ct − Cb) + FbCb, where F is the apparent fluorescence at each G-quadruplex concentration, F0 is the fluorescence intensity of free labeled i-motif, and Fb is the fluorescence intensity of the bound species. Ct and Cb are the molar concentrations or total and bound labeled i-motif, respectively. For 1:1 duplex formation it can be easily shown that Kx2 − x(KS0 + KD0 + 1) + KS0D0 = 0, where x = Cb, K is the association constant, S0 is the total G-rich strand concentration, and D0 is the total i-motif concentration. This equation is readily solved by using the quadratic formula. Data in the form of fluorescence response F as a function of total G-rich strand concentration at fixed concentration of i-motif may then be fit by nonlinear least-squares methods to get K, F0, and Fb.
2-Ap has been widely used as a fluorescent probe for nucleic acids (29). Fluorescence spectra of 2-Ap-labeled i-motif DNA were measured (29) by using an excitation wavelength of 305 nm and recorded from 320 nm to 500 nm in the absence or presence of different amount of SWNTs in pH 5.5 cacodylic buffer. The concentration of 2-Ap-labeled i-motif DNA was fixed at 4 μM in base.
Native PAGE experiments (8, 9, 23) were carried out in 0.045 M Tris·borate buffer. G-rich/C-rich duplex was formed by mixing an equimolar concentration of G-quadruplex and i-motif for 12 h at 4°C in 0.1 mM sodium cacodylate and 100 mM NaCl buffer (pH 7.0). Then, SWNT-COOH was added into the duplex at a 1 μg·ml−1:2 μM ratio and incubated for 12 h at 4°C. Electrophoresis was carried out by using 20% acrylamide at 200 V for 50 min at room temperature. The gels were silver-stained.
S1 nuclease digestion and PAGE using fluorescent oligonucleotide were carried out according to ref. 28. 5′-fluorescein-labeled oligomer in pH 5.5 buffer was annealed at 95°C for 5 min and then cooled slowly to room temperature. The mixtures of 5′-fluorescein-labeled oligomer and SWNT-COOH at different ratios were incubated for ≈24 h at 4°C. Digestion was performed at pH 5.5. Reaction mixtures contained S1 buffer and 5.5 units of S1 nuclease. The digestion procedure was according to ref. 28. Briefly, the template, 5′-fluorescein-labeled oligomer (1.1 μg), and the complexes of 5′-fluorescein-labeled oligomer-SWNT-COOH were incubated overnight at 4°C in buffer before initiating digestion at 37°C by adding S1 nuclease and Zn2+. After 5 min digestions were stopped by adding 4 μl of stop buffer (70% formamide/57 mM EDTA, pH 7.5) and then freezing. The frozen samples were treated with 1 μl of formamide, heated at 95°C for 1 min, and centrifuged to remove SWNTs before they were loaded on a 20% polyacrylamide gel and electrophoresed (70 min in 1× TBE running buffer) at room temperature and 20 V/cm.
Supplementary Material
Acknowledgments
We are grateful for the referees' helpful comments on the manuscript. We thank the National Natural Science Foundation of China (Grants 20225102, 20331020, 20325101, and 20473084) and the Hundred People program from the Chinese Academy of Sciences and Jilin Province for support.
Abbreviations
- SWNT
single-walled carbon nanotube
- 2-Ap
2-aminopurine
- TAMRA
3′-tetramethylrhodamine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at http://www.pnas.org/cgi/content/full/0607245103/DC1.
References
- 1.Gehring K, Leroy JL, Gueron M. Nature. 1993;363:561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- 2.Gueron M, Leroy JL. Curr Opin Struct Biol. 2000;10:326–331. doi: 10.1016/s0959-440x(00)00091-9. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn EH. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 4.Mergny JL, Helene C. Nature. 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- 5.Hurley LH. Nat Rev Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 6.Parkinson GN, Lee MP, Neidle S. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Patel DJ. Structure (London) 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 8.Li W, Wu P, Ohmichi T, Sugimoto N. FEBS Lett. 2002;526:77–81. doi: 10.1016/s0014-5793(02)03118-6. [DOI] [PubMed] [Google Scholar]
- 9.Ren J, Qu X, Trent JO, Chaires JB. Nucleic Acids Res. 2002;30:2307–2315. doi: 10.1093/nar/30.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 11.Fedoroff OY, Rangan A, Chemeris VV, Hurley LH. Biochemistry. 2000;39:15083–15090. doi: 10.1021/bi001528j. [DOI] [PubMed] [Google Scholar]
- 12.Alberti P, Ren J, Teulade-Fichou MP, Guittat L, Riou JF, Chaires JB, Helene C, Vigneron JP, Lehn JL, Mergny JL. J Biomol Struct Dyn. 2001;19:505–513. doi: 10.1080/07391102.2001.10506758. [DOI] [PubMed] [Google Scholar]
- 13.Wong SS, Joselevich E, Woolley AT, Cheung CL, Lieber CM. Nature. 1998;394:52–55. doi: 10.1038/27873. [DOI] [PubMed] [Google Scholar]
- 14.Zheng M, Jagota A, Strano MS, Santos AP, Barone P, Chou SG, Diner BA, Dresselhaus MS, McLean PS, Onoa GB, et al. Science. 2003;302:1545–1548. doi: 10.1126/science.1091911. [DOI] [PubMed] [Google Scholar]
- 15.Kam NWS, O'Connell M, Wisdom JA, Dai HJ. Proc Natl Acad Sci USA. 2005;102:11600–11605. doi: 10.1073/pnas.0502680102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh R, Pantarotto D, McCarthy D, Chaloin O, Hoebeke J, Partidos C, Briand J-P, Prato M, Bianco A, Kostarelos K. J Am Chem Soc. 2005;127:4388–4396. doi: 10.1021/ja0441561. [DOI] [PubMed] [Google Scholar]
- 17.Lin Y, Taylor S, Li H, Fernando KAS, Qu L, Wang W, Gu L, Zhou B, Sun Y-P. J Mater Chem. 2004;14:527–541. [Google Scholar]
- 18.Heller DA, Jeng ES, Yeung T-K, Martinez BM, Moll AE, Gastala JB, Strano MS. Science. 2006;311:508–511. doi: 10.1126/science.1120792. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Peng Y, Qu X. Nucleic Acids Res. 2006;34:3670–3676. doi: 10.1093/nar/gkl513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mergny JL, Lacroix L. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 21.Miyoshi D, Matsumura S, Nakano S, Sugimoto N. J Am Chem Soc. 2004;126:165–169. doi: 10.1021/ja036721q. [DOI] [PubMed] [Google Scholar]
- 22.Qu X, Trent JO, Fokt I, Priebe W, Chaires JB. Proc Natl Acad Sci USA. 2000;97:12032–12037. doi: 10.1073/pnas.200221397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H, Yu H, Ren J, Qu X. Biophys J. 2006;90:3203–3207. doi: 10.1529/biophysj.105.078402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qu X, Wan C, Becker H, Zhong D, Zewail AH. Proc Natl Acad Sci USA. 2001;98:14212–14217. doi: 10.1073/pnas.241509698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, Yu H, Ren J, Qu X. FEBS Lett. 2006;580:3726–3730. doi: 10.1016/j.febslet.2006.05.046. [DOI] [PubMed] [Google Scholar]
- 26.Phan AT, Mergny JL. Nucleic Acids Res. 2002;30:4618–4625. doi: 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berger I, Egli M, Rich A. Proc Natl Acad Sci USA. 1996;93:12116–12121. doi: 10.1073/pnas.93.22.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar P, Verma A, Maiti S, Gargallo R, Chowdhury S. Biochemistry. 2005;44:16426–16434. doi: 10.1021/bi051452x. [DOI] [PubMed] [Google Scholar]
- 29.Kimura T, Kawai K, Fujitsuka M, Majima T. Chem Commun. 2006:401–402. doi: 10.1039/b514526k. [DOI] [PubMed] [Google Scholar]
- 30.Steuerman DW, Star A, Narizzano R, Choi H, Ries RS, Nicolini C, Stoddart JF, Heath JR. J Phys Chem B. 2002;106:3124–3130. [Google Scholar]
- 31.Chen J, Hamon MA, Hu H, Chen Y, Rao AM, Eklund PC, Haddon RC. Science. 1998;282:95–98. doi: 10.1126/science.282.5386.95. [DOI] [PubMed] [Google Scholar]
- 32.Qu X, Ren J, Riccelli PV, Benight AS, Chaires JB. Biochemistry. 2003;42:11960–11967. doi: 10.1021/bi0347813. [DOI] [PubMed] [Google Scholar]
- 33.Tikhomirova A, Chalikian TV. J Mol Biol. 2004;341:551–563. doi: 10.1016/j.jmb.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 34.Xia T, Frankel A, Ren J, Takahashi TT, Roberts RW. Nat Struct Biol. 2003;10:812–819. doi: 10.1038/nsb983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.