Abstract
Divergent natural selection acting on ecological traits, which also affect mate choice, is a key element of ecological speciation theory, but has not previously been demonstrated at the molecular gene level to our knowledge. Here we demonstrate parallel evolution in two cichlid genera under strong divergent selection in a gene that affects both. Strong divergent natural selection fixed opsin proteins with different predicted light absorbance properties at opposite ends of an environmental gradient. By expressing them and measuring absorbance, we show that the reciprocal fixation adapts populations to divergent light environments. The divergent evolution of the visual system coincides with divergence in male breeding coloration, consistent with incipient ecological by-product speciation.
The rapid evolution of African cichlid fish driven by strong divergent selection is revealed in a gene that is responsible for ecological adaptation. This gene also affects mate choice and is thus consistent with incipient ecological by-product speciation.
Introduction
Adaptive radiation, the generation of ecological diversity in a rapidly multiplying lineage, is receiving much attention by evolutionary biologists and ecologists because it is thought to be a force able to quickly generate large numbers of new species and ecological diversity through multiple episodes of ecological speciation [1]. Divergent natural selection (selection on ecologically relevant traits that favors different alleles in different environments) is thought to be its main driver [1,2]. It can potentially cause ecological differentiation and speciation simultaneously when its action on ecological traits in contrasting environments affects mate choice as a by-product [1]. The concept of by-product speciation is theoretically and mathematically well established [1,3], and experimental evidence supports that the mechanism works [2,4,5]. However, because the genes responsible for variation in ecological and mating traits are rarely identified, the hypothesis has remained largely untested at the gene level. In particular, that selection has fixed alleles with opposite effects on an adaptive trait in different closely related populations has rarely been demonstrated [6,7], and to our knowledge, not for traits that affect adaptation and mating preferences simultaneously, even though chromosomal regions with effects on both have been singled out by quantitative trait locus mapping [4,5]. Yet, without such demonstration, the discussion about the role of divergent natural selection in speciation remains somewhat speculative.
African cichlid fish are becoming a model system for the genetics of vertebrate speciation and adaptive radiation [8]. Lake Victoria, the largest of the African great lakes, hosts the youngest of the large cichlid radiations. Geological evidence even suggests that Lake Victoria dried up at the end of the Pleistocene and refilled only 15,000 years ago [9]. Levels of polymorphism in mitochondrial DNA suggest that the neutral genetic diversity contained in the radiation is less than 200,000 y old [10] and may indeed be younger [11]. Surprisingly, despite 10-fold lower mitochondrial DNA diversity, the phenotypic diversity among Lake Victoria cichlids is similar to that in the several-million-y-old-Lake Malawi [12]. This implies rapid selection-driven diversification in Lake Victoria but remained untested at the gene level.
Male nuptial coloration is one of the most amazingly variable phenotypic traits among Lake Victoria cichlid fish. Water is a dense medium that generates highly heterogeneous light environments that fish have to adapt their visual systems to. Visual pigments in the photoreceptor cells of the retina consist of a light-absorbing component—the chromophore—and a protein moiety, the opsin [13]. The light sensitivity of a visual pigment is determined by the chromophore (A1 [11-cis-retinal] or A2 [11-cis-3, 4-dehydroretinal] pigments) and by its interaction with the amino acid residues coating the retinal binding pocket of the opsin in which the chromophore lies [14].
In haplochromine cichlids, eight different opsins have been found [15–18]. As predicted by adaptive evolution, the most variable between species are those that absorb at the extreme ends of the light spectrum where most environmental variation in light transmission is found: short wavelength–sensitive opsin 1 and long wavelength–sensitive opsin (LWS) [17].
Within Lake Victoria, LWS, which has a peak value of light absorption (λmax) at long (red) wavelengths [16], is by far the most variable opsin [17]. LWS is a candidate for a gene under strong divergent selection in Lake Victoria, because it entails five times more variation in the cichlids of Lake Victoria than in the at least 10-fold older Lake Malawi cichlid radiation [19,20]. The photic environment in Lake Victoria is characterized by much steeper gradients in water clarity, light intensity, and spectral composition [21]. Given the importance of vision in food acquisition, predator avoidance, territorial defence, and mate choice of cichlid fish, these gradients are likely exerting strong selection on opsin genes and on associated mating traits. In several pairs of closely related species of Lake Victoria cichlids, the species with the larger LWS λmax has red male breeding colors and lower detection thresholds for red light, whereas the species with smaller LWS λmax has blue breeding colors and is less sensitive to red light and more sensitive to blue light, consistent with sensory drive (divergence in male coloration evolving as a consequence of divergent visual sensitivities) [20,22,23]. Hence, the LWS gene may be involved in ecological adaptation and mate choice simultaneously. Here we demonstrate evolution in LWS under strong divergent ecological selection along an environmental gradient, and we show correlated divergence in male nuptial coloration, implicating population divergence through sensory drive.
Results/Discussion
Population Divergence at the LWS Gene
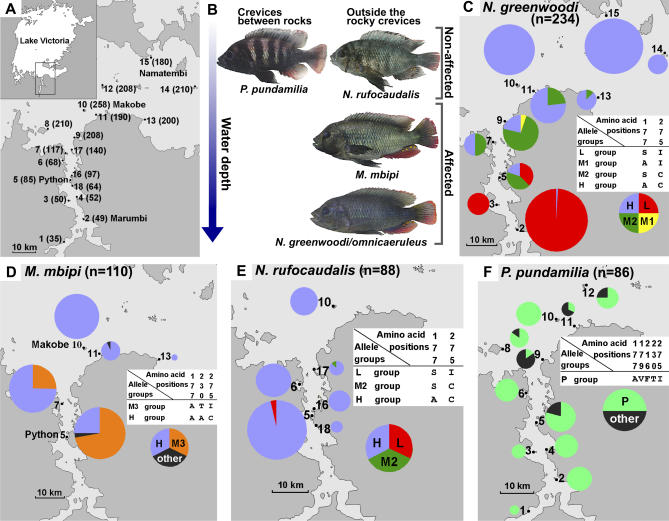
We studied multiple populations of each of four different Lake Victoria cichlid species along a 100-km-long cline in water clarity (Figure 1A). All four have lake-wide but patchy distributions, being restricted to rocky islands and headlands: Neochromis greenwoodi (including its offshore incipient species N. omnicaeruleus [24]), Neochromis rufocaudalis, Mbipia mbipi, and Pundamilia pundamilia [24]. We determined the sequences of exons 2–5 of LWS (872 base pairs [bp]) that code the trans-membrane region from 117 individuals (234 alleles) of N. greenwoodi/omnicaeruleus (10 populations), 44 individuals (88 alleles) of N. rufocaudalis (6 populations), 55 individuals (110 alleles) of Mbipia mbipi (5 populations), and 43 individuals (86 alleles) of Pundamilia pundamilia (11 populations). For this analysis, we considered it appropriate to treat N. greenwoodi/omnicaeruleus as one taxon (i.e., superspecies), because N. omnicaeruleus replaces N. greenwoodi at the clear water islands in the Speke Gulf and differs only in male coloration (light blue) from neighbouring N. greenwoodi populations along the mainland (blue-black) [24]. The four taxa occupy different water depths and microhabitats, living either mostly outside or inside the rocky interstices (Figures 1B and S1). N. rufocaudalis and P. pundamilia are everywhere restricted to very shallow waters. N. rufocaudalis grazes on algae on the outside of rocks, whereas P. pundamilia lives in crevices between rocks, feeding on insect larvae. M. mbipi has a deeper modal depth and lives predominantly outside the rocky crevices. N. greenwoodi/omnicaeruleus has the widest depth range, a modal depth slightly deeper than M. mbipi, and also lives mostly outside the rocky crevices [25] (Figures 1B and S1). Both are omnivorous feeders. We predicted that N. greenwoodi/omnicaeruleus and M. mbipi should experience strong divergent selection on the visual system between populations, exerted by differences in light transmission between islands with different water clarity, whereas species that live exclusively in very shallow water (and in rocky interstices) would be less affected.
Figure 1. Maps and Frequencies of LWS Alleles.
(A) The study area in southern Lake Victoria. Arabic numerals indicate stations at which cichlids were collected. The Secchi disk water transparency (cm) at each station is shown in parentheses. The stations are: 1, Buyago Rocks; 2, Marumbi Island; 3, Matumbi Island; 4, Luanso Island; 5, Python (Nyamatala) Islands; 6, Kissenda Island; 7, Hippo Island; 8, Juma Island; 9, Bwiru Point; 10, Makobe Island; 11, Igombe Island; 12, Ruti Island; 13, Senga Point; 14, Sozihe Islands; 15, Namatembi Island; 16, Nyamatala (Nyameruguyu) Island; 17, Gabalema Islands; 18, north end of Luanso Bay. Where the local name differs from that used in previous publications, the local name is indicated in parenthesis.
(B) A representation of the microhabitat distribution of the studied species by water depth and between, as opposed to outside, the rocky boulders. Photos show males of the blue morph in nuptial coloration. “Affected” and “Non-affected” indicate the species with the LWS allele frequencies that are strongly affected and not strongly affected by variation in water transparency, respectively.
(C) LWS allele group frequencies in the populations of N. greenwoodi/omnicaeruleus.
(D) LWS allele group frequencies in the populations of M. mbipi.
(E) LWS allele group frequencies in the populations of N. rufocaudalis.
(F) LWS allele group frequencies in the populations of P. pundamilia.
In (B–E), Arabic numerals correspond to those in (A). The size of a pie indicates the number of haplotypes sequenced: N. greenwoodi: n = 58 at station 2; n = 10 at 3; n = 14 at 5; n = 10 at 7; n = 14 at 9; n = 22 at 11; n = 8 at 13; n = 8 at 14; n = 50 at 15, and N. omnicaeruleus, n = 40 at 10. M. mbipi: n = 36 at station 5; n = 32 at 7; n = 30 at 10; n = 10 at 11; n = 2 at 13. N. rufocaudalis: n = 42 at station 5; n = 16 at 6; n = 10 at 10; n = 10 at 16; n = 6 at 17; n = 4 at 18. P. pundamilia: n = 2 at station 1; n = 10 at 2; n = 4 at 3; n = 8 at 4; n = 18 at 5; n = 8 at 6; n = 6 at 8; n = 6 at 9; n = 10 at 10; n = 6 at 11; n = 8 at 12. The color of the sections of the pie indicates the frequency of allele groups L (red), M1 (yellow), M2 (green), H (blue), M3 (orange), P (blue-green), and other alleles (black). The amino acid differences among allele groups are shown for every species in the corresponding white panels.
We report the results on N. greenwoodi/omnicaeruleus first and then compare results on the three other species against them. We observed ten polymorphic sites (one synonymous, nine nonsynonymous) among the LWS sequences of N. greenwoodi/omnicaeruleus (Figure S2). From the bovine rhodopsin crystal structure [26], we inferred that two of the variable amino acid positions, 177 and 275, are located in the retinal binding pocket, and the others are distant from retinal. We focused on positions 177 (nucleotide site 529) and 275 (sites 823 and 824) and divided all observed alleles into four groups based on these two amino acid positions: The L group includes all alleles with 177S (529T) and 275I (823A and 824T). The H group includes all alleles with 177A (529G) and 275C (823T and 824G). The M1 group includes all alleles with 177A (529G) but 275I (823A and 824T). The M2 group includes all alleles with 177S (529T) but 275C (823T and 824G). M1 and M2 alleles can be considered recombinants of L and H alleles or intermediate alleles. The frequencies of allele groups in the ten populations are shown in Figure 1C.
In populations that live in very turbid water (<80 cm Secchi disk transparency), the L group alleles appeared to be fixed or almost fixed (station 2: number of L alleles [L] = 57, 98.3% and station 3: L = 10, 100%) (Figures 1C and S2). On the other hand, the H group alleles (H) appeared to be fixed in all the offshore island populations with high water transparency (≥180 cm; station 14: H = 8, 100%, station 15: H = 50, 100%, station 10: H = 40, 100%) (Figures 1C and S2). With one exception (one H group allele at station 2), the L and H allele groups were observed only in populations that live in waters of equal to or less than, and equal to or more than 85 cm Secchi transparency, respectively (Figure 1C). Mantel tests [27] of the significance of correlations between matrices of (i) pairwise population differentiation at the LWS locus, (ii) pairwise difference in water transparency, and (iii) pairwise geographical distance, revealed a highly significant positive correlation between LWS divergence and transparency (cross matrix correlation 0.80, p = 0.0008), but only a weak correlation between LWS divergence and geographical distance (0.55, p = 0.04), and an even weaker correlation between transparency and geographical distance (0.42, p = 0.08). The clinal change from L-dominated to H-dominated populations through populations dominated by the recombinant/intermediate allele groups M1 and M2 is consistent with gene flow between populations in a stepping stone fashion. We estimated the number of migrants per generation (M) from neutral marker FST (Table S1) [M = (1/ FST − −1)/4] as M = 5.6 between the most turbid inshore station 2 and the most clear offshore station 10, M = 7.6 between station 2 and the offshore Namatembi Island (station 15), and M = 14.5 between the two clear offshore stations 10 and 15. However, FST-derived estimates of M should be taken with great caution because they rely on the assumptions of Wright's island model [28] (all populations have the same population size and migration rate, are in migration/drift equilibrium, equally likely to give and receive migrants from all other populations, and the number of alleles is infinite) [29]. Our estimates of gene flow between the most distant and the most environmentally distinct islands may be taken as a clear indication that significant differentiation at the LWS locus between any two geographically closer populations on our transect requires divergent selection on LWS to overcome gene flow.
Divergent Selection Acting on LWS Gene
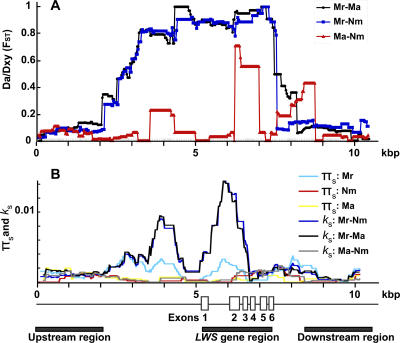
To search for the molecular signature of divergent natural selection between populations, we analyzed sequences up- and downstream of LWS. The DNA fragments including the LWS gene and its 5-kilobase (kb) upstream and 3.5-kb downstream flanking sequences (total 11 kb) were amplified by long PCR, and 10.5 kb were determined. We sequenced 18 alleles (from nine individuals of N. greenwoodi/omnicaeruleus) from each of the Marumbi (station 2), Makobe (station 10), and Namatembi (station 15) populations. Sliding window analysis revealed that population differentiation in the LWS gene region between populations from clear and turbid waters (FST > 0.8) (Figure 2A, Makobe-Marumbi, black line; Namatembi-Marumbi, blue line) was at least four times larger than in the regions of up- and downstream sequences (FST < 0.2) and 17 times larger than in unlinked neutral polymorphic loci (FST < 0.046: Table S1). Populations in clear and turbid waters are strongly differentiated from one another only in the sequences of the LWS gene region. The contrast between differentiation in LWS gene region versus flanking sequences was shown to be statistically significant by applying the McDonald test [30] (Makobe: p < 0.0001, Namatembi: p < 0.0001, Marumbi: p < 0.024, with the recombination parameter set to 2, 4, 10, 32, and 1000 replicates) and was further supported by HKA tests [31] revealing a significantly smaller ratio of within-population polymorphism to between-population divergence in the LWS gene region than in up- and downstream flanking regions (Table S2). The larger FST value can be caused either by stronger differentiation between populations or by reduced variation within populations [32]. In the present case, the differentiation is caused by larger divergence of the region between exon 1 and exon 3 compared to those in other regions (Figure 2B, Makobe-Marumbi, black line; Namatembi-Marumbi, blue line). This result strongly suggests action of divergent selection on the LWS gene, possibly caused by the difference between clear and turbid waters.
Figure 2. Detection of Selection Pressure on the LWS Gene.
The genome structure of the LWS gene and its flanking region and (A) sliding-window analysis of FST between Marumbi (Mr., station 2: N. greenwoodi), Namatembi (Nm., station 15: N. greenwoodi), and Makobe (Ma., station 10: N. omnicaeruleus) populations (Marumbi versus Makobe, Marumbi versus Namatembi, and Makobe versus Namatembi indicated by black, blue, and red lines, respectively).
(B) Sliding-window analysis of silent polymorphism (πs) in Marumbi (Mr: light blue), Namatembi (Nm: red), and Makobe (Ma: yellow), and silent divergence (ks) between Marumbi, Namatembi, and Makobe populations (Marumbi versus Namatembi, Marumbi versus Makobe, and Makobe versus Namatembi indicated by blue, black, and gray lines, respectively). π s and k s were calculated for segments of 700 bp in 25-bp intervals. The solid lines under the genome structure of LWS indicate the three regions used in HKA tests (Table S2). The LWS gene region is defined as the sequence between initiation codon and stop codon (2205 bp) of LWS. The up- and downstream regions are defined as the 5′ and 3′ ends of sequences of the same length as the LWS gene region.
In contrast, only weak population differentiation was observed throughout the sequences when we compared populations from similar water transparencies (Figure 2A, red line, Figure 2B, gray line: Makobe-Namatembi), and no evidence for divergent selection (HKA tests, Table S2). Hence, divergent selection has acted on the LWS alleles only between populations from different water transparencies, strongly implicating divergent adaptation to the different photic environments. The FST values in the LWS gene region imply fewer than one migrant every ten generations (<0.06 per generation), compared to about one migrant per generation in the up- and downstream regions, and six to eight at unlinked neutral loci. Hence, strong divergent selection effectively reduces migration in the LWS region compared to the rest of the genome by more than an order of magnitude. The FST values in 2–3 kb of sequences upstream of LWS were also higher than further up- and downstream, suggesting the possibility of genetic hitchhiking caused by the divergent selection on the downstream LWS gene, or further divergent selection on this region itself (Figure 2A, black and blue lines).
Parallel Divergent Adaptation of the LWS Gene
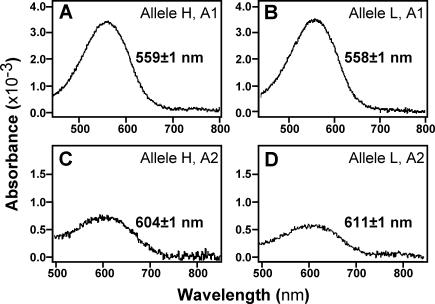
To determine the adaptive significance of the LWS divergence, we reconstituted the LWS pigments from both L and H alleles in vitro with both A1- and A2-derived retinal and measured their absorption spectra. The ratio of A1 to A2 chromophores in haplochromine cichlids was reported to be about ten to one [16]. We confirmed this ratio from laboratory-bred Lake Victoria cichlids and detected both chromophores in N. greenwoodi from the Mwanza Gulf by the method described previously [16]. Replacing an A1- by an A2-derived retinal shifts the λmax value to a longer wavelength [33]. Even though in other vertebrates a 7-nm shift was reported even with A1 pigments for the amino acid replacement at the position corresponding to cichlid LWS position 177 (position 180 in human red and green pigments [14]), we did not observe any difference between A1 pigments of H and L alleles (Figure 3A and 3B). However, we found that the peak spectral sensitivity (λmax) of the A2 pigment of the L allele was red shifted by 7 nm compared to that of the H allele (Figure 3C and 3D).
Figure 3. Absorption Spectra of the LWS Pigments Evaluated by the Dark–Light Difference Spectra.
The LWS pigments were reconstituted from (A) H allele with A1 retinal, (B) L allele with A1 retinal, (C) H allele with A2 retinal, and (D) L allele with A2 retinal. The λmax values are indicated with their standard errors.
The transmission light spectra at different water transparencies in Lake Victoria were measured previously. Highly transparent waters transmit broad spectra, whereas the dissolved and dispersed organic matter in turbid waters selectively scatters and absorbs light of short wavelengths, leading to a shift in spectral composition towards longer wavelengths [21]. Although we do not rule out other functional differences between H and L alleles [13], the fixation in red-shifted turbid waters of the LWS alleles of the L group, from which we reconstituted a red shifted A2 pigment, is likely an adaptation to a photic environment enriched in longer wavelengths.
Our results on the three other species corroborate this conclusion. In M. mbipi, the species that is ecologically most similar to N. greenwoodi (Figures 1B and S1), populations from high (258 cm Secchi, station 10, n = 30 alleles) and low (85 cm Secchi, station 5, n = 36 alleles) transparency were strongly differentiated in their LWS sequences too (Figure 1D, FST = 0.54, p < 0.001). Just like in N. greenwoodi, the clear water population of M. mbipi was fixed for H group alleles. Going into more turbid waters, the frequencies of these alleles stayed somewhat higher than in the somewhat deeper living sympatric N. greenwoodi. In contrast with N. greenwoodi, the M. mbipi population from the most turbid water in which this species was found (station 5) was dominated by a group of M3 alleles including all alleles with amino acid positions 177A (529G), 230T (688A), and 275I (823A and 824T) (Figures 1D and S3).
In N. rufocaudalis, a species that lives exclusively in very shallow waters (Figures 1B and S1), the allele of the H group that in the deeper living N. greenwoodi and M. mbipi dominated only in clear water populations dominated in all populations. One allele of the M2 group was found at a place of intermediate transparency, and the only two alleles of the L group came from the turbid end of this species' distribution range (Figures 1E and S4). In the cave-dwelling P. pundamilia, populations from high and low water transparency were identical and a different allele group P (including all alleles with 177A, 179V, 216F, 230T, and 275I) dominated (Figures 1F and S5). Hence, (i) strong divergent evolution at the LWS locus between populations occurred in two species from photic habitats that are strongly affected by variation in water transparency, but not in two species whose habitats are not strongly affected; (ii) the transparency-associated geographical cline in allele frequencies observed within two species was mirrored by a depth-associated cline in allele frequencies between the three species—N. rufocaudalis, M. mbipi, and N. greenwoodi—that live outside the rocky crevices (Figure 1B). The parallel divergent adaptation in N. greenwoodi and M. mbipi is likely to include some true parallel evolution at the gene level, because visual adaptation to turbid waters is achieved by substituting the clear water LWS allele H with different alleles in the two species (L and M3, Figure S6), which cannot be explained by very recent introgressive hybridization.
Correlation between Frequencies of LWS Allele and Male Nuptial Coloration
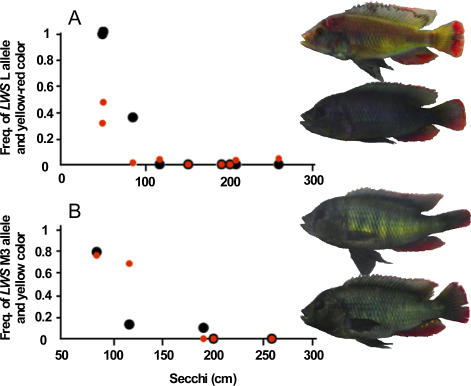
Nuptial color display is important in mate choice of Lake Victoria cichlids [21,34,35]. Female mating preferences may be affected by color vision [22,36], and closely related species with red versus blue male nuptial coloration possess LWS alleles with larger and smaller λmax respectively, where the difference in λmax is similar to the one observed here between L and H alleles [20]. It is therefore conceivable that the reciprocal fixation of LWS alleles with small and large λmax would cause populations at the ends of the water transparency cline to diverge in nuptial coloration, a first step towards parapatric ecological by-product speciation [37]. We collected and photographed a large number of males in breeding dress from all populations of N. greenwoodi/omnicaeruleus and M. mbipi and determined the frequencies of male nuptial color morphs in each population. Globally, most males of these species are blue or blue-black, but yellow-red (N. greenwoodi/omnicaeruleus) or yellow (M. mbipi) males occur at low, but variable frequencies in much of their ranges. In the relatively turbid waters of Lake Victoria, yellow and red light travel further than blue light, and yellow and red colors may hence be perceived as brighter than blue at long path length (deeper water) [22]. This effect is stronger the more turbid the water [21].
In populations of N. greenwoodi/omnicaeruleus, the frequency of yellow-red males ranged from 0%–47% (Figure 4A) and was strongly positively correlated with the frequency of relatively longer wavelength–shifted LWS alleles (cross matrix correlation between population differentiation at the LWS locus and in color morph frequency 0.87, Mantel test p = 0.019), and less strongly with water transparency (cross matrix correlation 0.58, p = 0.023). Both relationships were also strong in M. mbipi (Figure 4B, cross matrix correlation between difference in the frequency of yellow males and differentiation at the LWS locus 0.83, p < 0.001, with water transparency 0.93, p < 0.001), and populations at the extremes of the cline were almost fixed for the different colors. Hence we have evidence for parallel correlated divergent evolution in a visual gene and in male nuptial coloration in two species along the same environmental cline. Divergence in male nuptial coloration may be a consequence of divergent adaptation in the LWS gene. Although long wave–shifted male nuptial coloration became common, it has not become fixed in populations in which long wave–shifted LWS alleles got fixed. This may suggest that population divergence at the LWS locus alone is not necessarily sufficient for complete population divergence in male nuptial coloration. This may not be surprising, because several selection forces are known to operate on male nuptial coloration in haplochromine cichlids [21,34,35,38]. Our data suggest that evolution at opsin genes may influence the balance between these. It is possible that complete fixation of alternative nuptial colors requires reinforcement of mating preferences in sympatry, something we are currently testing with another data set.
Figure 4. The Relationship between the Frequency of LWS Alleles, Male Nuptial Color Morphs, and Water Transparency.
(A) The relationship between the frequency of long wavelength–sensitive allele group L at the LWS locus (black), yellow-red male nuptial color (red), and water transparency (Secchi disk [cm]) in N. greenwoodi. (B) The relationship between the frequency of allele group M3 at the LWS locus (black), yellow male nuptial color (red), and water transparency in M. mbipi.
The parallel divergence in LWS along the same environmental cline in two unrelated cichlid species suggests rapid divergent evolution under natural and sexual selection, may not be uncommon along the strong gradients in light environment that characterize Lake Victoria. The nucleotides in L, H, and M3 alleles at 529, 688, 823, and 824 also occur in other cichlid species in Lake Victoria [19], and the different alleles are not monophyletic within the species that we studied here (Figure S6). This suggests that the alleles originated prior to establishment of the species, and certainly prior to establishment of the intraspecific population structure that we investigated here, consistent with the fact that the island populations that we sampled cannot be older than 15,000 y independent of whether the lake was entirely dry during its last major low stand [9]. Hence, the intraspecific allelic diversity must have been acquired either from a single polymorphic ancestral population or through hybridization between species. Persistence of L/H allele polymorphisms seems rare in contemporaneous populations, and we found no evidence for L/M3 polymorphisms at all (Figures 1 and S6). It seems therefore more likely that locally favored LWS alleles are acquired by occasional hybridization in overlap zones between species adapted to different light environments (i.e., different water depths), a hypothesis that is consistent with our finding that the most common H group alleles in clear water populations of the unrelated species M. mbipi, N. greenwoodi, and N. rufocaudalis are sequence-identical (Figure S6). It is also consistent with the observation that P. pundamilia, the species with quite different habitat use, shares none of the LWS alleles with the other species.
Here, we have for the first time, to our knowledge, shown rapid evolution in African cichlid fish driven by strong divergent selection in a gene responsible for ecological adaptation, which also affects mate choice, consistent with incipient ecological by-product speciation that occurs in parallel in several unrelated species. The evolutionary history of the mutations involved, and the relevance of reticulate evolution in swapping locally advantageous mutations between species during the rapid adaptive radiation of Lake Victoria cichlids deserve the attention of evolutionary geneticists in the future.
Materials and Methods
Samples.
Specimens of N. greenwoodi, N. omnicaeruleus, M. mbipi, and P. pundamilia were collected by OS in 1995 and 1996, except for 17 of the 25 individuals from Namatembi, which were collected in 2005 by TS, HM, and SM, 11 of 18 of M. mbipi from Python, and 12 of 16 from Hippo Island, which were collected in 2005 by NK and HM, and eight of 15 M. mbipi from Makobe, which were collected in 2004 by HM, SM, and S Mzighani. Specimens of N. rufocaudalis were collected in 2004 and 2005 by SM, KT, and S. Mzighani. Identification of all specimens was verified by OS.
DNA sequencing.
Determination of the cichlid LWS gene was as described [19]. To determine the LWS flanking sequences, the BAC clone including the LWS gene was screened and isolated from a Lake Victoria cichlid BAC library [39]. The DNA of BAC clone was digested with HindIII and subsequently subcloned into pUC 19 plasmid vector. The HindIII fragment including the LWS gene was screened, and the sequence determined by the shot-gun sequencing method described elsewhere [40]. Based on the sequence of the HindIII fragment, we designed primers for PCR amplification and performed direct sequencing. The positions of primers are described above and below the schematic HindIII fragment (Figure S7). The DNA fragment including LWS gene and both flanking regions was amplified by LA (long and accurate) PCR (TaKaRa, Shiga, Japan) using primers LWSB_LF and LWSB_LR with genomic DNA (∼50 ng) as templates. Amplifications were carried out in the PTC-100 Programmable Thermal Controller (MJ Research, Massachusetts, United States). The PCR program consisted of a denaturation step for 3 min at 94 °C, followed by 30 cycles, each cycle consisting of 20 s denaturation at 98 °C, 15 min annealing and extension at 68 °C. The amplification product was then used as a template to amplify and sequence four overlapping fragments using the primers for upstream (LWSB_LF and LWSB_R3), (LWSB_F3 and LWSB_R5), LWS gene (LWSB_F5 and LWSB_R8), and downstream (LWSB_F8 and LWSB_LR) regions. These PCR products were purified and determined by direct sequencing with all primers described above and below the schematic HindIII fragment (Figure S7). Once determined, the sequences were connected by using the GENETYX-MAC Version 10.1 program. The sequences of primers are described in Figure S8.
Measurement of absorption spectra of cichlid LWS pigments.
The H allele sequence was amplified via RT-PCR using total RNA extracted from eyes of Lake Victoria cichlid as template with a pair of PCR primers (Eco6LWS and LWS_Flag_stop_NotI, Figure S8) designed to produce fusion protein with FLAG-tag (Sigma-Aldrich, St. Louis, Missouri, United States) in its C terminus. The amplified DNA fragments were digested with EcoRI and NotI and cloned into the EcoRI/NotI-digested (removing ID4) pMT5 expression vector [41]. In vitro mutagenesis of the LWS for construction of other sequences of alleles, expression, reconstitution, purification, and measurement were performed as described previously [16] with minor modifications. After pigment reconstitution, these experiments were performed under infrared light (>900 nm) with the vision of a digital video-camera in “night shot” mode (SONY, Tokyo, Japan) or complete darkness.
Matrix correlations.
We calculated the mean pairwise LWS sequence distance between populations as Dxy values (the average number of nucleotide substitutions per site between populations) using only the sites that were used for the identification of allele groups. We measured geographical distance as the shortest waterway distance between sites. We calculated the pairwise difference in water transparency and in the frequency of yellow-red or yellow males between sites. To evaluate the significance of correlations between these variables we used Mantel tests [27].
Population genetic analysis.
The DNA fragment including the LWS gene and its flanking sequence (10560 bp) was split into two putative alleles for the analysis by the program DnaSP 4.0 [42]. We performed sliding-window analysis to calculate approximate values for FST (D a/D xy [43]), πs , and k s in 700 bp windows, sliding the window in steps of 25 bp throughout the total 10.5-kb sequences. We performed the McDonald test for heterogeneity across a region of a DNA sequence in the ratio of polymorphism to divergence [30].
Supporting Information
The distribution of (A) P. pundamilia, (B) N. rufocaudalis, (C) M. mbipi, and (D) N. greenwoodi studied along a cline of water transparency (x-axis) and water depth (y-axis). Arrowheads indicate transparencies at which we sampled. Ranges of occurrence are hatched.
(231 KB PDF)
The nucleotide sites and amino acid positions are shown on top and below the alignment, respectively. Dots indicate where nucleotides are identical with those in the top line. The nucleotide sites 529, 823, and 824 are highlighted. Amino acids in the row directly under the amino acid positions indicate the amino acids translated from the sequence in the top line. Amino acids in the bottom line indicate amino acids substituted from the top line. The sampling station numbers are described in Figure 1A.
(294 KB PDF)
The nucleotide sites are shown on top of the alignment. n and s indicate nonsynonymous and synonymous sites, respectively. Dots indicate where nucleotides are identical with those in the top line. The sequences of N. omnicaeruleus from Makobe (N. omnicaeruleus Ma) and N. greenwoodi from Marumbi (N. greenwoodi Mr) are aligned at the bottom. The sampling station numbers are described in Figure 1A.
(52 KB PDF)
The nucleotide sites are shown on top of the alignment. n and s indicate nonsynonymous and synonymous sites, respectively. Dots indicate where nucleotides are identical with those in the top line. The sequences of N. omnicaeruleus from Makobe (N. omnicaeruleus Ma) and N. greenwoodi from Marumbi (N. greenwoodi Mr) are aligned at the bottom. The sampling station numbers are described in Figure 1A.
(50 KB PDF)
The nucleotide sites are shown on top of the alignment. n and s indicate nonsynonymous and synonymous sites, respectively. Dots indicate where nucleotides are identical with those in the top line. The sequences of N. omnicaeruleus from Makobe (N. omnicaeruleus Ma) and N. greenwoodi from Marumbi (N. greenwoodi Mr) are aligned at the bottom. The sampling station numbers are described in Figure 1A.
(64 KB PDF)
Maximum-likelihood analysis was performed with MOLPHY version 2.3 [44]. An NJ tree was used as the starting tree for a local rearrangement search for the ML tree with the HKY85 model [45]. The scale bar indicates the number of substitutions per site. Bootstrap values are shown at the branches. An alignment of all informative sites of alleles and the frequencies (%) of each allele are shown in right panel. Dots indicate where nucleotides are identical with those in the top line. The frequency of “other” alleles is not shown.
(41 KB PDF)
Arrows indicate the primers. The flanking sequences of the LWS gene were determined from a BAC clone.
(33 KB PDF)
(15 KB PDF)
(81 KB PDF)
(34 KB PDF)
Accession Numbers
The GenBank http://www.ncbi.nlm.nih.gov/Genbank) accession numbers for DNA sequences discussed in this paper are: AB221133–AB221343 and AB240064–AB240138.
Acknowledgments
We thank Mr. Mhoja Kayeba and Mr. Mohamed Haluna for technical assistance during fieldwork of OS, HM, and NK; Mr. A. Fujimoto (Kyushu University) for help with population genetic analysis; Dr. H. Nishihara (Tokyo Institute of Technology) for help with construction of phylogenetic tree of LWS alleles; Ms. K. Sakai (Kyoto University) for technical assistance; and the Tanzania Fisheries Research Institute (Professor POJ Bwathondi, Mr. EFB Katunzi, and S Mzighani) for research permits and hospitality. We also thank Dr. S Kawamura (Tokyo University) for pMT5 vector; Dr. RSH Liu (Hawaii University) for A2 retinal; and Dr. M Maan and Dr. MV Schneider for comments on the manuscript.
Abbreviations
- A1 chromophore
11-cis-retinal
- A2 chromophore
11-cis-3, 4-dehydroretinal
- bp
base pair
- kb
kilobase
Footnotes
Competing interests. The authors have declared that no competing interests exist.
Author contributions. Y. Terai was responsible for most of the laboratory work, data analysis, designing experiments, and writing. O. Seehausen was responsible for study conception, collection of most fish samples and ecological data, identification of all fish samples, data analysis, designing experiments, and writing. T. Sasaki was responsible for the determination of LWS sequences of N. rufocaudalis. K. Takahashi contributed the collection of N. greenwoodi at Namatembi and M. mbipi at Makobe and determination of LWS sequences of N. rufocaudalis. S. Mizoiri contributed the collection of N. greenwoodi at Namatembi and M. mbipi at Makobe. T. Sugawara provided development of the method for measurement of absorbance of reconstituted LWS pigments with Y. Terai. T. Sato assisted with collection of N. greenwoodi at Namatembi and M. mbipi at Makobe. M. Watanabe contributed the construction of a BAC library. N. Konijnendijk provided collection of M. mbipi at Python and Hippo islands; HDJ Mrosso performed collection of M. mbipi at Python and Hippo islands. H. Tachida supervised the population genetic analyses. H. Imai developed the method for measurement of absorbance of reconstituted LWS pigments with Y. Terai. Y. Shichida supervised measurement of absorbance of reconstituted opsin pigments. N. Okada provided study conception, supervision, and writing.
Funding. The Ministry of Education, Culture, Sports, Science and Technology of Japan (to NO), Tetra Germany and the Netherlands Foundation for the Advancement of Tropical Research (WOTRO), and the Swiss Science Foundation (to OS) supported this work.
References
- Schluter D. Ecology and the origin of species. Trends Ecol Evol. 2001;16:372–380. doi: 10.1016/s0169-5347(01)02198-x. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sunderland (Massachusetts): Sinauer Associates; 2004. 545 [Google Scholar]
- Gavrilets S. Fitness landscapes and the origin of species. Princeton: Princeton University Press; 2004. 476 [Google Scholar]
- Hawthorne DJ, Via S. Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature. 2001;412:904–907. doi: 10.1038/35091062. [DOI] [PubMed] [Google Scholar]
- Kronfrost MR, Young LG, Kapan DD, McNeely C, O'Neill RJ, et al. Linkage of butterfly mate preference and wing color preference cue at the genomic location of wingless. Proc Natl Acad Sci U S A. 2006;103:6575–6580. doi: 10.1073/pnas.0509685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder JA, Dyreson EG, O'Neill RJ, Spangler ML, Gupta R, et al. Contrasting modes of natural selection acting on pigmentation genes in the Drosophila dunni subgroup. J Exp Zoolog B Mol Dev Evol. 2004;302:469–482. doi: 10.1002/jez.b.21012. [DOI] [PubMed] [Google Scholar]
- Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G, Jr, Dickson M, et al. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- Kocher TD. Adaptive evolution and explosive speciation: The cichlid fish model. Nat Rev Genet. 2004;5:288–298. doi: 10.1038/nrg1316. [DOI] [PubMed] [Google Scholar]
- Johnson TC, Kelts K, Odada E. The Holocene history of Lake Victoria. Ambio. 2000;29:2–11. [Google Scholar]
- Nagl S, Tichy H, Mayer WE, Takezaki N, Takahata N, et al. The origin and age of the haplochromine species flock in Lake Victoria, East Africa. Proc Biol Sci. 2000;267:1049–1061. doi: 10.1098/rspb.2000.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SYW, Larson G. Molecular clocks: When times are a-changin. Trends Genet. 2006;22:79–83. doi: 10.1016/j.tig.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Joyce DA, Lunt DH, Bills R, Turner GF, Katongo C, et al. An extant cichlid fish radiation emerged in an extinct Pleistocene lake. Nature. 2005;435:90–95. doi: 10.1038/nature03489. [DOI] [PubMed] [Google Scholar]
- Shichida Y. Visual pigment: Photochemistry and molecular evolution. In: Toyoda J, editor. The retinal basis of vision. Amsterdam: Elsevier Science; 1999. 290 [Google Scholar]
- Yokoyama S. Molecular evolution of vertebrate visual pigments. Prog Retin Eye Res. 2000;19:385–419. doi: 10.1016/s1350-9462(00)00002-1. [DOI] [PubMed] [Google Scholar]
- Carleton KL, Harosi FI, Kocher TD. Visual pigments of African cichlid fishes: Evidence for ultraviolet vision from microspectrophotometry and DNA sequences. Vision Res. 2000;40:879–890. doi: 10.1016/s0042-6989(99)00238-2. [DOI] [PubMed] [Google Scholar]
- Sugawara T, Terai Y, Imai H, Turner GF, Koblmuller S, et al. Parallelism of amino acid changes at the RH1 affecting spectral sensitivity among deep-water cichlids from Lakes Tanganyika and Malawi. Proc Natl Acad Sci U S A. 2005;102:5448–5453. doi: 10.1073/pnas.0405302102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spady TC, Seehausen O, Loew ER, Jordan RC, Kocher TD, et al. Adaptive molecular evolution in the opsin genes of rapidly speciating cichlid species. Mol Biol Evol. 2005;6:1412–1422. doi: 10.1093/molbev/msi137. [DOI] [PubMed] [Google Scholar]
- Parry JW, Carleton KL, Spady T, Carboo A, Hunt DM, et al. Mix and match color vision: Tuning spectral sensitivity by differential opsin gene expression in Lake Malawi cichlids. Curr Biol. 2005;15:1734–1739. doi: 10.1016/j.cub.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Terai Y, Mayer WE, Klein J, Tichy H, Okada N. The effect of selection on a long wavelength-sensitive (LWS) opsin gene of Lake Victoria cichlid fishes. Proc Natl Acad Sci U S A. 2002;99:15501–15506. doi: 10.1073/pnas.232561099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton KL, Parry JW, Bowmaker JK, Hunt DM, Seehausen O. Colour vision and speciation in Lake Victoria cichlids of the genus Pundamilia. Mol Ecol. 2005;14:4341–4353. doi: 10.1111/j.1365-294X.2005.02735.x. [DOI] [PubMed] [Google Scholar]
- Seehausen O, van Alphen JJM, Witte F. Cichlid fish diversity threatened by eutrophication that curbs sexual selection. Science. 1997;277:1808–1811. [Google Scholar]
- Maan ME, Hofker KD, van Alphen JJ, Seehausen O. Sensory drive in cichlid speciation. Am Nat. 2006;167:947–954. doi: 10.1086/503532. [DOI] [PubMed] [Google Scholar]
- Boughman JW. How sensory drive can promote speciation. Trends Ecol Evol. 2002;17:571–577. [Google Scholar]
- Seehausen O, Lippitsch E, Bouton N, Zwennes H. Mbipi, the rock-dwelling cichlids of Lake Victoria: Description of three new genera and fifteen new species (Teleostei). Ichthyol. Explor. Freshw. 1998;9:129–228. [Google Scholar]
- Seehausen O, Bouton N. Microdistribution and fluctuations in niche overlap in a rocky shore cichlid community in Lake Victoria. Ecology of Freshwater Fish. 1997;6:161–173. [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Wright S. Evolution in a Mendelian population. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock MC, McCauley DE. Indirect measures of gene flow and migration: FST≠1/(4Nm+1) Heredity. 1999;82:117–125. doi: 10.1038/sj.hdy.6884960. [DOI] [PubMed] [Google Scholar]
- McDonald JH. Improved tests for heterogeneity across a region of DNA sequence in the ratio of polymorphism to divergence. Mol Biol Evol. 1998;15:377–384. doi: 10.1093/oxfordjournals.molbev.a025934. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. Measures of divergence between populations and the effect of forces that reduce variability. Mol Biol Evol. 1998;15:538–543. doi: 10.1093/oxfordjournals.molbev.a025953. [DOI] [PubMed] [Google Scholar]
- Harosi FI. An analysis of two spectral properties of vertebrate visual pigments. Vision Res. 1994;34:1359–1367. doi: 10.1016/0042-6989(94)90134-1. [DOI] [PubMed] [Google Scholar]
- Maan ME, Seehausen O, Söderberg L, Johnson L, Ripmeester EAP, et al. Intraspecific sexual selection on a speciation trait, male coloration, in the Lake Victoria cichlid Pundamilia nyererei. Proc Biol Sci. 2004;271:2445–2452. doi: 10.1098/rspb.2004.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maan ME, van der Spoel M, Quesada Jimenez P, van Alphen JJM, Seehausen O. Fitness correlates of male coloration in a Lake Victoria cichlid fish. Behav Ecol. 2006. In press.
- Boughman JW. Divergent sexual selection enhances reproductive isolation in sticklebacks. Nature. 2001;411:944–948. doi: 10.1038/35082064. [DOI] [PubMed] [Google Scholar]
- Endler JA. Geographic variation, speciation, and clines. Princeton: Princeton University Press; 1977. 246. [PubMed] [Google Scholar]
- Dijkstra PD, Seehausen O, Gricar BLA, Maan ME, Groothuis TGG. Can male-male competition stabilize speciation? A test in Lake Victoria haplochromine cichlid fish. Behav Ecol Sociobiol. 2006. In press.
- Watanabe M, Kobayashi N, Fujiyama A, Okada N. Construction of a BAC library for Haplochromis chilotes, a cichlid fish from Lake Victoria. Genes Genet Syst. 2003;78:103–105. doi: 10.1266/ggs.78.103. [DOI] [PubMed] [Google Scholar]
- Murata Y, Nikaido M, Sasaki T, Cao Y, Fukumoto Y, et al. Afrotherian phylogeny as inferred from complete mitochondrial genomes. Mol Phylogenet Evol. 2003;28:253–260. doi: 10.1016/s1055-7903(03)00035-6. [DOI] [PubMed] [Google Scholar]
- Khorana HG, Knox BE, Nasi E, Swanson R, Thompson DA. Expression of a bovine rhodopsin gene in Xenopus oocytes: Demonstration of light-dependent ionic currents. Proc Natl Acad Sci U S A. 1988;85:7917–7921. doi: 10.1073/pnas.85.21.7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sanchez-Del Barrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Inbreeding coefficients and coalescence time. Genet Res. 1991;58:159–175. doi: 10.1017/s0016672300029827. [DOI] [PubMed] [Google Scholar]
- Adachi J, Hasegawa M. MOLPHY version 2.3: Programs for molecular phylogenetics based on maximum likelihood. Tokyo: Institute of Statistical Mathematics; 1996. 150 [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The distribution of (A) P. pundamilia, (B) N. rufocaudalis, (C) M. mbipi, and (D) N. greenwoodi studied along a cline of water transparency (x-axis) and water depth (y-axis). Arrowheads indicate transparencies at which we sampled. Ranges of occurrence are hatched.
(231 KB PDF)
The nucleotide sites and amino acid positions are shown on top and below the alignment, respectively. Dots indicate where nucleotides are identical with those in the top line. The nucleotide sites 529, 823, and 824 are highlighted. Amino acids in the row directly under the amino acid positions indicate the amino acids translated from the sequence in the top line. Amino acids in the bottom line indicate amino acids substituted from the top line. The sampling station numbers are described in Figure 1A.
(294 KB PDF)
The nucleotide sites are shown on top of the alignment. n and s indicate nonsynonymous and synonymous sites, respectively. Dots indicate where nucleotides are identical with those in the top line. The sequences of N. omnicaeruleus from Makobe (N. omnicaeruleus Ma) and N. greenwoodi from Marumbi (N. greenwoodi Mr) are aligned at the bottom. The sampling station numbers are described in Figure 1A.
(52 KB PDF)
The nucleotide sites are shown on top of the alignment. n and s indicate nonsynonymous and synonymous sites, respectively. Dots indicate where nucleotides are identical with those in the top line. The sequences of N. omnicaeruleus from Makobe (N. omnicaeruleus Ma) and N. greenwoodi from Marumbi (N. greenwoodi Mr) are aligned at the bottom. The sampling station numbers are described in Figure 1A.
(50 KB PDF)
The nucleotide sites are shown on top of the alignment. n and s indicate nonsynonymous and synonymous sites, respectively. Dots indicate where nucleotides are identical with those in the top line. The sequences of N. omnicaeruleus from Makobe (N. omnicaeruleus Ma) and N. greenwoodi from Marumbi (N. greenwoodi Mr) are aligned at the bottom. The sampling station numbers are described in Figure 1A.
(64 KB PDF)
Maximum-likelihood analysis was performed with MOLPHY version 2.3 [44]. An NJ tree was used as the starting tree for a local rearrangement search for the ML tree with the HKY85 model [45]. The scale bar indicates the number of substitutions per site. Bootstrap values are shown at the branches. An alignment of all informative sites of alleles and the frequencies (%) of each allele are shown in right panel. Dots indicate where nucleotides are identical with those in the top line. The frequency of “other” alleles is not shown.
(41 KB PDF)
Arrows indicate the primers. The flanking sequences of the LWS gene were determined from a BAC clone.
(33 KB PDF)
(15 KB PDF)
(81 KB PDF)
(34 KB PDF)