Abstract
Spectrin is a multidomain cytoskeletal protein, the component three-helix bundle domains are expected to experience mechanical force in vivo. In thermodynamic and kinetic studies, neighboring domains of chicken brain α-spectrin R16 and R17 have been shown to behave cooperatively. Is this cooperativity maintained under force? The effect of force on these spectrin domains was investigated using atomic force microscopy. The response of the individual domains to force was compared to that of the tandem repeat R1617. Importantly, nonhelical linkers (all-β immunoglobulin domains) were used to avoid formation of nonnative helical linkers. We show that, in contrast to previous studies on spectrin repeats, only 3% of R1617 unfolding events gave an increase in contour length consistent with cooperative two-domain unfolding events. Furthermore, the unfolding forces for R1617 were the same as those for the unfolding of R16 or R17 alone. This is a strong indication that the cooperative unfolding behavior observed in the stopped-flow studies is absent between these spectrin domains when force is acting as a denaturant. Our evidence suggests that the rare double unfolding events result from misfolding between adjacent repeats. We suggest that this switch from cooperative to independent behavior allows multidomain proteins to maintain integrity under applied force.
INTRODUCTION
Spectrin is a multidomain protein that is an important component of the membrane-associated cytoskeleton, where it binds actin molecules. In erythrocytes, this membrane network is responsible for the generation and maintenance of cell shape. To fulfill this role, the spectrin matrix will undergo repeated cycles of deformation as the cell repeatedly passes through small blood vessels and then promptly regains its original biconcave shape; thus, it is likely that spectrin has the ability to resist unfolding under mechanical stress (1). Spectrin is composed of two large subunits, α-spectrin and β-spectrin, which associate in an antiparallel manner to form a heterodimer. Two heterodimers associate in a head-to-head interaction to form a tetramer (2). Both α- and β-spectrin are principally composed of multiple 106-amino-acid repeats (spectrin domains) that fold into a three-helix bundle. These repeating units are thought to allow spectrin to function in maintaining the flexibility and integrity of the erythrocyte membrane (3,4). This serial repeat structure is also apparent in the homologous proteins dystrophin and α-actinin (5,6). Isolated spectrin domains have been shown to be folded and stable in solution (4,7–10). They are joined by a short “linker” region. Recent structural studies have demonstrated that the last helix of one domain forms a continuous, well-structured helix with the first helix of the next domain (6,11–14). NMR studies of isolated chicken-brain α-spectrin domain R16 have shown that the last six residues at the C-terminal end are unstructured (10), whereas in the tandem repeat R1617 this region makes up a continuous helix with R17 (11) (Fig. 1).
FIGURE 1.
Structure of R1617. Ribbon diagram showing the crystal structure of the 16th and 17th α-helical repeats from chicken brain α-spectrin. Each helix is colored differently from the first helix of R16 (blue, N-terminus) to the third helix of R17 (red, C-terminus). The third helix of R16 and the first helix of R17 are colored green and they make up the linker, which appears helical in this structure. The figure was made using Deep View Swiss-PDB Viewer, version 3.7, and Pov-Ray, version 3.5, using the coordinates 1cun (11).
As spectrin domains have a mechanical function, how they respond to applied force has been the subject of both experimental studies and MD simulations (15–23). The results have been somewhat contradictory. The first AFM studies of forced unfolding of spectrin used a naturally occurring multidomain construct comprising domains R13–R18 from chicken brain α-spectrin. Domains unfolded independently at low forces, between 25 and 35 pN (at pulling speeds of 0.08–0.8 nm/ms), giving a gain in contour length (ΔLc) of ∼31 nm. The unfolding events were “all or none” processes (15). A few double-length cooperative unfolding events were observed and it was suggested that these might result from two domains unfolding at once due to stabilizing effects of adjacent domains. However, using an engineered construct of four R16 domains in tandem, two different elongation lengths were observed, the longest equaling the fully unfolded length of one spectrin domain and the shorter being half this length. The shorter elongation length was attributed to the presence of an intermediate, proposed to arise from the local unfolding of structure around a kink in helix B (16,17). It has been suggested that in this study the absence of cooperative unfolding events was due to the insertion of a long and flexible linker between each spectrin R16 module (18). Discher and co-workers studied the forced unfolding of tandem repeats of α-spectrin (α18–21) and β-spectrin (β1–4) and concluded that although there was no evidence of intermediate species in the unfolding of each single domain, there were long intervals between some force peaks. The length of these intervals corresponded to the unfolding of two domains in tandem (18). These cooperative events were attributed to the natural helical linker region between the domains. The same researchers showed that at low temperatures (10°C) the number of cooperative events between the domains increased, whereas at temperatures near the Tm of the protein (42°C) this number decreased (24). They suggested that the linker region is destabilized at high temperatures and without this being fully structured, the domains behave as independent units. This mirrors what was found in thermodynamic studies for a different domain pair, chicken brain α-spectrin R1617 (25). Recently, the possibility of two forced unfolding pathways existing for spectrin domains has been proposed from analysis of MD simulations; the distinction between them being whether the linker region between domains is structured or not (20).
This study examines the mechanical unfolding of a tandem pair of spectrin domains, R16 and R17 (R1617). No other domain pair of spectrin has been subject to such rigorous thermodynamic and kinetic analysis, so this domain pair seems an ideal candidate for examining cooperativity in mechanical unfolding in detail. Both domains alone are stable and fold by a two-state mechanism at equilibrium (26). The folding pathways have been investigated by protein-engineering Φ-value analysis and, in the case of R17, by MD simulations (27–29). The tandem repeat of spectrin, R1617, is more stable than either constituent domain alone and unfolds cooperatively, as a single unit, at 25°C (9), although this cooperativity is lost at higher temperatures (45°C), in the presence of salt and where the linker region is altered by mutation (25). The domains fold more rapidly and unfold more slowly in the tandem construct (30).
None of the previous studies of spectrin domain forced unfolding have compared the unfolding of tandem pairs with the unfolding of individual domains. In this study, we compare the forced unfolding of the individual domains R16 and R17 with the unfolding of the tandem pair R1617. Since the helical nature of the linker is believed to be important in maintaining cooperativity between the domains (18,24,25,31), care was taken when making the multidomain AFM constructs to ensure that the spacers between the domains were not helical, as nonnatural helical linkers could, possibly, act equally well. Thus, each spectrin domain, or pair of spectrin domains, was separated by an all-β immunoglobulin domain, TI I27. Our results show unequivocally that although R16 and R17 unfold cooperatively in folding experiments in the absence of force, there is no evidence that specific domain-domain interactions, mediated by the helical linker, induce cooperativity in forced unfolding. We suggest that absence of cooperativity in forced unfolding of multidomain proteins will be physiologically advantageous.
MATERIALS AND METHODS
Protein preparation
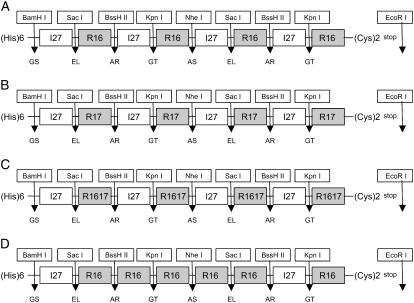
A modified version of an existing versatile cloning system (32) was used as the basis for making spectrin-I27 polyproteins. Four spectrin modules (of R16, R17, or R1617) were introduced into the constructs, each flanked by I27 domains to provide a nonhelical linker between the spectrin domains (Fig. 2). All eight modules had different restriction sites incorporated at their ends, to allow specific arrangement of the modules within the construct. Modules 1–4 and 5–8 were constructed separately, sequenced, and then ligated together. A fourth polyprotein was constructed with five R16 domains adjacent to each other in tandem array (Fig. 2).
FIGURE 2.
AFM polyprotein constructs. Schematic diagram showing how the polyproteins were constructed. (A–C) Each spectrin domain is flanked by TI I27 domains (I27). The spectrin domains were either R16, R17, or tandem R1617. (D) In the polyprotein with repeating R16 domains, (R16)5, domains 2–6 were all R16 domains (see text). Each domain is separated by a two-residue linker (shown in single-letter code below the domains) encoded by the unique restriction sites (shown above the domains). Modules 1–4 and 5–8 were cloned and sequenced separately before being ligated together (32,39).
Protein expression was carried out in Escherichia coli C41 cells (33). Transformed cells were grown at 37°C to an A600 of 0.4–0.6 before induction and growth overnight at 26°C. Cells were harvested and lysed by sonication. Proteins were isolated from the soluble fraction after centrifugation by affinity chromatography on Ni2+-agarose resin. Bound proteins were subsequently eluted from the resin in 250 mM imidazole and further purified by size-exclusion chromatography on a G200-superdex column (Pharmacia Biotech, Uppsala, Sweden). Proteins were either used immediately or stored for short periods of time at 4°C in 0.01% sodium azide and filtered before use.
Stability measurements
Urea and guanidinium chloride-induced equilibrium denaturation was performed to ensure that the spectrin domains were folded in the AFM polyproteins as described previously (26,34,35). Unfolding was monitored by fluorescence spectroscopy using a Perkin Elmer spectrometer. The protein concentration was between 1 and 2 μM (total domain concentration), and 5 mM DTT was added for all studies of R17-containing proteins. Proteins were excited at a wavelength of 280 nm with the emission being monitored from 320 to 350 nm. All experiments were carried out at 25°C in 50 mM phosphate, pH 7.0. The data were fitted to a two-state transition as described by Pace (36).
Atomic force microscopy experiments
AFM experiments were all conducted in phosphate buffer (pH 7.0) at ambient temperature (20–25°C) on an Asylum Research (Santa Barbara, CA) molecular force probe. Gold-coated silicon nitride Biolever cantilevers (Olympus, Tokyo, Japan) were used to measure the unfolding forces of spectrin domains as they are specifically manufactured for the force measurement of biosamples, being small and soft with low spring constants. Helical spectrin domains have been shown to have much lower mechanical stability than titin, unfolding at forces in the region of 30 pN (15), and so are more easily studied using these small cantilevers rather than longer v-shaped silicon nitride cantilevers, where the unfolding peaks for spectrin are barely distinguishable from the background thermal fluctuations (results not shown).
The cantilevers were calibrated in the experimental buffer with the spring constant determined by the standard thermal fluctuation method (37). The spring constants for the cantilevers used were between 4.5 and 7.5 pN/nm. We allowed 100 μl of freshly prepared and filtered protein solution (with an OD280 of 0.5 cm−1) in buffer to adsorb onto a freshly evaporated gold-coated slide for 10 min. The excess was then washed off and the bound protein covered by a fresh layer of phosphate buffer.
Force traces for all the polyproteins were collected at a range of pulling speeds from 300 nm/s to 2500 nm/s to determine the pulling-speed dependence of the force at which the domains unfolded. At least 40 “good” peaks at each speed were collected and analyzed using three different cantilevers to ensure sufficient sampling. The data were analyzed using Igor Pro software (Wavemetrics, Lake Oswego, OR). The final “pull-off” baseline was used to determine the baseline for analyzing the force traces (38).
RESULTS
Spectrin and titin domains are folded and stable within the multidomain polyproteins. Equilibrium denaturation experiments were conducted in both urea and guanidinium chloride to verify that the spectrin and the I27 domains, respectively, were folded and stable in the polyproteins. Analysis of the denaturation curves showed that the spectrin domains in the polyproteins were slightly stabilized compared to their monomeric counterparts, manifested by a small increase in [D]50%. I27 domains in all multimers had the same stability, within error, as isolated domains (data not shown.)
Spectrin unfolds at much lower forces than titin
Force measurements were taken at pulling speeds of 300 nm/s, 600 nm/s, 1000 nm/s, and 2500 nm/s. Traces to be analyzed were selected according to standard criteria (38,39), and all the peaks from these traces were used to determine the unfolding force and the change in contour length upon each spectrin domain unfolding. Sample force-extension curves collected for each protein, showing domain unfolding events, are shown in Fig. 3.
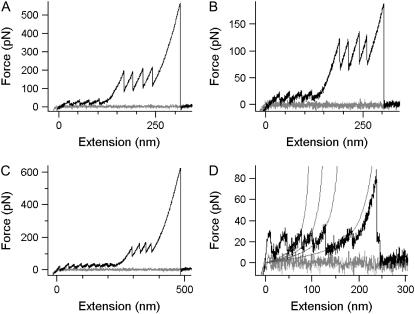
FIGURE 3.
Representative force extension curves. (A) R16 polyprotein, recorded at 1000 nm/s. (B) R17 polyprotein, recorded at 600 nm/s. (C) R1617 polyprotein, recorded at 1000 nm/s. (D) (R16)5 polyprotein recorded at 1000 nm/s. In A–C, both spectrin unfolding events (low force) and TI I27 unfolding events (high force) can be seen. The soft Biolever cantilevers do not allow full relaxation to the baseline after the high-force unfolding of the TI I27 domains. (D) The peaks are shown fitted to the WLC model. One of the unfolding events resulted in an increase in length of double the normal contour length (ΔLc).
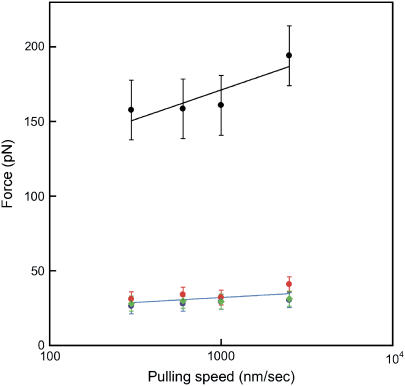
Spectrin domains were clearly distinguishable from the mechanically stronger titin domains, unfolding at less than half the force that I27 can withstand. In Fig. 4 the modal unfolding force is plotted versus the log of the pulling speed. The slope of this plot gives an indication of the position of the transition state relative to the ground states (xu). The xu for spectrin domains is significantly larger than for TI I27 (∼18 vs. 3 Å), as indicated by the shallower slope of the spectrin unfolding forces in the force spectrum (Fig. 4).
FIGURE 4.
Force spectra. The means of the modal unfolding force (from at least 3 days of different measurements) are shown for spectrin peaks R16 (red), R17 (blue), and R1617 (green), and for TI I27 (black). The error bars show ±1 standard deviation. The spectrin unfolding forces are the same for all proteins, within error.
Most importantly, the unfolding forces of R16, R17, and R1617 were the same, within error, at all pulling speeds (Fig. 4).
Spectrin domains unfold as single units
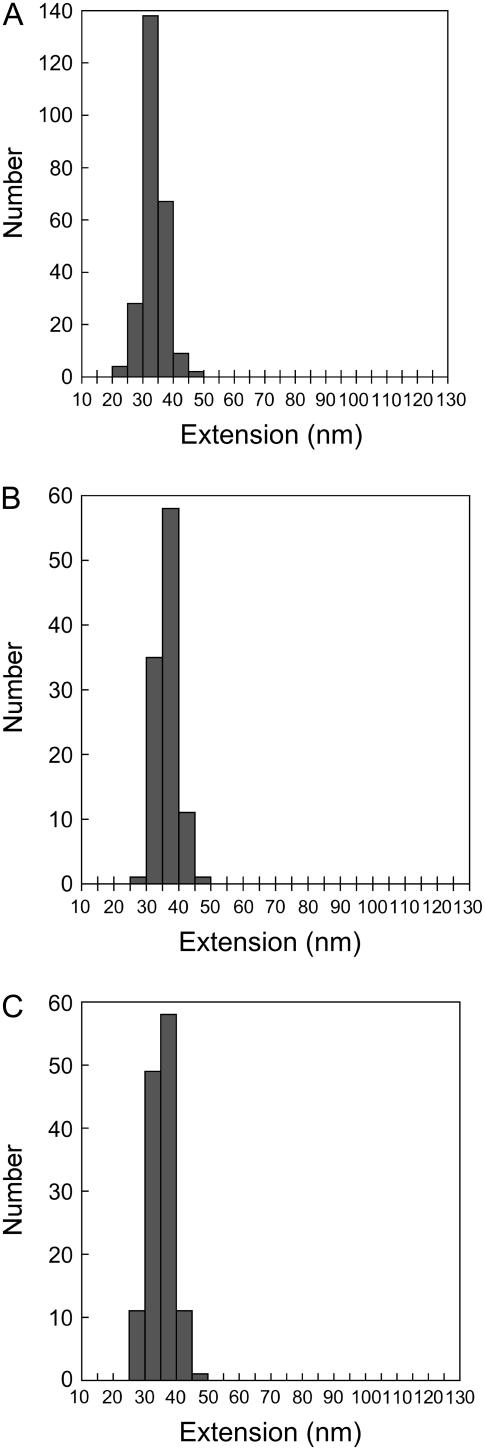
The change in the contour length upon each domain unfolding was determined by fitting the WLC model of polymer elasticity to each of the spectrin peaks. Unlike TI I27, spectrin is not easily modeled by the WLC, as spectrin domains unfold at much lower forces and so have a very short baseline to fit. We found that when we used a persistence length (p) of 0.38 nm, the fits to the WLC were better than at p = 0.8 nm, the length used by Rief et al. (15). Nonetheless, the unfolding of the spectrin domains in all of the multimers caused a modal increase in contour length (ΔLc) of ∼34 nm (see Fig. 5), similar to that observed previously (15). This change in contour length is what would be expected for the full unfolding of a single 116-amino-acid spectrin domain (note that our single spectrin domains are 10 residues longer than the defined boundaries of MacDonald and Pozharski (9) to ensure that the entire domain was incorporated (26)). Although there were no double-length unfolding events in R16 and R17, a few were observed in R1617; however, these accounted for only 3% of the total (only 14 in a total of 478 unfolding events).
FIGURE 5.
Changes in contour length (ΔLc) on unfolding the spectrin domains. The distribution of ΔLc on the unfolding of spectrin domains (A) R16, (B) R17, and (C) R1617. The modal contour length is the same, within error, for all three polyproteins. All data shown were collected at a single pulling speed (1000 nm/s).
A few “double” unfolding events are observed in a polyprotein where R16 domains are expressed in tandem
To investigate whether the few double unfolding events observed in R1617 were the result of specific interactions between the two domains, a fourth polyprotein was constructed (Fig. 2) with five spectrin R16 domains in tandem. Data were acquired for this protein at 1000 nm/s only. The spectrin unfolding forces were the same as for all the other proteins in this study. There were, however, a few longer unfolding events (∼8% of unfolding events, 21 out of 278) (see, for example, Fig. 3). These events occurred at forces similar to those of the other unfolding events.
DISCUSSION
It is important, in analyzing specific cooperative interactions between neighboring domains, that the behavior of a tandem domain protein is compared to each domain in isolation. Our polyprotein constructs were carefully designed to ensure that the linkers between the spectrin domains were nonhelical. Since the spectrin domains always unfold at significantly lower forces than the TI I27 domains, we are completely sure that each spectrin domain is separated from another by a fully structured all-β domain in the AFM experiments. The equilibrium studies showed clearly that the spectrin domains were fully folded and stable in the polyproteins.
Cooperativity is not seen under force
In traditional thermodynamic and kinetic studies of R1617, compared to R16 and R17 alone, both domains are seen to be stabilized by their neighbors and much of this stabilization arises from a significant reduction in the rate of unfolding. i.e., the height of the transition state barrier to unfolding, relative to the native state, is significantly higher in the tandem construct than in either domain alone (25,30). This could be due to specific stabilization of the native state or, more unlikely, to destabilization of the transition-state structures. Only a single unfolding phase is seen in these stopped-flow experiments on R1617, because unfolding of the first domain (R17) precipitates the rapid unfolding of the second domain (R16) (30). Thus, if there is cooperative unfolding in the AFM experiments, due to specific domain-domain interactions, possibly propagated through the natural linkers, we would expect to see two signatures: 1) an increase in the modal unfolding forces of R1617 compared to R16 or R17 alone; and, if two domains unfold simultaneously, 2) an increase in contour length equivalent to the unfolding of two domains (as was observed for α-spectrin 18–21 and β-spectrin 1–4 in the experiments of Discher and co-workers (18)).
Our results demonstrate unequivocally that neither of these signatures was observed. The unfolding forces of R1617 were the same as those of R16 and R17 alone and the number of double unfolding events in R1617 was negligible (3% of unfolding events). Moreover dependence of the force on pulling speed was the same for all three proteins. There is no indication that the pathway of forced unfolding is altered in R1617.
Rare double unfolding events may indicate misfolding
There were a very few (14 out of 478) apparently double unfolding events in R1617—all at forces within the range of those observed for the single unfolding events. (i.e., the few “double” unfolding events that were observed cannot “reflect a certain stabilizing effect of [the] adjacent domain,” as suggested by Rief et al. (15), since they occurred at the same unfolding forces as the domains on their own.) Such double unfolding events have been observed at similar frequencies in the unfolding of TI I27 and tenascin (40,41). These have been associated with interdomain misfolding after release of force. This misfolding is more possible in the R1617 polyprotein than in R16 or R17 because the unfolded spectrin domains are adjacent. Is this the origin of these rare events? It has been shown that such misfolding in a tandem repeat protein is more likely to occur if the adjacent domains are identical in sequence (42). To investigate this possibility we examined the folding of a polyprotein with five spectrin domains in tandem, (R16)5. We would expect to see more misfolding in this construct than in the R1617 construct, since now the neighboring domains are identical (42). This is indeed what we observed. Where there were a number of R16 domains in tandem, there were a small but significant number of unfolding events with larger-than-expected ΔLc measurements, ∼8% (21 out of 278), consistent with more than one domain unfolding at a time, and consistent with a hypothesis that interdomain misfolding might explain the very few double unfolding events seen in R1617.
Spectrin is a complex molecule
In contrast to the results presented here, Discher and co-workers (18) identified cooperative behavior (tandem repeats unfolding together) in two different systems: α-spectrin repeats 18–21 and β-spectrin repeats 1–4. They found that these events were frequent, with the probability of a tandem unfolding event occurring almost the same as that of a single unfolding event occurring. This result is inconsistent with the misfolding hypothesis presented for spectrin R1617—unless these domains are remarkably prone to interdomain misfolding, the systems do indeed appear to behave differently. Interestingly, although they did not compare their results with domains in isolation, the force required to unfold these tandem repeats was found to be the same as for unfolding of a single repeat, which is not what we would have per se predicted for a cooperative system; we would expect the stabilization of one domain by another to result in a higher unfolding force. Law et al. address the question of possible different behavior in different spectrin repeats in their manuscript. They note that both their constructs comprise domains from the ends of the spectrin molecule; they suggest that “the cooperativity seen in the end domains … may prove more atypical than typical of spectrin.” (18). Our results would certainly support this hypothesis, and they indicate that the mechanical properties of an entire protein might not be simply represented by looking at just one or two domains.
CONCLUSION
R1617 is the most well characterized of all tandem repeats in spectrin, with thermodynamic, kinetic, and AFM data on both single and tandem proteins. Our single-molecule AFM results make it clear that kinetic cooperativity does not extend to forced unfolding. Titin domains have similarly been shown to unfold independently under force (43,44) (although in this case they are also independent (noncooperative) in kinetic and thermodynamic studies (45)).
This independent behavior under applied force offers an obvious advantage to a mechanical protein in vivo: the forced unfolding of one domain will not promote the unfolding of adjacent domains, resulting in an unfolding cascade. Such extensive unfolding events in intact spectrin would leave the protein vulnerable to misfolding once the force is released, and would compromise its functionality. This is in contrast to the cooperativity observed for spectrin repeats in the stopped-flow experiments in the absence of force. These show that should a single domain of spectrin unfold under force, it will recover much more rapidly once the force is released than if it folded independently. In the case of R1617, for instance, a folded R16 domain accelerates the folding of R17 some 30-fold (30). Interestingly, it has also been estimated that the folded R16 domain will slow the (thermal) unfolding of R17 to increase the half-life from ∼30 min to ∼15 h, which may be important for a protein of the red blood cell, which has a lifetime of ∼120 days (30,46).
Thus, we suggest that the capacity of spectrin domains to switch between cooperative and independent behavior is important for its in vivo role as a structural protein. The cooperative switching observed could, potentially, allow rapid and repeated work cycles of the spectrin matrix without compromising its mechanical properties.
Acknowledgments
We thank Julia Forman and Sean Ng for technical advice.
This work was supported by the Wellcome Trust and the Biotechnology and Biological Sciences Research Council (BBSRC). J.C. is a Wellcome Trust Senior Research Fellow. L.G.R. holds a BBSRC research studentship.
Abbreviations used: R1617, the 16th and 17th α-helical repeats from chicken brain α-spectrin in a tandem construct; R16, the 16th α-helical repeat from chicken brain α-spectrin; R17, the 17th α-helical repeat from chicken brain α-spectrin; TI I27, the 27th Ig-like domain from the I-band of human cardiac titin, a muscle protein; AFM, atomic force microscopy; MD, molecular dynamics; WLC, wormlike chain.
References
- 1.Elgsaeter, A., B. T. Stokke, A. Mikkelsen, and D. Branton. 1986. The molecular basis of erythrocyte shape. Science. 234:1217–1223. [DOI] [PubMed] [Google Scholar]
- 2.McGough, A. M., and R. Josephs. 1990. On the structure of erythrocyte spectrin in partially expanded membrane skeletons. Proc. Natl. Acad. Sci. USA. 87:5208–5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Speicher, D. W., and V. T. Marchesi. 1984. Erythrocyte spectrin is comprised of many homologous triple helical segments. Nature. 311:177–180. [DOI] [PubMed] [Google Scholar]
- 4.DeSilva, T. M., S. L. Harper, L. Kotula, P. Hensley, P. J. Curtis, L. Otvos, Jr., and D. W. Speicher. 1997. Physical properties of a single-motif erythrocyte spectrin peptide: a highly stable independently folding unit. Biochemistry. 36:3991–3997. [DOI] [PubMed] [Google Scholar]
- 5.Koenig, M., A. P. Monaco, and L. M. Kunkel. 1988. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 53:219–226. [DOI] [PubMed] [Google Scholar]
- 6.Ylanne, J., K. Scheffzek, P. Young, and M. Saraste. 2001. Crystal structure of the α-actinin rod reveals an extensive torsional twist. Structure. 9:597–604. [DOI] [PubMed] [Google Scholar]
- 7.Winograd, E., D. Hume, and D. Branton. 1991. Phasing the conformational unit of spectrin. Proc. Natl. Acad. Sci. USA. 88:10788–10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ursitti, J. A., L. Kotula, T. M. DeSilva, P. J. Curtis, and D. W. Speicher. 1996. Mapping the human erythrocyte β-spectrin dimer initiation site using recombinant peptides and correlation of its phasing with the α-actinin dimer site. J. Biol. Chem. 271:6636–6644. [DOI] [PubMed] [Google Scholar]
- 9.Macdonald, R. I., and E. V. Pozharski. 2001. Free energies of urea and of thermal unfolding show that two tandem repeats of spectrin are thermodynamically more stable than a single repeat. Biochemistry. 40:3974–3984. [DOI] [PubMed] [Google Scholar]
- 10.Pascual, J., M. Pfuhl, D. Walther, M. Saraste, and M. Nilges. 1997. Solution structure of the spectrin repeat: a left-handed antipararallel triple-helical coiled-coil. J. Mol. Biol. 273:740–751. [DOI] [PubMed] [Google Scholar]
- 11.Grum, V. L., D. Li, R. I. Macdonald, and A. Mondragon. 1999. Structures of two repeats of spectrin suggest models of flexibility. Cell. 98:523–535. [DOI] [PubMed] [Google Scholar]
- 12.Djinovic-Carugo, K., P. Young, M. Gautel, and M. Saraste. 1999. Structure of the alpha-actinin rod: molecular basis for cross-linking of actin filaments. Cell. 98:537–546. [DOI] [PubMed] [Google Scholar]
- 13.Kusunoki, H., R. I. MacDonald, and A. Mondragon. 2004. Structural insights into the stability and flexibility of unusual erythroid spectrin repeats. Structure. 12:645–656. [DOI] [PubMed] [Google Scholar]
- 14.Kusunoki, H., G. Minasov, R. I. Macdonald, and A. Mondragon. 2004. Independent movement, dimerization and stability of tandem repeats of chicken brain α-spectrin. J. Mol. Biol. 344:495–511. [DOI] [PubMed] [Google Scholar]
- 15.Rief, M., J. Pascual, M. Saraste, and H. E. Gaub. 1999. Single molecule force spectroscopy of spectrin repeats: low unfolding forces in helix bundles. J. Mol. Biol. 286:553–561. [DOI] [PubMed] [Google Scholar]
- 16.Lenne, P. F., A. J. Raae, S. M. Altmann, M. Saraste, and J. K. H. Horber. 2000. States and transitions during forced unfolding of a single spectrin repeat. FEBS Lett. 476:124–128. [DOI] [PubMed] [Google Scholar]
- 17.Altmann, S. M., R. G. Grunberg, P. F. Lenne, J. Ylanne, A. Raae, K. Herbert, M. Saraste, M. Nilges, and J. K. H. Horber. 2002. Pathways and intermediates in forced unfolding of spectrin repeats. Structure. 10:1085–1096. [DOI] [PubMed] [Google Scholar]
- 18.Law, R., P. Carl, S. Harper, P. Dalhaimer, D. W. Speicher, and D. E. Discher. 2003. Cooperativity in forced unfolding of tandem spectrin repeats. Biophys. J. 84:533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhasin, N., R. Law, G. Liao, D. Safer, J. Ellmer, B. M. Discher, H. L. Sweeney, and D. E. Discher. 2005. Molecular extensibility of mini-dystrophins and a dystrophin rod construct. J. Mol. Biol. 352:795–806. [DOI] [PubMed] [Google Scholar]
- 20.Ortiz, V., S. O. Nielsen, M. L. Klein, and D. E. Discher. 2005. Unfolding a linker between helical repeats. J. Mol. Biol. 349:638–647. [DOI] [PubMed] [Google Scholar]
- 21.Paci, E., and M. Karplus. 2000. Unfolding proteins by external forces and temperature: the importance of topology and energetics. Proc. Natl. Acad. Sci. USA. 97:6521–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paramore, S., G. S. Ayton, D. T. Mirijanian, and G. A. Voth. 2006. Extending a spectrin repeat unit. I. Linear force-extension response. Biophys. J. 90:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paramore, S., G. S. Ayton, and G. A. Voth. 2006. Extending a spectrin repeat unit. II. Rupture behavior. Biophys. J. 90:101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Law, R., G. Liao, S. Harper, G. L. Yang, D. W. Speicher, and D. E. Discher. 2003. Pathway shifts and thermal softening in temperature-coupled forced unfolding of spectrin domains. Biophys. J. 85:3286–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Batey, S., L. G. Randles, A. Steward, and J. Clarke. 2005. Cooperative folding in a multi-domain protein. J. Mol. Biol. 349:1045–1059. [DOI] [PubMed] [Google Scholar]
- 26.Scott, K. A., S. Batey, K. A. Hooton, and J. Clarke. 2004. The folding of spectrin domains I: wild-type domains have the same stability but very different kinetic properties. J. Mol. Biol. 344:195–205. [DOI] [PubMed] [Google Scholar]
- 27.Scott, K. A., L. G. Randles, and J. Clarke. 2004. The folding of spectrin domains II: φ-value analysis of R16. J. Mol. Biol. 344:207–221. [DOI] [PubMed] [Google Scholar]
- 28.Scott, K. A., and J. Clarke. 2005. Spectrin R16: broad energy barrier or sequential transition states? Protein Sci. 14:1617–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott, K. A., L. G. Randles, S. J. Moran, V. Daggett, and J. Clarke. 2006. The folding pathway of spectrin R17 from experiment and simulation: using experimentally validated MD simulations to characterize states hinted at by experiment. J. Mol. Biol. 359:159–173. [DOI] [PubMed] [Google Scholar]
- 30.Batey, S., K. A. Scott, and J. Clarke. 2006. Complex folding kinetics of a multidomain protein. Biophys. J. 90:2120–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacDonald, R. I., and J. A. Cummings. 2004. Stabilities of folding of clustered, two-repeat fragments of spectrin reveal a potential hinge in the human erythroid spectrin tetramer. Proc. Natl. Acad. Sci. USA. 101:1502–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steward, A., J. L. Toca-Herrera, and J. Clarke. 2002. Versatile cloning system for construction of multimeric proteins for use in atomic force microscopy. Protein Sci. 11:2179–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289–298. [DOI] [PubMed] [Google Scholar]
- 34.Rounsevell, R. W. S., A. Steward, and J. Clarke. 2005. Biophysical investigations of engineered polyproteins: implications for force data. Biophys. J. 88:2022–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fowler, S. B., R. B. Best, J. L. Toca Herrera, T. J. Rutherford, A. Steward, E. Paci, M. Karplus, and J. Clarke. 2002. Mechanical unfolding of a titin Ig domain: structure of unfolding intermediate revealed by combining AFM, molecular dynamics simulations, NMR and protein engineering. J. Mol. Biol. 322:841–849. [DOI] [PubMed] [Google Scholar]
- 36.Pace, C. N. 1986. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 131:266–280. [DOI] [PubMed] [Google Scholar]
- 37.Hutter, J. L., and J. Bechhoefer. 1993. Calibration of atomic force microscope tips. Rev. Sci. Instrum. 64:1868–1873. [Google Scholar]
- 38.Rounsevell, R. W. S., J. R. Forman, and J. Clarke. 2004. Atomic force microscopy: mechanical unfolding of proteins. Methods. 34:100–111. [DOI] [PubMed] [Google Scholar]
- 39.Ng, S. P., L. G. Randles, and J. Clarke. 2006. Single molecule studies of protein folding using atomic force microscopy. In Protein Folding Protocols (Methods in Molecular Biology series). Y. Bai, and R. Nussinov, editors. Humana Press, Totowa, NJ. 139–167. [DOI] [PubMed]
- 40.Oberhauser, A. F., P. E. Marszalek, M. Carrion-Vazquez, and J. M. Fernandez. 1999. Single protein misfolding events captured by atomic force microscopy. Nat. Struct. Biol. 6:1025–1028. [DOI] [PubMed] [Google Scholar]
- 41.Oberhauser, A. F., P. E. Marszalek, H. P. Erickson, and J. M. Fernandez. 1998. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 393:181–185. [DOI] [PubMed] [Google Scholar]
- 42.Wright, C. F., S. A. Teichmann, J. Clarke, and C. M. Dobson. 2005. The importance of sequence diversity in the aggregation and evolution of proteins. Nature. 438:878–881. [DOI] [PubMed] [Google Scholar]
- 43.Rief, M., M. Gautel, F. Oesterhelt, J. M. Fernandez, and H. E. Gaub. 1997. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 276:1109–1112. [DOI] [PubMed] [Google Scholar]
- 44.Li, H. B., A. F. Oberhauser, S. B. Fowler, J. Clarke, and J. M. Fernandez. 2000. Atomic force microscopy reveals the mechanical design of a modular protein. Proc. Natl. Acad. Sci. USA. 97:6527–6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scott, K. A., A. Steward, S. B. Fowler, and J. Clarke. 2002. Titin: a multidomain protein that behaves as the sum of its parts. J. Mol. Biol. 315:819–829. [DOI] [PubMed] [Google Scholar]
- 46.Shemin, D., and D. Rittenberg. 1946. The life span of the human red blood cell. J. Biol. Chem. 166:627–636. [PubMed] [Google Scholar]