Abstract
Metal ions are crucial for nucleic acid folding. From the free energy landscapes, we investigate the detailed mechanism for ion-induced collapse for a paradigm system: loop-tethered short DNA helices. We find that Na+ and Mg2+ play distinctive roles in helix–helix assembly. High [Na+] (>0.3 M) causes a reduced helix–helix electrostatic repulsion and a subsequent disordered packing of helices. In contrast, Mg2+ of concentration >1 mM is predicted to induce helix–helix attraction and results in a more compact and ordered helix–helix packing. Mg2+ is much more efficient in causing nucleic acid compaction. In addition, the free energy landscape shows that the tethering loops between the helices also play a significant role. A flexible loop, such as a neutral loop or a polynucleotide loop in high salt concentration, enhances the close approach of the helices in order to gain the loop entropy. On the other hand, a rigid loop, such as a polynucleotide loop in low salt concentration, tends to de-compact the helices. Therefore, a polynucleotide loop significantly enhances the sharpness of the ion-induced compaction transition. Moreover, we find that a larger number of helices in the system or a smaller radius of the divalent ions can cause a more abrupt compaction transition and a more compact state at high ion concentration, and the ion size effect becomes more pronounced as the number of helices is increased.
INTRODUCTION
Nucleic acid (DNA and RNA) structures and structural changes are critical for biological functions (1,2). Due to the polyanionic nature of the nucleic acid molecules, the thermal stability and folding kinetics of nucleic acids are strongly dependent on the ionic condition of the solution (1–14). A rudimentary question in nucleic acid folding is how the negatively charged helices pack against each other to form a compact 3D structure, and how different ions and ion concentrations affect the structure and stability of the helix assembly. Experiments have suggested that multivalent ion-mediated force plays an important role in DNA condensation (15–28) and compaction of other polyelectrolyte molecules (29–32). Moreover, extensive experiments have demonstrated the particularly efficient role of multivalent ions in RNA compaction (33–39). For example, it was found that [Mg2+] in millimolar (mM) or [Na+] in molar (M) can stabilize a compact intermediate state (33–44), and the compact state induced by Mg2+ is more compact and ordered (39,41).
Recent experimental studies have significantly advanced our understanding of RNA-ion interactions. For example, several experiments demonstrated that cations with higher valency and smaller size are more efficient in RNA-folding and such ions could also slow down the folding by stabilizing possible misfolded states (8,33–36). Moreover, a series of experiments indicated that charge neutralization of the nucleotide backbone may be responsible for the initial compaction of RNA (39–43). A recent small angle X-ray scattering (SAXS) measurement for a simple two-helix system suggested the existence of an attractive inter-helix force at high [Mg2+] and a repulsive force at very low [Mg2+] (45). Furthermore, a different SAXS experiment for loop-tethered helix system showed that the inter-helix force at high [Mg2+] is not sufficient to cause a collapsed state for the system (46). Based on the (mean-field) non-linear Poisson-Boltzmann (PB) theory, a thermodynamic framework for Mg2+–RNA interaction suggested that the predominant electrostatic contribution to RNA-folding stability comes from the diffusively bound ions (9,46–48).
Inspired by the experiments on the ion effect in nucleic acid folding (15–43,45,49), we investigate the ion-dependence of the folding free energy landscape for a paradigm system: two or three loop-tethered helices immersed in an ionic solution. The motivation of the present study is to quantitatively understand (i) the physical mechanism of the ion-induced compaction of the helix assembly and (ii) how the length and the electrostatic properties of loops, the multiplicity of the helices, and the ion size determine the stability and structural changes of the system. The free energy landscape, which gives the complete free energy distribution in the conformational space, enables detailed study of the physical mechanism of the ion-induced helix–helix assembly, such as the stability and the compactness of the collapsed state and the cooperativity of the compaction transition.
One of the key issues in the present study is the ion-mediated helix–helix interactions (19,20,28,33). Previous computer simulations (50–54) and analytical calculations (55–60) for similar systems reveal the existence of helix–helix attraction in the presence of multivalent ions (31,51). However, these previous studies are mostly based on simplified nucleic acid models and do not take into account the effects of the tether loops and the added salt in the solution. The mean-field PB theory (61–65) predicts no helix–helix attraction even for multivalent ion solutions. The counterion condensation theory (66) can predict the inter-helix attraction arising from the increased ion entropy (67–69) for both monovalent and multivalent ions.
Recently, we developed a statistical mechanical model, the tightly bound ion (TBI) model (70–72). The advantage of the model is that it can account for correlations and fluctuations, i.e. different ion-binding modes, for the bound ions. The model is based on realistic helical structure of nucleic acids and takes into account the effect of the added salt in the supporting solutions and the realistic ion sizes (70–72).
MATERIALS AND METHODS
Structural model
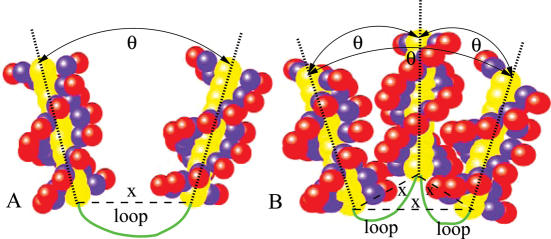
We focus on two model systems: two helices tethered by a semiflexible loop [Figure 1A and (49)] and three helices tethered by two loops (Figure 1B). To simplify the calculation, we treat only symmetric helix–helix configurations and assume that the axes of the helices are co-planar (2D). As a result, the system can be characterized by two structural parameters: the angle θ and the distance x between the helices. The helices are assumed to adopt the B-form structure and are described by the grooved primitive model (70–73), where the charged phosphate groups, the neutral groups and the ion-inaccessible helix core regions are represented by charged spheres, neutral spheres and ion-inaccessible cylinders, respectively. The coordinates and sizes of the spheres and the cylinders are determined from the experimental atomic coordinates of a B-form helix; see Supplementary Data. We choose the above described two- (or three-) helix system because (i) the system is simple thus ideal for detailed analysis and (ii) there are available experimental results for a similar system (49).
Figure 1.
Symmetric configurations for two (A) and three (B) DNA helices tethered by one (or two) semiflexible loop(s) can be characterized by two structural parameters: the angle θ between the helical axises and the end-end distance x. The DNA helices are produced from the grooved primitive model (see Materials and Methods) (70,72,73).
Free energy landscape
For a configuration described by (x, θ) for the nucleic acid model, the free energy G (x, θ) is calculated as
| 1 |
where GE (x, θ) is the electrostatic free energy for the helices immersed in ion solutions and Gloop (x, L) (L = loop length) is the entropic free energy of the loop(s).
We compute GE (x, θ) from TBI model (70–72), which explicitly accounts for the correlation and fluctuation of the bound ions, and we obtain Gloop (x, L) from analytical expressions from polymer theories (74):
| 2 |
where Ploop (x, L), the probability of finding a loop with end-end distance x is given by an analytical expression derived from the polymer theory (74): where A is thenormalization factor. We note that calculations based on a different analytical formula for Ploop (x, L) (75) would give very close results for our calculations.
A loop of polyethylene or PEG chain used in the experiment (49) is neutral and its persistence length is ion-independent. In contrast, a realistic polynucleotide loop is polyanionic and its persistence length lP (and the loop entropy) is sensitive to the ionic condition (76–79). We account for the ion-dependence of the entropy of a polynucleotide loop through an empirical ion-dependent persistence length derived from the experimental measurements (76–79).
The distribution of free energy G (x, θ) (Equation 1) for different (x, θ) states gives the free energy landscape of the system. We explore the landscape for conformations described by x ∈ [22 Å, 60 Å] and θ ∈ [0°, 180°]. Such an ensemble of conformations would include a broad range of different types of conformations, including the compact, the random and the extend conformations. We choose 22 Å as the lower limit for x because this is the closest inter-axis distance of approach between two helices. In the actual depiction of the landscape, we scan the (x, θ) space with increments of 2 Å and 20° for x and θ, respectively. For each given (x, θ) state, we enumerate all the accessible 3D conformations through the rotation of the planar rigid body with the fixed x and θ. We use such an ensemble of 3D conformations to compute the free energy G (x, θ).
TBI model
For a given helix–helix configuration (x, θ), we calculate the electrostatic free energy using the TBI model. The development of the TBI model is motivated by the need to accurately account for Coulombic and excluded volume correlations between bound ions and between bound ions and helices as well as the fluctuations of the ion-binding modes (70–72); see Supplementary Data and Refs (70–72) for details.
In the TBI model, according to the ion–ion correlation strength, we classify the tightly bound (= strongly correlated) and the diffusive (= weakly correlated) ions/regions (70–72). We treat diffusive ions using non-linear PB and treat the tightly bound ions separately by exhaustively enumerating all the possible distributions of the tightly bound ions (= ion-binding modes). Each mode M is described by a set of numbers with each number mi equal to the number of the tightly bound ions surrounding the ith phosphate. For each mode, we compute the partition function ZM by evaluating the energy and entropy of the diffusive ions, the tightly bound ions, and the interactions between the diffusive and the tightly bound ions (70–72). In the calculation, the many-particle Coulombic and excluded volume correlations between the tightly bound ions and between the tightly bound ions and the helices are accounted for. Sum over all the possible ion-binding modes gives the total partition function for a given nucleic acid structure, from which the electrostatic free energy is obtained.
| 3 |
We assume Na+ and Mg2+ ions are hydrated and have radii 3.5 Å and 4.5 Å (70–72), respectively. When solving the non-linear PB, we add a thin charge-free layer of one cation radius to the molecular surface to account for the volume exclusion of the cations (61,70–72). The dielectric constant ɛ is assumed to be 12 [experimental value (80)] for the helix interior and 78 (of bulk water at 25°C) elsewhere in the solution.
SAXS profile
SAXS can effectively reveal the shape and size of macromolecules (42–49,81,82). To compute the SAXS profile I (Q) with the grooved primitive helix model, we use the following equation for I (Q) (83):
| 4 |
where M0 is the total number of the charged and neutral spheres in the model, and rij is the distance between two spheres. The scattering vector Q is equal to 4πsin(θ)/λ, where θ is the Bragg angle and λ is the wavelength of the radiation. f is the scattering factor (84) , where Q is in the range between 0 and Qmax, the largest scattering vector in reciprocal space (84). For simplicity, we use the same scattering factor f (Q) for all spheres in the model (84). Qmax is taken as 3 Å−1 in the calculations. Our tests with different Qmax (≳2 Å−1) give quite robust results.
RESULTS AND DISCUSSIONS
We consider a system of loop-tethered DNA helices immersed in an ionic solution. Each helix is assumed to have a finite length of 10 bp. To simplify the analysis, we assume that the helices are symmetrically oriented, so the system can be characterized by two structural parameters: angle θ and distance x between the helices (see Figure 1). The stability of a state described by (x, θ) is determined by its free energy G (x, θ) (see Equation 1). G (x, θ) for all the possible (x, θ) states with x ∈ [20 Å, 60 Å] and θ ∈ [0°, 180°] gives the free energy landscape.
From the free energy landscape G (x, θ), we can identify all the possible stable states, which are represented by the minima on the landscape, and the transitions between the different stable states. We characterize the compactness of the helix assembly by a Boltzmann-weighted mean value for the distance rg between the centers of the helix pair:
| 5 |
As described in Methods, in our calculation, we choose x ∈ [22 Å, 60 Å] and θ ∈ [0°, 180°] with increments of 2 Å and 20° for x and θ, respectively.
According to the compactness of the structure, we distinguish three types of states: (i) the extended state E for configurations with large x and large θ, (ii) the random fluctuation state R for configurations with random x and θ and (iii) the collapsed state C for configurations with x ∈ [22, 26]Å and θ ∈ [0°, 40°] (69). Our calculation shows that the collapsed state C has a mean compactness of , which agrees with the upper limit for the helix–helix separation for a collapsed (aggregated) DNA array (18–28). It is important to note that such an electrostatically collapsed state is less compact than the final RNA native structure (44), where further compaction can be induced by specific tertiary interactions that are ignored in the present study.
The key to understanding ion-induced structural transition is the free energy landscape G (x, θ). As shown in Equation 1 in ‘Methods’, G (x, θ) of the system consists of two parts: (i) the electrostatic free energy GE (x, θ) defined as the loop-free free energy for the helices and the ionic solution and (ii) the loop-free energy Gloop (x) for the loop conformational entropy. Before including the loop effect, we first focus on the effect of the electrostatic free energy landscape GE (x, θ).
Without the tether loop
Na+ ions
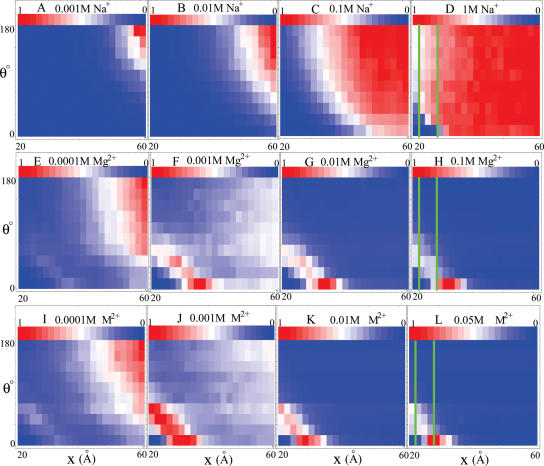
In Figure 2A–D we show density plots for the electrostatic free energy landscapes GE (x, θ) for two helices immersed in a Na+ solution. At very low [Na+] (∼1 mM), the favorable low-free energy state is the extended state E with the largest x and θ so that the repulsive helices can maximally avoid each other. As [Na+] is increased, the favorable state would ‘smear out’ to reach the region of smaller x and θ as shown in Figure 2B and C for 0.01 M and 0.1 M NaCl, respectively. When [Na+] becomes very high, such as 1 M, the favorable conformations can spread over nearly the whole region of x and θ. In Figure 3A, the solid lines show the compactness as a function of [Na+]. We find that the increase of [Na+] can induce the compaction process. The rapid decrease in occurs for [Na+] 0.3 M. For [Na+] >0.3 M, the helix–helix system approaches the random fluctuation state R. Physically, as [Na+] increases, with the decrease in the entropic penalty for ion-binding, more Na+ ions can bind to the helices, resulting in a weakened repulsion between the helices. The predictions show that Na+ can cause a random fluctuation state at high [Na+] (42–49), and the random fluctuation state is more compact than the electrostatically repulsive unfolded state at low [Na+]. Such high concentration Na+-induced compaction has been observed in experiments for RNAs (33–43). For larger RNA molecules, experiments also show that Na+ can induce the partial formation of some native tertiary contacts (33–43).
Figure 2.
The normalized probability exp[−GE (x, θ)/kBT] for loop-free helix–helix system. Here GE (x, θ) is the electrostatic free energy landscape for different (x, θ) configurations in a solution of NaCl (A–D), MgCl2 (E–H), or divalent ion (M2+) with (small) radius = 3.5 Å (I–L). The red and blue colors represent the low and high free energies, respectively.
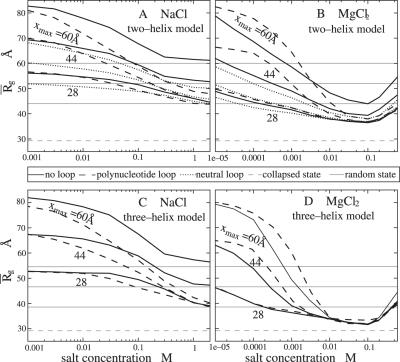
Figure 3.
(A and B) Ion concentration-dependence of the compactness (= the mean distance between helix centers) for two helices without loop (solid lines), with neutral polyethylene loop (dotted lines) and polynucleotide loop (dashed lines) in NaCl (A) and MgCl2 (B) solutions. (C and D) The compactness for three helices. Results for different end-to-end distance maximum (= loop length L): xmax = 60 Å, 44 Å and 28 Å (from the upper to lower) are plotted, respectively. In A–D, the gray solid lines denote the compactness of the random fluctuation states R for different xmax's and gray dashed lines denote the compactness of the collapsed state C. The compactness is computed from the random averaging over x ∈ [22,26]Å and θ ∈ [0°, 40°] (69) for state C and x ∈ [22 Å, xmax] and θ ∈ [0, θmax] (θmax = 180° for a two-helix system and 120° for a three-helix system) for state R. We note that for state C, and the result is in agreement with the helix–helix separation for an (least compact) aggregated DNA array (18–28).
In each panel of Figure 3, we show three curves, each corresponding to a different constraint for the maximum helix–helix distance xmax. Physically, xmax is determined by the length of the linker (loop) that tethers the helices. The results for different xmax's indicate how the spatial constraint from the finite loop length affects the two-helix electrostatic free energies and the resultant compactness of the system. In general, for low ion concentration, the two-helix configuration tends to become extended. So the compactness is more sensitive to xmax (= the upper limit of the extension). Larger xmax causes a more extended state. For high ion concentration, the system becomes more compact and thus the compaction is less sensitive to xmax. As a result, larger xmax causes a more abrupt (ion-induced) change in the compactness, i.e. a larger xmax causes a sharper compaction transition.
Mg2+ ions
For [Mg2+] below 1 mM, as [Mg2+] is increased, due to the stronger charge screening and consequently the weakened repulsion, helices become more compact as shown by the shift of the free energy minima from Figure 2E to F.
For [Mg2+] above 1 mM, more ions are bound and the correlation effect becomes strong. The bound ions can self-organize to reach correlated low-free energy configurations. Such correlated energies decrease rapidly as helices approach each other, leading to an inter-helix attractive force (45,72). Higher [Mg2+] causes more bound ions and stronger inter-helix attraction (72), and hence a more compact state (as shown by the shift of the free energy minima from Figure 2F to H and the change of the compactness in Figure 3B).
Because the inter-helix attraction is strongest for two parallel helices (53), the free energy minima occur at θ = 0 in Figure 2F–H. As shown in Figure 3B, the Mg2+-induced compact state is more compact than the random fluctuation state R but less compact than the collapsed state C. Such effect is more notable for larger xmax; see Figure 3 for the curves for three different xmax. Moreover, for very close approach between the helices, the parallel helices would repel each other due to the Coulombic repulsions between helices and crowded bound ions (72). In such close-packing case, the helices could be stabilized through rotation in order to minimize the repulsion; see the (rotated) local minimum at (θ, x) ≃ (40°, 22 Å) in Figure 2H. As [Mg2+] exceeds 0.1 M, the helices may be over-neutralized by the Mg2+ ions and the compaction becomes saturated and close to the random fluctuation state (R) (especially for smaller xmax), due to the Coulombic and excluded volume repulsions between the bound Mg2+ ions (27,28,72).
Both Na+ and Mg2+ can facilitate helix–helix compaction, but through different mechanisms. High concentration of Na+ causes compaction through strong ionic screening for helix charges, while Mg2+ ions can cause the compaction through helix–helix attraction (72). Moreover, Na+-induced compact state is heterogeneous as shown in Figure 2D while Mg2+-induced compact states are ordered as shown in Figure 2G and H.
The overwhelming efficiency of Mg2+ over Na+ is much more pronounced for tertiary than secondary structure folding. For example, we find that for a 9 bp single helix, 10 mM Mg2+ and 1 M Na+ can provide the same stability (71). For helix–helix assembly as a tertiary fold, however, our results show that a 10-fold dilute [Mg2+], i.e. 1 mM [Mg2+], would be sufficient to provide the same stability as 1 M Na+. This suggests that Mg2+ tends to promote the formation of the tertiary structure so the free energy can be more efficiently lowered by the Mg2+ ions. The result is in accordance with the experiments on Na+ and Mg2+-mediated RNA compaction (33–43). It is important to note that the ‘anomalous’ efficiency of the Mg2+ cannot be merely attributed to the more efficient charge screening, as shown by the PB-predicted electrostatic free energy landscape for Mg2+ in Supplementary Figure 9, which gives a disordered and much less compact state even at high [Mg2+].
It is important to note that, however, the Mg2+-mediated helix–helix interaction is not strong enough and x0, the inter-helix distance at the free energy minimum, is not small enough to cause a collapsed state; see Supplementary Figure 10. In Supplementary Figure 10, we show the magnitude of ion-mediated helix–helix interaction in NaCl and MgCl2 solutions. For example, ΔGE = GE − GE (extended state) for 0.1 M Mg2+ is the lowest at (x0, θ) ≅ (32 Å, 0°) with ΔGE ∼ −3.3kBT. As the helices further approach each other (with x < x0), ΔGE for the parallel helices increases, and the minimum ΔGE is shifted from parallel helices (θ = 0) to a rotated helix–helix configuration with θ ≠ 0 in order to avoid electrostatic repulsion between the helices. For example, for x = 24 Å, the minimum ΔGE (∼−2.4kBT) occurs for a V-shaped conformation with θ = 40°. The rotation angle is larger for smaller x. For example, the minimum ΔGE (most stable configuration) occurs at θ ≃ 60° for x = 20 Å. When Mg2+ exceeds 0.1 M, the Mg2+-mediated attraction becomes weaker due to the increased likelihood for over-neutralization (70,72). For example, for [Mg2+] = 0.6 M, the free energy minimum occurs at (x0, θ0) ≃ (32 Å, 0°) with ΔGE ∼ −2.7kBT, which is weaker than −3.3kBT, the minimum free energy for 0.1 M Mg2+. Similar multivalent ion-induced destabilization has been observed experimentally (22,27,28,30,53). For example, packed DNA assembly has been found to be dissolved by high multivalent ion concentration (22,27,28). The predicted ΔGE is slightly stronger than the experimentally derived value (49). The slightly overestimated stability in the model may be caused by the restriction of (2D) co-planar and symmetric conformations for the two-helix assembly and the possible change in the hydration state of the helices and ions.
With a neutral tether loop
To investigate the loop effect, we consider a system of two helices tethered by a polyethylene glycol (PEG) loop with persistence length lP ∼ 3.8 Å (49,85). We obtain the total free energy G(x, θ) from Equation 1 with the loop-free energy Gloop(x, L) determined from Equation 2 for each given configuration (x, θ) for two helices tethered by the loop. We first compare our prediction with the experimental measurements for a similar nucleic acid model (49). To model the short loop in the experiment (49), we assume loop length L = 26 Å. The loop size defines the maximum helix–helix end-end distance: x ≤ xmax = 26 Å. We calculate the SAXS profiles from Equation 4 for each given (x, θ). The Boltzmann-weighted average by factor e−G(x,θ)/kBT for all the possible states (x, θ) gives the mean SAXS profile:
| 6 |
where the conformational sampling covers the full conformational space described by x ∈ [22 Å, 60 Å] and θ ∈ [0°, 180°] with increments of 2 Å and 20°, respectively.
Short neutral loop and comparisons with experiment
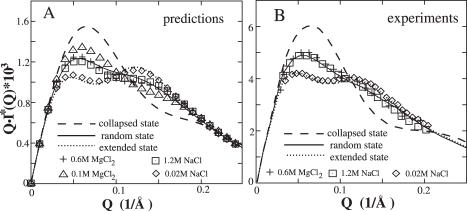
As shown in Figure 4, the predicted SAXS profiles for low (20 mM) and high (1.2 M) [Na+] and for high (0.6 M) [Mg2+] are close to the experimental profiles for a similar nucleic acid model (49). For comparison, in Figure 4A we also show the predicted profile for 0.1 M Mg2+. The state in 0.1 M Mg2+ is (slightly) more compact than that the random state in 0.6 M Mg2+ but much less compact than the collapse state.
Figure 4.
The SAXS profiles for the two-helix model with neutral PEG loop for different ionic conditions: diamond 20 mM NaCl; plus 0.6 M MgCl2 and square 1.2 M NaCl. (A) The predicted SAXS profiles weighted by the free energy G(x, θ) (see Materials and Methods); (B) The experimental SAXS profiles (49). The dashed lines, solid lines and dotted lines represent the standard SAXS profiles for the collapsed C, random R, and extended E states of the tethered helices, respectively.
Figure 4 shows close SAXS profiles for 1.2 M [Na+] and 0.6 M [Mg2+], while Figure 2 shows very different free energy landscape for Na+ and Mg2+ ionic conditions. How can such seemingly contrasting results be consistent with each other? The SAXS profile I*(Q) is a weighted average over all the available conformations (see Equation 5). As shown in Figure 2D and H as examples, for a given loop length of 26 Å, the accessible conformational space on the free energy landscape is limited to the region with x ≤ 26 Å. Though the full (unrestricted) landscape would certainly yield drastically different I*(Q), the restricted accessible region of the landscape may give similar I*(Q). Physically, within the restricted conformational space, the conformational distribution for high [Na+] and high [Mg2+] are both quite inhomogeneous over a wide-range of the angle θ between the helices.
It is important to note that the above prediction-experiment comparisons involve several limitations of the present theory: (i) The two-helix configuration is assumed to be symmetric and planar. Including the non-planar/non-symmetric configurations would likely weaken the predicted helix packing stability and compactness. (ii) The loop (linker) in the present model connects two helices at the helical axes instead of the helix edges in the experimental model (49). Such difference causes no significant effects for long loops (linkers) but can play a role for shorter loops.
Furthermore, the length of L = 26 Å used in the theoretical model and the experimental system may cause additional effects. As shown in Figure 2A–H, the most stable (lowest free energy) configuration occurs at inter-helix distance >26 Å. As a result, all the configurations considered in the model and in experiment (with x ≳ xmax = L = 26 Å) would result in ΔGE higher than the lowest free energy. Therefore, the short loop would effectively reduce the electrostatic force in the compaction process. Moreover, short loops would significantly restrict the accessible conformations of the extended state, resulting in a reduced conformational difference between the compact and the extended states, and hence a reduced sharpness of the compaction transition. For the same reason, longer loops can lead to sharper compaction transition; see Figure 3.
In summary, our results agree with the experimental finding that the Mg2+-mediated electrostatic interaction cannot induce the electrostatically collapsed state C for the two-helix system. Although our predicted helix–helix attraction is slightly stronger than the value derived from experiments, the predicted compact state is only slightly more compact than the random state but much less compact than the collapsed state C. A longer loop causes a sharper compaction transition. Furthermore, as we discuss below, a polynucleotide loop (instead of an electrically neutral loop), which has an ion-dependent flexibility and entropy, would also lead to a sharper transition. Nevertheless, despite the different types of loops considered, we find the compact state is much less compact than the collapsed state.
Long neutral loop
To further investigate the loop effect, we consider longer polyethylene (-CH2-) tether loops (49) with lP ∼ 6 Å (86,87). As shown in Figure 3 for different loop length (maximum helix–helix end-to-end distance), the loop promotes compaction in both Na+ and Mg2+ solutions in order to gain larger loop entropy, which occurs for smaller x. For example, at 10 mM [Mg2+] in Figure 2F, our free energy landscape shows that the inter-helix distance x for the most stable state decreases from 34 Å (without loop) to 32 Å with a loop of length L = 60 Å. As the loop length is decreased, the loop entropy effect is weakened because of the reduced conformational entropy of the loop (74,75).
As a caveat, we note that if the loop is too short, the helices would be restricted to the small x region to form a rotated state, which is less compact (larger ) than the parallel state, as discussed above.
With a polynucleotide loop
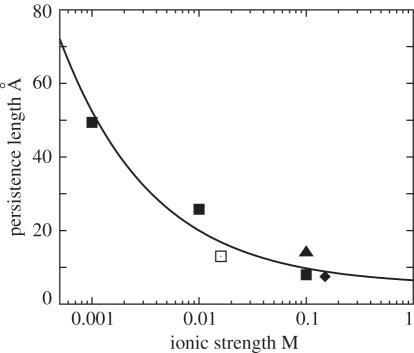
As described in Materials and Methods, a realistic polynucleotide loop is polyanionic. Unlike neutral loops, such as polyethylene or PEG chains, polynucleotide persistence length lP and the loop entropy are sensitive to the ionic condition (76–79). Based on the experimental data for the ion-dependence of lP (76–79), we obtain an empirical expression for lP as a function of the ionic strength I (in M) in the solutions (see Figure 5).
| 7 |
The above expression is mainly derived from the experimental data for Na+ due to the limited experimental data for lP in Mg2+ solution. As an approximation, we use Equation 7 for both Na+ and Mg2+ solutions.
Figure 5.
The persistence length lP of a single-stranded polynucleotide chain as a function of ionic strength I. The solid line is the fitted function Equation 7. The symbols are the experimental data: filled square (76), filled diamond (77) and filled triangle (78) are for Na+ solutions and open square (79) is for mixed Na+/Mg2+ solution.
As shown in Figure 3A and B for the ion-dependence of the compactness, for low ion concentration the loop is stiff (with large lP, so the system is much more extended than the neutral loop case, especially for large loops. For high ion concentration, the loop is flexible (with small lP) (76), so the helices collapse to a state as compact as the neutral polyethylene loop case. Therefore, the polynucleotide loop greatly enhances the sharpness of the compaction transition. Such effect is much more pronounced for Mg2+ due to the stronger ion concentration-dependence of lP. The Mg2+-induced compaction transition occurs in the [Mg2+] range of 1–10 mM. In addition, since Equation 7, which is fitted from the experimental data mainly for Na+, may underestimate the [Mg2+]-dependence of lP (79), the realistic [Mg2+]-induced transition could be even sharper than that predicted here.
With more helices and loops
Since realistic RNA-folding often involves multiple (>2) helices, it would be useful to investigate how the multiplicity of helices affects the compaction process. We consider a simplified system consisting of three helices tethered by two loops as shown in Figure 1B. A rigorous treatment for the system requires a full account of all the possible bound ion configurations for the three-helix system for all the possible (x, θ) configurations, which would be extremely demanding in computational time. So we parse the problem into two simplified sub-problems. We first treat the full (x, θ) configurational ensemble by assuming pairwise additive electrostatic free energy for the three-helix system: , where is the electrostatic free energy for a system of two helices i and j. We then examine the effect of the three-helix correlation by considering the system of three parallel helices (θ = 0), where the enumeration for all the possible triple-helix (rather than pairwise) correlated bound ion modes can be performed.
Pairwise approximation
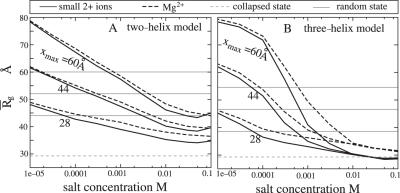
With the pairwise (additivity) approximation, for each given symmetric three-helix configuration (x, θ), the ion-dependence of the electrostatic free energy as well the loop-free energy Gloop(x, θ) ≃ 2Gsingle loop(x) are ‘amplified’. Therefore, the three-helix system shows a qualitatively similar but quantitatively much stronger ion-dependence than the two-helix system; see Figure 3A–D. For example, at low [Mg2+], the extended state becomes more extended due to the enhanced helix–helix repulsion and the effect from the extended rigid loops, and at high [Mg2+], the compact state becomes more compact due to the stronger electrostatic attraction and the enhanced loop entropy effect.
Triple-helix correlation
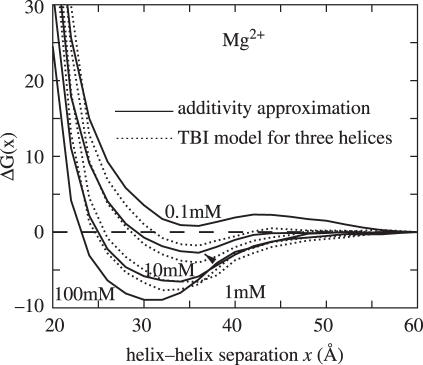
By considering the interactions between the three helices (and their associated bound ions), we compute the triple-helix correlated electrostatic free energy landscape ΔGE(x, 0°) = GE(x, 0°) − GE(60 Å, 0°). As shown in Figure 6, the pairwise approximation slightly underestimates and overestimates the stability of the free energy minima for [Mg2+] below 1 mM and above 10 mM, respectively.
Figure 6.
The electrostatic free energy ΔGE(x, 0°) = GE(x, 0°)−GE(60 Å, 0°) (in kBT) as functions of the helix–helix separation x for three parallel DNA helices (θ = 0) at different Mg2+ concentrations: 0.1, 1, 10 and 100 mM (from the upper to lower). Solid lines: calculated from the pairwise additivity approximation. Dotted lines: calculated with three-helix correlation in the TBI model.
For high [Mg2+], the helices are highly neutralized. For example, at 0.1 M Mg2+, tightly bound Mg2+ can neutralize about 92% of the phosphate charges for separated DNA helices. Compared to the two-helix case, the freedom of the bound ions for the helix triad is much more restricted due to the ion–helix and ion–ion Coulombic and excluded volume interactions. Such triple-helix restriction tends to prevent the (frustrated) bound ions from self-organizing to reach the lowest energy states that would be realized for a two-helix system. As a result, the resultant most compact state is less stable and slightly less compact than that predicted from the pairwise additive model. On the other hand, for lower [Mg2+], the number of bound ions is smaller and the charge neutralization is weaker, so the bound Mg2+ ions can self-organize relatively freely to reach the correlated low-energy states (88). In such case, due to the higher charge density for the three-helix case, the system can reach an energy lower than that predicted from the additive two-helix model (88).
Our above predictions agree with the previous analysis based on simplified models (68,88), which showed that the non-additivity effect is not strong for three-body system, and the non-additivity can result in slightly stronger attraction than the pairwise approximation for low ion concentration (68,88).
Ion size effect
Ion size has been shown experimentally to play important roles in RNA compaction (35). To investigate the size effect, we compare the free energy landscapes and the compactness for two divalent ions of different sizes: Mg2+ with radius ∼4.5 Å (70,72) and divalent ion (denoted by M2+) with a smaller radius 3.5 Å. Due to the lack of the information about the ion size-dependence of the loop persistence length, we would focus on the loop-free system.
Two-helix system
By comparing the free energy landscapes in Figure 2E–L and the compactness in Figure 7A for Mg2+ and the small divalent ions, we find that the smaller ions can induce stronger helix–helix attraction and the resultant stable states are more compact (72). Therefore, small ions are more efficient to induce the compaction. Physically, smaller ions can bind to the helices at a smaller ion-helix distance. Stronger ion–helix interaction leads to a higher ion-binding affinity and hence a larger number of bound ions, which, as a result, causes a stronger helix–helix attraction (72) and thus a more compact state.
Figure 7.
Compactness of two loop-free helices in Mg2+ (radius ≃ 4.5 Å) and smaller divalent ion (M2+, radius = 3.5 Å) solutions, respectively. Results for different end-to-end distance maximum: xmax = 60 Å, 44 Å and 28 Å (from the upper to lower) are plotted separately. The gray lines denote the compactness of the random fluctuation states R for different xmax's.
Three-helix system
As shown in Figure 7B for the ion-dependence of the compactness, the higher efficiency of the smaller ions is much more pronounced for the three-helix system. Notably, the three-helix system with high concentration of smaller ions can reach a very compact state—the collapsed state C. Such a collapsed state can never be reached by a two-helix system or a three-helix system in the solution of the (more bulky) Mg2+ ions. Physically, this is caused by the stronger helix–helix attraction for smaller ions and the effect is further ‘amplified’ by the larger number of helices.
CONCLUSIONS AND DISCUSSIONS
Using the TBI model (70–72) to account for the correlation and fluctuation effects of bound ions, we explore the electrostatic free energy landscape for nucleic acid helix–helix assembly for several paradigm systems: two and three helices with and without tether loop(s). The computed free energy landscapes lead to several predictions for the ion-dependent compaction of the helices.
Adding Na+ to the solution would induce the nucleic acid helices to undergo a transition from an extended state to a more compact random fluctuation state. However, Na+-mediated electrostatic interactions cannot cause the helices to form the collapsed state even at very high concentrations. This prediction is in accordance with the experiments on Na+-mediated RNA compaction (42–49).
Mg2+ can mediate helix–helix attraction, which causes the helix–helix assembly to form a state more compact than the random fluctuation state but less compact than the collapsed state. Such effect would be reduced for shorter helix–helix linker. Mg2+ ions are much more efficient than Na+ to stabilize nucleic acid helix–helix assembly. For example, 1 mM Mg2+ can induce a state equally compact as that induced by 1 M Na+. The maximum compact state occurs at [Mg2+ around 0.1 M.
The tether loop(s) can facilitate the compaction transition. While an electrically neutral loop enhances compaction for both low and high ion concentrations, a polynucleotide loop can significantly enhance the sharpness of the ion-induced compaction transition due to the ion-dependence of loop flexibility. Such effect is more pronounced for Mg2+ (than Na+) and for larger loops.
Multiplicity of the helices and the loops can significantly enhance the sharpness of the compaction transition and the compactness of the compact state.
Smaller ions are more efficient to induce the electrostatic compaction and such effect is much more pronounced for a multiple helix assembly.
Our treatment here is limited by several simplifications and approximations. First, we have neglected the possible partially or completely (dehydrated) ion site-binding (9,89) and possible preferential interactions between ions and specific groups of the nucleic acids (1). Ion site-binding to specific pockets with very strong negative electric potential can help stabilize specific tertiary structures (46,47) and can be essential for RNA catalytic function (90). The preferential ion-binding to specific groups, e.g. Mg2+ binds to phosphate groups or Mn2+ binds to both phosphates and bases (1), would affect the spatial distribution of ions on nucleic acid surface and contribute to the ion-mediated helix–helix interactions. However, the overall contribution of ion site-binding to the global stability may be small, because the large dehydration penalty may reduce the likelihood of specific binding of the dehydrated ions (46,47).
Second, in order to simplify the computation, we assume each helix pair to be symmetric and planar. Such a system covers a full spectrum of typical configurations, including compact, random and extended states. Thus, our predicted free energy landscape reveals the main feature of the ion-dependent helix assembly. We have also ignored the effect of different rotational phase of the helices about their axes. Previous analysis showed that the free energy landscape is not sensitive to such rotations if the helix–helix separation is large (e.g. x ≥ 28 Å) (91). However, for two closely approached helices (x ≤ 24 Å), such rotation would play an important role because of the detailed geometrical compatibility between helices and the related charge distributions (69,92). For very closely approached helices, parallel configuration is unfavorable because of the strong electrostatic repulsion. In the present model, the helices would be stabilized through a rotation to form a V-shaped structure. In realistic full 3D conformational space, the two helices can be stabilized through off-plane rotations to form a typical X-shaped structure (69,92,93) with two possible packing modes: groove-into-groove mode where the major groove of one helix fits in the major groove of the other helix (69), and backbone-into-groove mode where the nucleotide strand of a helix fits in the major groove of the other helix (92). Helix rotation about its axis is important for the formation of such X-shaped structures (69,92,93).
Third, we have ignored the sequence effect and specific tertiary interactions, which are essential for RNAs to form specific functional structures. In this study, we only investigated the ion electrostatic effects on the electrostatic collapse process. The quantitative modeling of further folding steps requires the inclusion of the sequence-dependent tertiary interactions.
Finally, we have neglected the detailed loop structures and possible deformation and unfolding of helices at different ionic conditions. Nevertheless, the predicted free energy landscapes, the detailed compaction mechanisms for Na+ and Mg2+ ions, and the effects of different loops, multiplicity of helices and ion size may be useful for understanding the ion-dependent nucleic acid helix–helix assembly.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Supplementary Material
Acknowledgments
This research was supported by NIH through grant GM063732 (to S.-J.C.). Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bloomfield V.A., Crothers D.M., Tinoco I., Jr . Nucleic Acids: Structure, Properties and Functions. Sausalito, CA: University Science Books; 2000. [Google Scholar]
- 2.Tinoco I., Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 3.Anderson C.F., Record M.T., Jr Salt-nucleic acid interactions. Annu. Rev. Phys. Chem. 1995;46:657–700. doi: 10.1146/annurev.pc.46.100195.003301. [DOI] [PubMed] [Google Scholar]
- 4.Brion P., Westhof E. Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 5.Strobel S.A., Doudna J.A. RNA seeing double: close-packing of helices in RNA tertiary structure. Trends Biochem. Sci. 1997;22:262–266. doi: 10.1016/s0968-0004(97)01056-6. [DOI] [PubMed] [Google Scholar]
- 6.Pyle A.M. Metal ions in the structure and function of RNA. J. Biol. Inorg. Chem. 2002;7:679–690. doi: 10.1007/s00775-002-0387-6. [DOI] [PubMed] [Google Scholar]
- 7.Sosnick T.R., Pan T. RNA folding: models and perspectives. Curr. Opin. Struct. Biol. 2003;13:309–316. doi: 10.1016/s0959-440x(03)00066-6. [DOI] [PubMed] [Google Scholar]
- 8.Woodson S.A. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 2005;9:104–109. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Draper D.E., Grilley D., Soto A.M. Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 10.Rook M.S., Treiber D.K., Williamson J.R. An optimal Mg2+ concentration for kinetic folding of the Tetrahymena ribozyme. Proc. Natl Acad. Sci. USA. 1999;96:12471–12476. doi: 10.1073/pnas.96.22.12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serra M.J., Baird J.D., Dale T., Fey B.L., Retatagos K., Westhof E. Effects of magnesium ions on the stabilization of RNA oligomers of defined structures. RNA. 2002;8:307–323. doi: 10.1017/s1355838202024226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Auffinger P., Bielecki L., Westhof E. Symmetric K+ and Mg2+ ion-binding sites in the 5S rRNA loop E inferred from molecular dynamics simulations. J. Mol. Biol. 2004;335:555–571. doi: 10.1016/j.jmb.2003.10.057. [DOI] [PubMed] [Google Scholar]
- 13.Sorin E.J., Rhee Y.M., Pande V.S. Does water play a structural role in the folding of small nucleic acids? Biophys. J. 2005;88:2516–2524. doi: 10.1529/biophysj.104.055087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thirumalai D., Hyeon C. RNA and protein folding: common themes and variations. Biochemistry. 2005;44:4957–4970. doi: 10.1021/bi047314+. [DOI] [PubMed] [Google Scholar]
- 15.Bloomfield V.A. DNA condensation by multivalent cations. Biopolymers. 1997;44:269–282. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 16.Wilson R.W., Bloomfield V.A. Counterion-induced condensation of deoxyribonucleic acid. a light-scattering study. Biochemistry. 1979;18:2192–2196. doi: 10.1021/bi00578a009. [DOI] [PubMed] [Google Scholar]
- 17.Widom J., Baldwin R.L. Cation-induced toroidal condensation of DNA studies with Co3+(NH3)6. J. Mol. Biol. 1980;144:431–453. doi: 10.1016/0022-2836(80)90330-7. [DOI] [PubMed] [Google Scholar]
- 18.Schellman J.A., Parthasarathy N. X-ray diffraction studies on cation-collapsed DNA. J. Mol. Biol. 1984;175:313–329. doi: 10.1016/0022-2836(84)90351-6. [DOI] [PubMed] [Google Scholar]
- 19.Rau D.C., Parsegian V.A. Direct measurement of the intermolecular forces between counterion-condensed DNA double helices. Evidence for long range attractive hydration forces. Biophys. J. 1992;61:246–259. doi: 10.1016/S0006-3495(92)81831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rau D.C., Parsegian V.A. Direct measurement of temperature-dependent solvation forces between DNA double helices. Biophys. J. 1992;61:260–271. doi: 10.1016/S0006-3495(92)81832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma C., Bloomfield V.A. Condensation of supercoiled DNA induced by MnCl2. Biophys. J. 1994;67:1678–1681. doi: 10.1016/S0006-3495(94)80641-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pelta J., Livolant F., Sikorav J.L. DNA aggregation induced by polyamines and cobalthexamine. J. Biol. Chem. 1996;271:5656–5662. doi: 10.1074/jbc.271.10.5656. [DOI] [PubMed] [Google Scholar]
- 23.Raspaud E., de la Cruz M.O., Sikorav J.L., Livolant F. Precipitation of DNA by polyamines: a polyelectrolyte behavior. Biophys. J. 1998;74:381–393. doi: 10.1016/S0006-3495(98)77795-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koltover I., Wagner K., Safinya C.R. DNA condensation in two dimensions. Proc. Natl Acad. Sci. USA. 2000;97:14046–14051. doi: 10.1073/pnas.97.26.14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vijayanathan V., Thomas T., Shirahata A., Thomas T.J. DNA condensation by polyamines: a laser light scattering study of structural effects. Biochemistry. 2001;40:13644–13651. doi: 10.1021/bi010993t. [DOI] [PubMed] [Google Scholar]
- 26.Hud N.V., Vilfan I.D. Toroidal DNA condensates: unraveling the fine structure and the role of nucleation in determining size. Annu. Rev. Biophys. Biomol. Struct. 2005;34:295–318. doi: 10.1146/annurev.biophys.34.040204.144500. [DOI] [PubMed] [Google Scholar]
- 27.Raspaud E., Durand D., Livolant F. Interhelical spacing in liquid crystalline spermine and spermidine-DNA precipitates. Biophys. J. 2005;88:392–403. doi: 10.1529/biophysj.104.040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J., Rau D.C. Incomplete ion dissociation underlies the weakened attraction between DNA helices at high spermidine concentrations. Biophys. J. 2005;89:1932–1940. doi: 10.1529/biophysj.105.065060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang J.X., Janmey P.A. The polyelectrolyte nature of F-actin and the mechanism of actin bundle formation. J. Biol. Chem. 1996;271:8556–8563. doi: 10.1074/jbc.271.15.8556. [DOI] [PubMed] [Google Scholar]
- 30.Tang J.X., Janmey P.A., Lyubartsev A., Nordenskiold L. Metal ion-induced lateral aggregation of filamentous viruses fd and M13. Biophys. J. 2002;83:566–581. doi: 10.1016/S0006-3495(02)75192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angelini T.E., Liang H., Wriggers W., Wong G.C. Like-charge attraction between polyelectrolytes induced by counterion charge density waves. Proc. Natl Acad. Sci. USA. 2003;100:8634–8637. doi: 10.1073/pnas.1533355100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Butler J.C., Angelini T., Tang J.X., Wong G.C. Ion multivalence and like-charge polyelectrolyte attraction. Phys. Rev. Lett. 2003;91:028301. doi: 10.1103/PhysRevLett.91.028301. [DOI] [PubMed] [Google Scholar]
- 33.Heilman-Miller S.L., Thirumalai D., Woodson S.A. Role of counterion condensation in folding of the Tetrahymena ribozyme I. equilibrium stabilization by cations. J. Mol. Biol. 2001;306:1157–1166. doi: 10.1006/jmbi.2001.4437. [DOI] [PubMed] [Google Scholar]
- 34.Heilman-Miller S.L., Pan J., Thirumalai D., Woodson S.A. Role of counterion condensation in folding of the Tetrahymena ribozyme II. counterion-dependence of folding kinetics. J. Mol. Biol. 2001;309:57–68. doi: 10.1006/jmbi.2001.4660. [DOI] [PubMed] [Google Scholar]
- 35.Koculi E., Lee N.-K., Thirumalai D., Woodson S.A. Folding of the Tetrahymena ribozyme by polyamines: importance of counterion valence and size. J. Mol. Biol. 2004;341:27–36. doi: 10.1016/j.jmb.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 36.Koculi E., Thirumalai D., Woodson S.A. Counterion charge density determines the position and plasticity of RNA folding transition states. J. Mol. Biol. 2006;359:446–454. doi: 10.1016/j.jmb.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 37.Perez-Salas U.A., Rangan P., Krueger S., Briber R.M., Thirumalai D., Woodson S.A. Compaction of a bacterial group I ribozyme coincides with the assembly of core helices. Biochemistry. 2004;43:1746–1753. doi: 10.1021/bi035642o. [DOI] [PubMed] [Google Scholar]
- 38.Buchmueller K.L., Webb A.E., Richardson D.A., Weeks K.M. A collapsed non-native RNA folding state. Nature Struct. Biol. 2000;7:362–366. doi: 10.1038/75125. [DOI] [PubMed] [Google Scholar]
- 39.Russell R., Millett I.S., Doniach S., Herschlag D. Small angle X-ray scattering reveals a compact intermediate in RNA folding. Nature Struct. Biol. 2000;7:367–370. doi: 10.1038/75132. [DOI] [PubMed] [Google Scholar]
- 40.Russell R., Millett I.S., Tate M.W., Kwok L.W., Nakatani B., Gruner S.M., Mochrie S.G., Pande V., Doniach S., Herschlag D., et al. Rapid compaction during RNA folding. Proc. Natl Acad. Sci. USA. 2002;99:4266–4271. doi: 10.1073/pnas.072589599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takamoto K., He Q., Morris S., Chance M.R., Brenowitz M. Monovalent cations mediate formation of native tertiary structure of the Tetrahymena thermophila ribozyme. Nature Struct. Biol. 2002;9:928–933. doi: 10.1038/nsb871. [DOI] [PubMed] [Google Scholar]
- 42.Das R., Kwok L.W., Millett I.S., Bai Y., Mills T.T., Jacob J., Maskel G.S., Seifert S., Mochrie S.G.J., Thiyagarajan P., et al. The fastest global events in RNA folding: electrostatic relaxation and tertiary collapse of the Tetrahymena ribozyme. J. Mol. Biol. 2003;332:311–319. doi: 10.1016/s0022-2836(03)00854-4. [DOI] [PubMed] [Google Scholar]
- 43.Takamoto K., Das R., He Q., Doniach S., Brenowitz M., Herschlag D., Chance M.R. Principles of RNA compaction: insights from the equilibrium folding pathway of the P4-P6 RNA domain in monovalent cations. J. Mol. Biol. 2004;343:1195–1206. doi: 10.1016/j.jmb.2004.08.080. [DOI] [PubMed] [Google Scholar]
- 44.Kwok L.W., Shcherbakova I., Lamb J.S., Park H.Y., Andresen K., Smith H., Brenowitz M., Pollack L. Concordant exploration of the kinetics of RNA folding from global and local perspectives. J. Mol. Biol. 2006;355:282–293. doi: 10.1016/j.jmb.2005.10.070. [DOI] [PubMed] [Google Scholar]
- 45.Qiu X., Kwok L.W., Park H.Y., Lamb J.S., Andresen K., Pollack L. Measuring inter-DNA potentials in solution. Phys. Rev. Lett. 2006;96:138101. doi: 10.1103/PhysRevLett.96.138101. [DOI] [PubMed] [Google Scholar]
- 46.Bai Y., Das R., Millett I.S., Herschlag D., Doniach S. Probing counterion modulated repulsion and attraction between nucleic acid duplexes in solution. Proc. Natl Acad. Sci. USA. 2005;102:1035–1040. doi: 10.1073/pnas.0404448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Misra V.K., Draper D.E. A thermodynamic framework for Mg2+ binding to RNA. Proc. Natl Acad. Sci. USA. 2001;98:12456–12461. doi: 10.1073/pnas.221234598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Misra V.K., Draper D.E. The linkage between magnesium binding and RNA folding. J. Mol. Biol. 2002;317:507–521. doi: 10.1006/jmbi.2002.5422. [DOI] [PubMed] [Google Scholar]
- 49.Misra V.K., Shiman R., Draper D.E. A thermodynamic framework for the magnesium-dependent folding of RNA. Biopolymers. 2003;69:118–136. doi: 10.1002/bip.10353. [DOI] [PubMed] [Google Scholar]
- 50.Lyubartsev A.P., Nordenskiold L. Monte Carlo simulation study of ion distribution and osmotic pressure in hexagonally oriented DNA. J. Phys. Chem. 1995;99:10373–10382. [Google Scholar]
- 51.Gronbech-Jensen N., Mashl R.J., Bruinsma R.F., Gelbart W.M. Counterion-induced attraction between rigid polyelectrolytes. Phys. Rev. Lett. 1997;78:2477–2480. [Google Scholar]
- 52.Allahyarov E., Gompper G., Lowen H. Attraction between DNA molecules mediated by multivalent ions. Phys. Rev. E. 2004;69:041904. doi: 10.1103/PhysRevE.69.041904. [DOI] [PubMed] [Google Scholar]
- 53.Lee K.C., Borukhov I., Gelbart W.M., Liu A.J., Stevens M.J. Effect of mono- and multivalent salts on angle-dependent attractions between charged rods. Phys. Rev. Lett. 2004;93:128101. doi: 10.1103/PhysRevLett.93.128101. [DOI] [PubMed] [Google Scholar]
- 54.Ou Z., Muthukumar M. Langevin dynamics of semiflexible polyelectrolytes: rod-toroid-globule-coil structures and counterion distribution. J. Chem. Phys. 2005;123:074905. doi: 10.1063/1.1940054. [DOI] [PubMed] [Google Scholar]
- 55.Rouzina I., Bloomfield V.A. Macroion attraction due to electrostatic correlation between screening counterions. 1. Mobile surface-adsorbed ions and diffuse ion cloud. J. Phys. Chem. 1996;100:9977–9989. [Google Scholar]
- 56.Ha B.Y., Liu A.J. Counterion-mediated attraction between two like-charged rods. Phys. Rev. Lett. 1997;79:1289–1292. [Google Scholar]
- 57.Solis F.J., de la Cruz M.O. Attractive interactions between rodlike polyelectrolytes: polarization, crystallization, and packing. Phys. Rev. E. 1999;60:4496–4499. doi: 10.1103/physreve.60.4496. [DOI] [PubMed] [Google Scholar]
- 58.Arenzon J.J., Stilck J.F., Levin Y. Simple model for attraction between like-charged polyions. Eur. Phys. J. B. 1999;12:79–82. [Google Scholar]
- 59.Kornyshev A.A., Leikin S. Electrostatic zipper motif for DNA aggregation. Phys. Rev. Lett. 1999;82:4138–4141. [Google Scholar]
- 60.Naji A., Netz R.R. Attraction of like-charged macroions in the strong-coupling limit. Eur. Phys. J. E. 2004;13:43–59. doi: 10.1140/epje/e2004-00039-x. [DOI] [PubMed] [Google Scholar]
- 61.Gilson M.K., Sharp K.A., Honig B. Calculating the electrostatic potential of molecules in solution: method and error assessment. J. Comput. Chem. 1987;9:327–335. [Google Scholar]
- 62.Sharp K.A., Honig B. Calculating total electrostatic energies with the nonlinear Poisson-Boltzmann equation. J. Phys. Chem. 1990;94:7684–7692. [Google Scholar]
- 63.Nicholls A., Honig B. A rapid finite-difference algorithm, utilizing successive over-relaxation to solve the Poisson-Boltzmann equation. J. Comput. Chem. 1991;12:435–445. [Google Scholar]
- 64.You T.J., Harvey S.C. Finite element approach to the electrostatics of macromolecules with arbitrary geometries. J. Comput. Chem. 1993;14:484–501. [Google Scholar]
- 65.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2000;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manning G.S. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q. Rev. Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- 67.Ray J., Manning G.S. An attractive force between two rodlike polyions mediated by the sharing of condensed counterions. Langmiur. 1994;10:2450–2461. [Google Scholar]
- 68.Ray J., Manning G.S. Formation of loose clusters in polyelectrolyte solutions. Macromolecules. 2000;33:2901–2908. [Google Scholar]
- 69.Murthy V.L., Rose G.D. Is counterion delocalization responsible for collapse in RNA folding? Biochemistry. 2000;39:14365–14370. doi: 10.1021/bi001820r. [DOI] [PubMed] [Google Scholar]
- 70.Tan Z.J., Chen S.J. Electrostatic correlations and fluctuations for ion binding to a finite length polyelectrolyte. J. Chem. Phys. 2005;122:044903. doi: 10.1063/1.1842059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tan Z.J., Chen S.J. Nucleic acid helix stability: effects of salt concentration, cation valency and size, and chain length. Biophys. J. 2006;90:1175–1190. doi: 10.1529/biophysj.105.070904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tan Z.J., Chen S.J. Ion-mediated nucleic acid helix-helix interactions. Biophys. J. 2006;91:518–536. doi: 10.1529/biophysj.106.084285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Montoro J.C.G., Abascal J.L.F. Ionic distribution around simple DNA models. I. Cylindrically averaged properties. J. Chem. Phys. 1995;103:8273–8284. [Google Scholar]
- 74.Wilhelm J., Frey E. Radial distribution function of semiflexible polymers. Phys. Rev. Lett. 1996;77:2581–2584. doi: 10.1103/PhysRevLett.77.2581. [DOI] [PubMed] [Google Scholar]
- 75.Thirumalai D., Ha B.Y. Statistical mechanics of semiflexible chains. In: Grosberg A., editor. Theoretical and Mathematical Models in Polymer Research. Vol. 1. San Diego, CA: Academic Press; 1998. p. 135. [Google Scholar]
- 76.Tinland B., Pluen A., Sturm J., Weill G. Persistence length of single-stranded DNA. Macromolecules. 1997;30:5763–5765. [Google Scholar]
- 77.Smith S.B., Cui Y., Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 78.Kuznetsov S.V., Shen Y.Q., Benight A.S., Ansari A. A semiflexible polymer model applied to loop formation in DNA hairpins. Biophys. J. 2001;81:2864–2875. doi: 10.1016/S0006-3495(01)75927-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rivetti C., Walker C., Bustamante C. Polymer chain statistics and conformational analysis of DNA molecules with bends or sections of different flexibility. J. Mol. Biol. 1998;280:41–59. doi: 10.1006/jmbi.1998.1830. [DOI] [PubMed] [Google Scholar]
- 80.Dwyer J.J., Gittis A.G., Karp D.A., Lattman E.E., Spencer D.S., Stites W.E., Garcia-Moreno E.B. High apparent dielectric constants in the interior of a protein reflect water penetration. Biophys. J. 2000;79:1610–1620. doi: 10.1016/S0006-3495(00)76411-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sosnick T.R., Trewhella J. Denatured states of ribonuclease A have compact dimensions and residual secondary structure. Biochemistry. 1992;31:8329–8335. doi: 10.1021/bi00150a029. [DOI] [PubMed] [Google Scholar]
- 82.Prabhu V.M. Counterion structure and dynamics in polyelectrolyte solutions. Curr. Opin. Colloid Interface Sci. 2005;10:2–8. [Google Scholar]
- 83.Lattman E.E. Rapid calculation of the solution scattering profile from a macromolecule of known structure. Proteins. 1989;5:149–155. doi: 10.1002/prot.340050209. [DOI] [PubMed] [Google Scholar]
- 84.Lattman E.E., Fiebig K.M., Dill K.A. Modeling compact denatured states of proteins. Biochemistry. 1994;33:6158–6166. doi: 10.1021/bi00186a015. [DOI] [PubMed] [Google Scholar]
- 85.Kienberger F., Pastushenko V.P., Kada G., Gruber H.J., Riener C., Schindler H., Hinterdorfer P. Static and dynamical properties of single poly(ethylene glycol) molecules investigated by force spectroscopy. Single Mol. 2000;1:123–128. [Google Scholar]
- 86.Erman B., Flory P.J., Hummel J.P. Radial distribution function of semiflexible polymers. Macromolecules. 1980;13:484–491. [Google Scholar]
- 87.Allegra G., Meille S.V. Mesomorphic phases of flexible polymers: the self-compacting chain model. Macromolecules. 2004;37:3487–3496. [Google Scholar]
- 88.Ha B.Y., Liu A.J. Effect of non-pairwise-additive interactions on bundles of rodlike polyelectrolytes. Phys. Rev. Lett. 1998;81:1011–1014. [Google Scholar]
- 89.Mikulecky P.J., Feig A.L. Heat capacity changes associated with nucleic acid folding. Biopolymers. 2006;82:38–58. doi: 10.1002/bip.20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Piccirilli J.A., Vyle J.S., Caruthers M.H., Cech T.R. Metal ion catalysis in the Tetrahymena ribozyme reaction. Nature. 1993;361:85–88. doi: 10.1038/361085a0. [DOI] [PubMed] [Google Scholar]
- 91.Kornyshev A.A., Leikin S. Electrostatic interaction between long, rigid helical macromolecules at all interaxial angles. Phys. Rev. E. 2000;62:2576–2596. doi: 10.1103/physreve.62.2576. [DOI] [PubMed] [Google Scholar]
- 92.Timsit Y., Moras D. DNA self-fitting: the double helix directs the geometry of its supramolecular assembly. EMBO J. 1994;13:2737–2746. doi: 10.1002/j.1460-2075.1994.tb06567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lilley D.M. Structures of helical junctions in nucleic acids. Q. Rev. Biophys. 2002;33:109–159. doi: 10.1017/s0033583500003590. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.