Abstract
Tumor necrosis factor receptor superfamily members convey signals that promote diverse cellular responses. Receptor trimerization by extracellular ligands initiates signaling by recruiting members of the tumor necrosis factor receptor-associated factor (TRAF) family of adapter proteins to the receptor cytoplasmic domains. We report the 2.4-Å crystal structure of a 22-kDa, receptor-binding fragment of TRAF2 complexed with a functionally defined peptide from the cytoplasmic domain of the CD40 receptor. TRAF2 forms a mushroom-shaped trimer consisting of a coiled coil and a unique β-sandwich domain. Both domains mediate trimerization. The CD40 peptide binds in an extended conformation with every side chain in contact with a complementary groove on the rim of each TRAF monomer. The spacing between the CD40 binding sites on TRAF2 supports an elegant signaling mechanism in which trimeric, extracellular ligands preorganize the receptors to simultaneously recognize three sites on the TRAF trimer.
Specific transmembrane signaling represents a general problem for all cells. How does extracellular ligand binding selectively promote intracellular receptor recognition and signal propagation? Mechanisms including ligand-promoted, global conformational changes and receptor dimerization have been described. In the tumor necrosis factor (TNF) receptor superfamily, oligomerization of receptors by extracellular, trimeric ligands initiates a variety of growth, differentiation, and death signals in many cell types (1). Signaling through many TNF receptor superfamily members is mediated by intracellular binding of the receptor cytoplasmic domain to the TNF receptor-associated factor (TRAF) family of adapter proteins. Subsets of the six known TRAF family members have been shown to interact with TNF-R2, CD40, CD30, LTβR, ATAR, OX-40, CD27, RANK, and 4–1BB. As expected for a discriminatory component, TRAF binding to receptor cytoplasmic domains requires ligand-induced receptor oligomerization (2, 3).
TRAF oligomerization and receptor recognition are mediated by the TRAF domain, a bipartite, 28-kDa motif found in all six TRAF family members. The conserved, C-terminal, ≈170-aa, TRAF-C domain binds receptors and contributes to TRAF oligomerization (4–6). The TRAF-N domain, a less conserved region N-terminal to the TRAF-C domain, is a predicted coiled coil (4). The TRAF domains of TRAF1, TRAF2, TRAF3, and TRAF6 form homotrimers, and TRAF1/2 and TRAF3/5 hetero-oligomers have been described (1, 4, 7). Except for TRAF1, the N-terminal half of each TRAF protein contains a predicted RING motif and five or seven predicted zinc-finger motifs. These domains are required for TRAF2-mediated NF-κB and JNK activation and appear to determine signaling specificities of individual TRAF proteins (8, 9). The biological selectivity of signaling also relies on the oligomerization specificity and receptor affinity of the TRAFs.
The TNF receptor superfamily member, CD40, mediates diverse responses. In antigen-presenting cells that constitutively express CD40, it plays a critical role in T cell-dependent immune responses (10). TRAF1, TRAF2, TRAF3, and TRAF6 binding sites in the 62-aa, CD40 cytoplasmic domain have been mapped (7, 11). TRAF1, TRAF2, and TRAF3 bind to the same sequence, 250PVQET, and TRAF6 binds to the membrane-proximal sequence, 231QEPQEINF.
We report here the 2.4-Å-resolution crystal structure of a fragment of the TRAF domain of TRAF2 complexed with a peptide derived from CD40. While this manuscript was in preparation, the crystal structures of similar TRAF2 fragments alone and in complex with a TNF-R2 peptide were described in different crystal forms (12). Except for differences in the extent of order in the coiled coil, the TRAF2 protein displays the same overall, trimeric architecture in the separately determined structures. The divergent CD40 and TNF-R2 sequences make distinct contacts to a conserved groove on the rim of the TRAF monomer. In the crystalline TRAF2/TNF-R2 complex, however, four of six TRAF monomers did not bind receptor peptide. In contrast, the TRAF2/CD40 complex shows simultaneous receptor binding to all three TRAF subunits, strengthening the idea (12) that the spacing between receptor-binding sites on TRAF promotes selective recognition by matching the geometry of ligand-bound, trimeric receptors.
MATERIALS AND METHODS
Purification of Human TRAF2 and CD40-p1.
Cloning of the Flag–TRAF2–NC (TRAF2 residues 272–501) and the CA21-epitope-tagged fragment of TRAF2 (residues 311–501, designated TRAF2-311) have been described (7, 13). TRAF-domain proteins were expressed from recombinant baculoviruses in Spodoptera frugiperda (Sf21) cells and purified from cytoplasmic extracts (13). Selenomethionine-labeled TRAF2-311 protein was produced by incubating Sf21 cells for 1 h in modified Excel 401 medium lacking l-methionine (JRH Biosciences, Lenexa, KS) supplemented with 10% dialyzed, heat-inactivated fetal bovine serum (Life Technologies, Grand Island, NY) and 50 μg/ml gentamicin. After a 1-h infection, 100 mg/liter dl-selenomethionine (Fluka) was added. Mass spectrometry of the purified protein indicated an average of ≈40% Se occupancy.
The CD40-p1 peptide, Ac-TyrProIleGlnGluThr-Am, was synthesized by using fluorenylmethoxycarbonyl (Fmoc) solid-phase methods and purified by using reversed-phase HPLC (14). The mass measured by electrospray ionization mass spectrometry was within 0.1 Da of the expected mass. The sequence corresponds to CD40 Pro-250–Thr-254. Val-251 was replaced with Ile to increase binding affinity (11). The N-terminal Tyr did not alter binding (11).
Domain Mapping.
Because initial crystals of the TRAF domain were of poor quality, the Flag–TRAF2–NC protein (2 mg/ml) was mildly digested with trypsin (1:400 wt/wt) in 0.1 M Tris (pH 8.8) and 85 mM NaCl at 25°C. Reversed-phase HPLC, N-terminal sequence analysis, and mass spectrometry showed major stable fragments beginning at residues 305 and 308 and continuing to the C terminus. A fragment (TRAF2-311) containing TRAF2 Gln-311–Leu-501 fused to a C-terminal CA21 tag was expressed and used for structural studies.
Crystallization and Structure Determination.
Cocrystals were grown at 20°C by vapor diffusion from equimolar TRAF2-311 (10 mg/ml) and CD40-p1 equilibrated with 20% MME–PEG 2000, 0.1 M Bistris propane, and 0.2 M MgCl2. Crystals were crosslinked with 0.1% glutaraldehyde transferred through the vapor phase and quenched with 0.5% n-butylamine (15). The cryoprotectant 30% MME–PEG 2000, 0.1 M Bistris propane (pH 7.0), 0.2 M MgCl2 allowed flash-cooling in liquid N2. The crystals have the symmetry of space group P212121 (a = 87.1 Å, b = 90.1 Å, c = 92.6 Å) with a single trimeric complex in the asymmetric unit. For derivative screens, crystals were soaked for ≈48 h in cryoprotectant containing 0.1 mM metal compound and back-soaked for >1 h before freezing.
Phases were obtained by multiwavelength anomalous diffraction (MAD) methods (Table 1). Synchrotron x-ray data were collected from one Hg crystal and one selenomethionine crystal. Data were integrated with mosflm (16) and scaled with scala (17) by using the automation program elves (J.M.H., unpublished data). Six Hg atoms were located with solve (18), and phases were refined with mlphare (17). Difference Fourier maps revealed 18 selenium sites in the asymmetric unit. Phases were refined with mlphare including all data from both derivatives. Solvent flattening and histogram matching (19) produced a clear electron density map that could be traced unambiguously with the program o (20).
Table 1.
Data collection, phasing, and refinement statistics
| Thimerosal (6 sites per asymmetric unit)
|
SelenoMet (18 sites per asymmetric unit)
|
|||||
|---|---|---|---|---|---|---|
| f" | f′ | f (low) | f" | f′ | f (low) | |
| Wavelength, Å | 1.006 | 1.009 | 1.025 | 0.9694 | 0.9696 | 1.069 |
| Resolution, Å | 2.55 | 2.55 | 2.55 | 2.40 | 2.40 | 2.40 |
| Rsym* | 0.050 | 0.049 | 0.047 | 0.052 | 0.052 | 0.048 |
| Completeness, % | 98.8 | 99.9 | 98.6 | 98.9 | 98.9 | 98.7 |
| Multiplicity | 3.8 | 3.8 | 3.8 | 7.1 | 7.1 | 7.2 |
| I/SD† | 19 (4.3) | 20 (3.9) | 19 (3.5) | 22 (4.2) | 22 (4.2) | 24 (4.8) |
| Phasing power‡ | 2.55/0.82 | 1.83/0.40 | 2.29/0.31 | 1.01/0.59 | 1.09/0.48 | — |
| Mean figure of merit§ (25–2.4 Å resolution): 0.602 (0.714 after solvent flattening). | ||||||
| Rcryst/Rfree¶ (25.0–2.4 Å): 0.215/0.257. | ||||||
| rms Δbonds, rms Δangles‖: 0.006 Å, 1.34°. | ||||||
| Average B factor: 58.0 Å2, 52.7 Å2 for TRAF-C domains. | ||||||
*Rsym = Σ|I − 〈I〉|/ΣI; I, intensity.
I, intensity; SD, standard deviation. Parentheses denote I/SD for highest resolution bin.
Phasing power, (dis/ano) = [Σn|FH|2/Σn|E|2]1/2; FH, calculated heavy-atom scattering factor; E, lack of closure error.
Mean figure of merit = 〈||ΣαP(α)eiα/ΣαP(α) ||〉; α, phase; P(α), phase probability distribution.
Rcryst = Σ|Fobs − Fcalc|/ΣFobs; Fobs, observed structure-factor amplitude; Fcalc, calculated structure-factor amplitude.
rms deviations from ideal values.
The structure was refined with cns (21) by using all of the data (low-energy selenomethionine) from 25- to 2.4-Å resolution, excluding 5% for calculating the free R value. Procedures carried out with cns included torsion angle dynamics, simulated annealing with a maximum-likelihood target function, restrained individual B-factor refinement, conjugate gradient minimization, and bulk solvent correction. Throughout the refinement, noncrystallographic symmetry restraints were used on the TRAF-C domain, but not on the coiled coil, which did not obey noncrystallographic symmetry. The model was adjusted to fit Σ-A-weighted and phase-combined electron- density maps.
RESULTS
The TRAF2-311 fragment defined by limited proteolysis was cocrystallized with a peptide, CD40-p1, containing the TRAF2 binding site in the CD40 cytoplasmic domain. The structure of TRAF2-311/CD40-p1 was solved by using MAD analysis (Table 1). The refined model contains residues 323–501 in each of three TRAF2 chains, three copies of CD40-p1 hexapeptide, and 273 water molecules. The model has good geometry (Table 1), and all residues have allowed main-chain torsion angles.
Overall Architecture of the Complex.
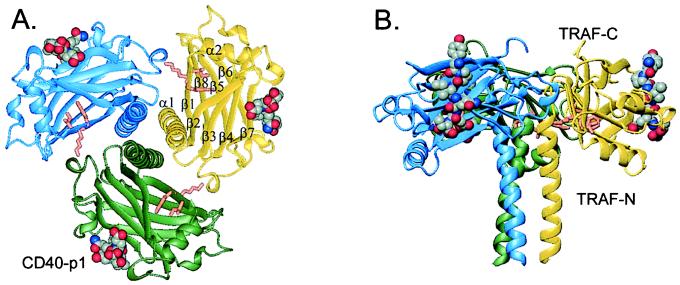
The TRAF-domain fragment folds as a mushroom-shaped trimer with overall dimensions 59 Å in height and 80 Å in diameter (Fig. 1). The trimeric architecture coincides with sedimentation measurements (12, 13). The TRAF-N-domain segment forms a triple-helical, parallel coiled coil that begins in the crystal near residue Gln-323. After the coiled coil, the TRAF-C domain (residues 348–501) forms a unique (22), eight-stranded β-sandwich containing two, twisted, four-stranded, antiparallel β-sheets. Sheet 1, forming the top of the mushroom cap, contains strands β1, β8, β5, and β6, and sheet 2, forming the underside of the mushroom cap, contains β2, β3, β4, and β7. The two sheets pack nearly parallel to each other, with the strands running tangentially around the trimer (Fig. 1). The longest helical segment in the TRAF-C domain links strands β1 and β2. Loops α1–β1, β2–β3, and β4–β5 form critical intersubunit contacts with the surface of the β-sandwich in the adjacent subunit.
Figure 1.
Three-dimensional structure of the TRAF–CD40-p1 complex. TRAF2-311 forms a trimer (blue, yellow, and green ribbons), and the CD40-p1 peptides (space-filling; atom colors) bind to the rim of all three TRAF2 monomers. TRAF-N domain sequences form a parallel coiled coil, followed by the TRAF-C domain, which adopts a topologically unique β-sandwich. (A) View along the trimer axis with the coiled coil in the front. The side chains of the conserved TrpLysIle motif implicated in recruitment of NF-κB inducing kinase, NIK (23), are shown in orange. The CD40 peptides are ≈38 Å from the trimer axis and ≈54 Å from each other. (B) Side view with the trimer axis vertical. The helix α2 is on the underside of the C domain, and the C terminus is at the top.
CD40-p1 binds in an extended conformation on the outer rim of each TRAF monomer (Fig. 1). The CD40 peptide backbones run parallel to the trimer three-fold axis, with the N terminus oriented toward the top of the mushroom. The peptide-binding sites are wholly within each monomer, not between TRAF2 subunits. The bound peptides make no contacts with each other (Fig. 1).
Surface Features of the TRAF2 Trimer.
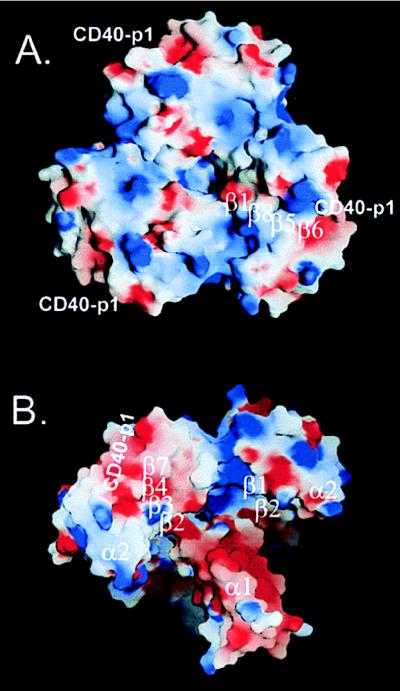
In addition to the CD40-p1 binding sites, several grooves, tunnels, and cavities mark the TRAF2-311 surface (Fig. 2). The top of the C domain contains a large bowl, 29 Å deep and >23 Å across. The sides of the bowl are formed by the β-sandwich motif. The C terminus forms the highest point on the rim of the bowl. The bottom of the bowl comprises a 12-Å-wide chamber on the top of the coiled coil. Three radial tunnels connect the bottom of the bowl to the underside of the cap, making a continuous solvent connection through the structure. Deep canyons exit the bowl along the subunit interfaces, forming a starfish pattern in the cap of the trimer. A deep moat is formed on the underside of the mushroom, at the connection between the TRAF-N and -C domains (Fig. 2).
Figure 2.
Surface features of TRAF2-311–CD40-p1. Electrostatic potential of the complex mapped on the molecular surface. The potential (−5 to +5 kT/e) was calculated with grasp (24) and displayed with blue (positive), white (neutral), and red (negative). Surface hydrophobicity is generally highest in the white regions. The position of CD40-p1 and the strands underlying the surface are indicated. (A) View along the trimer axis looking onto the “top” of the TRAF-C domain. Large indentations in the molecular profile mark the subunit interfaces. Overall, the trimer is polarized with positive potential facing the membrane. (B) Oblique view of the “underside” of the TRAF2-311–CD40-p1 complex with the coiled coil pointing toward the bottom right. Positive potential characterizes sides of the canyons that exit the bowl along the subunit interfaces. The coiled coil is ringed with negative potential.
Subunit Interactions.
The subunit interface in TRAF2-311 buries ≈1,290 Å2 of surface area in each monomer, with 809 Å2 contributed by the TRAF-C domain. A striking contact is made by Arg-385, which caps the coiled-coil helix of the neighboring subunit. Arg or Lys occupies this capping position in all of the TRAFs except TRAF3, where the corresponding residue is Tyr. The Arg-385 guanidino groups, by hydrogen bonding to the C-terminal carbonyl groups in the coiled coil, allow the backbone to turn away from the helix to initiate an open hairpin loop at the start of the C domain. The intersubunit interactions of the TRAF-C domain include the nonpolar clusters Ile-355/Tyr-386 and Leu-435/Leu-421/Phe-491. Hydrophilic interactions include the Arg-385 helix cap and a hydrogen bond from Lys-357 to the Pro-417 carbonyl. Other contact residues include Thr-349, Asn-418, Ala-420, and Gln-437.
The coiled coil displays classical, acute, knobs-into-holes packing except for Lys-331. This core residue is accommodated asymmetrically through contacts with Asp-332 and solvent. The residues in the coiled coil have higher than average B values, and no density is apparent for residues 311–322. These results suggest that the TRAF-N domain is flexible. Residues 332–347 in the coiled coil have a pitch of 179 Å, a crossing angle of 12.9° relative to the trimer axis, and a radius of 6.60 Å. These values are comparable to previously determined structures of coiled-coil trimers (25).
The CD40 Receptor-Binding Site.
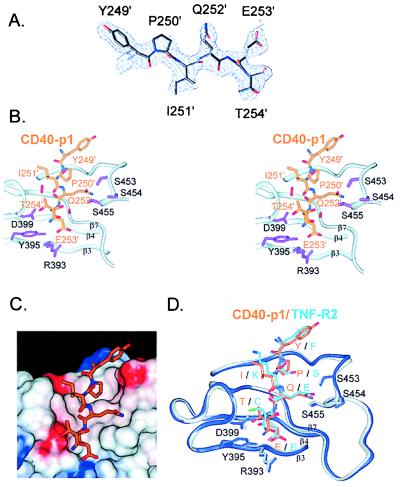
The CD40-p1 peptide is well defined in all three subunits (Fig. 3). The peptide binds nearly perpendicular to strands β3, β4, and β7, which form the floor of the receptor-binding groove. The irregular strand β7 (residues 463–470) plays key roles in peptide recognition. Ile-465 contacts CD40 Gln-252 and introduces a β-bulge that positions the succeeding residues, Ala-466, Ser-467, and Pro-470, to make receptor contacts. Pro-470 kinks β7 and contacts the side chain of CD40-p1 Ile-251. The buried hydroxyl of TRAF2 Ser-467 is over 3.7 Å from the nearest hydrogen-bonding partners. The sides of the peptide-binding site are formed by the β3–β4 and β6–β7 loops. These connecting segments contain conserved sequence elements—(GlyX)4 in β3–β4 and SerSerSer in β6–β7—containing Gly, Ser, and Thr residues that may facilitate the folding of the loops to form the binding groove.
Figure 3.
Receptor recognition by TRAF2. (A) Refined model of CD40-p1 (Tyr-249–Thr-254) superimposed on the solvent-flattened, MAD-phased, 2.4-Å-resolution electron density map (1σ). (B) Stereo view of the TRAF2–CD40-p1 contacts. The TRAF2 backbone is depicted with ribbons. Side chains of TRAF2 residues positioned to make hydrogen bonds to CD40-p1 are shown in purple. The serine tongs, in which conserved serines 453–455 form hydrogen bonds with CD40 Gln-252, are shown at the upper right. CD40 Glu-253 is at the bottom, within hydrogen-bonding distance of TRAF2 Arg-393 and Tyr-395. CD40 Thr-254 is within hydrogen-bonding distance of the conserved Asp-399. Primes (′) denote CD40 residues. (C) CD40-p1 (stick model) shown on the solvent accessible surface of TRAF2 colored by electrostatic potential (−8 to +8 kT/e; blue, positive; white, neutral; and red, negative). The side chains of CD40 Pro-250 and Ile-251 contact a hydrophobic region of the binding cleft, and the remaining CD40 side chains make polar or charged interactions. (D) Comparison of CD-40-p1 (orange) and TNF-R2 (light blue) bound to TRAF2 (CD40-p1 complex, gray; TNF-R2 complex, blue). Single letters denote the residues in CD40–TNF-R2. Three residues in the TNF-R2 peptide not present in CD40-p1 were omitted. The TRAF2 C domains in the two complexes were superimposed without reference to the receptor peptides. TRAF2 adopts similar conformations in the two complexes. The two receptor sequences make distinct side-chain contacts.
Each CD40 side chain makes contacts with TRAF2 (Fig. 3B, Table 2). The total surface area buried on binding is 969 Å2, with 535 Å2 contributed by the peptide and 434 Å2 by TRAF2. CD40 Pro-250, with the side chain fully buried, points into the binding pocket and orients the succeeding residues. CD40 Ile-251 points toward the side of the binding pocket, making van der Waals contacts with TRAF Pro-470 and main-chain hydrogen bonds with the amide and carbonyl groups of Gly-468. CD40 Gln-252 is in a position to form hydrogen bonds with three serines, 453–455, in the β6–β7 loop. Because the serine side chains surround CD40 Gln-252, we refer to this trifurcated interaction as the serine tongs. CD40 Glu-253 points away from solvent and forms a hydrogen-bonded ion pair with Arg-393 and a hydrogen bond with the side-chain hydroxyl group of Tyr-395. The TRAF2 Arg-393 side chain is extended and largely buried between Tyr-395 and Phe-377. The side-chain hydroxyl and main-chain amide of CD40 Thr-254 are within hydrogen-bonding distance of the carboxylate oxygens of TRAF2 Asp-399.
Table 2.
Analysis of CD40–TRAF2 interactions
| CD40-p1 residue | Area buried,* % | Putative H bonds
|
van der Waals
|
|
|---|---|---|---|---|
| CD40 atom | TRAF2 atom | TRAF2 residue | ||
| Tyr-249 | 26 | n/a | n/a | R448, P449, D450 |
| Pro-250 | 85 | n/a | n/a | F447, P449, F456, S467 |
| Ile-251 | 45 | N | G468: O | P470, S467, F410 |
| O | G468: N | |||
| Gln-252 | 61 | Nɛ2 | S453: Oγ | F410, I465, A466, S467 |
| Nɛ2 | S454: Oγ | |||
| Nɛ2 | S455: Oγ | |||
| Glu-253 | 83 | N | A466: O | F410, I465, A466 |
| Oɛ1 | A466: N | |||
| Oɛ2 | Y395: OH | |||
| Oɛ2 | R393: NH1 | |||
| Thr-254 | 28 | N | D399: Oδ1 | G400 |
| Oγ1 | D399: Oδ2 | |||
, % Solvent-accessible surface area buried on TRAF2 binding.
DISCUSSION
Implications for TRAF Oligomerization.
Available evidence suggests that all of the TRAFs form homotrimers, and heterotypic complexes are formed by TRAF1/2 and TRAF3/5 (4, 7, 13). The basis for this oligomerization specificity can be readily understood in terms of the structure of the trimeric TRAF2-311–CD40 complex. Both the TRAF-N coiled coil and the TRAF-C domain make intersubunit contacts. Of 12 TRAF-C domain contact residues in TRAF2, 5–7 are altered in each of the other TRAFs. Thus, the TRAF-C domains may help define the specificity of TRAF–TRAF associations. On the other hand, residues making key contacts between the TRAF-C domains (including the Arg-385 caps of the coiled coil and the hydrophobic cluster formed by Leu-421, Leu-435, and Phe-491) are conserved in TRAFs that fail to associate. The lack of a correlation between variations in the TRAF-C domain contacts and the oligomerization specificity suggests an important role for the coiled coil.
Sequence comparisons provide clear correlations between predicted coiled-coil structure and the observed oligomerization specificity. The N domains in the interacting proteins TRAF1 and TRAF2 contain characteristic, heptad-repeat regions of similar length with a break in the repeats at the same location. This break at TRAF2 304–310 encompasses the trypsin-sensitive sites in the TRAF domain. Compared with the other TRAFs, which contain >10 heptads, the N domain of TRAF4 contains only 3 heptads, providing a simple mechanism for disfavoring heterotypic associations. In TRAF6, the break in the heptad repeats occurs 21 residues farther from the TRAF-C domain than in TRAFs 1 and 2. In contrast, the N domains of TRAF3 and TRAF5 both contain a region of eight heptads with two homologous core histidines and a central stutter in the sequence register. Both of these features would be expected to inhibit associations with the other TRAFs. Moreover, strong TRAF3/5 interactions may require the unique Ile-zipper domains N-terminal to the TRAF domain (7). Not present in the other TRAFs, the Ile zipper is strongly predicted (26) to form a trimeric coiled coil in TRAF5 and weakly predicted to form a coiled coil in TRAF3. The many sequence differences in the coiled coil interfaces are consistent with core (a, d) and edge (g, e) residues mediating the observed specificity of TRAF oligomerization.
Implications for Receptor Recognition.
Each side chain in CD40-p1 contacts TRAF2, consistent with the high sensitivity of binding to mutations in this region of the receptor sequence. In a study of all 19 possible single amino acid changes at each position in the CD40 cytoplasmic domain, the only changes in the contact residues that preserved TRAF2 binding were Pro250His, Val251Ile and Thr254Ser (11). Such high selectivity argues for at least two specificity determinants in the interface. First, the invariant, buried, amino acids Pro-250, Gln-252, and Glu-253 anchor the sequence and organize the serine tong motif. With three simultaneous contacts to the side-chain amide of CD40 Gln-252, the Ser tongs may distinguish Glu, which would bury negative charge, and Asn, which is too short to contact all three Ser without conformational adjustments. Similarly, only CD40 Glu-253 is long enough to compensate for burial of the conserved TRAF Arg-393 in the binding site.
These anchor residues position the more exposed Ile-251 and Thr-254 of CD40-p1. The requirement in vitro for Val or Ile at position 251 implies that the β-branched side chain makes a critical contact with TRAF Pro-470. Similarly, the necessity in vitro for Thr or Ser at position 254 suggests an important role for the hydrogen bond to the TRAF2 Asp-399 carboxylate. Most replacements for the 250PVQET sequence in CD40 strongly reduce binding of TRAF1, TRAF2, and TRAF3 (11), suggesting a similar mode of binding to these TRAFs.
The conservation of the TRAF-interacting receptor sequences is mirrored in the amino acid conservation among the TRAFs. Except for Ser-454, all TRAF2 side chains within hydrogen-bonding distance of CD40-p1 are conserved in TRAFs 1, 2, and 3, which signal through the PVQET sequence in CD40 (1, 7). TRAFs 4 and 6, which do not recognize this sequence, contain significant changes in the peptide-binding site. This correlation affirms that the observed mode of TRAF recognition is a crucial determinant in TNF receptor signaling.
Comparison with TRAF2–TNF-R2 Complex.
The crystal structures of TRAF2 fragments in isolation and in complex with a nonapeptide from the TNF-R2 cytoplasmic domain (12) afford the opportunity to compare the modes of recognition of distinct receptors. The TRAF domain folds determined separately agree overall. The backbone rms deviation of the individual C domains in the CD40-p1 and TNF-R2 complexes averages 0.48 Å. Superposition of the C domain-trimer backbones gives a larger rms deviation of ≈0.78 Å, reflecting a small overall shift of the C domains relative to the coiled coil. The C domains undergo a subtle overall flexion on CD40-p1 and TNF-R2 binding. The coiled-coil domains show larger differences. In the CD40-p1 complex, the coiled coil is underwound and the N-terminal 12 residues are disordered compared with the continuous helices in the TRAF2-310–TNF-R2 complex. This difference may reflect flexibility in the coiled coil that is damped by intermolecular contacts in the TRAF2-310–TNF-R2 crystals.
The receptor sequences PVQET (CD40 and variants in OX40, CD27, CD30, and 4–1BB) and SKEEC (TNF-R2 and variants in HVEM and CD30) define two different consensus families that bind TRAF2 (1). Surface plasmon resonance measurements indicate that CD40-p1 and TNF-R2 peptides bind TRAF2-311 with similar Kd values (210 μM and 240 μM, respectively; data not shown). The CD40 and TNF-R2 peptides bind in similar conformations (backbone rms deviation of ≈0.3 Å) to the same groove on TRAF2. Despite similar backbone contacts, the two peptides are slightly shifted in the binding site (Fig. 3D). Every CD40 residue makes complementary contacts with TRAF2. By contrast, in the TNF-R2 complex, only the side chains of Ser in position one and Glu in position four are within hydrogen-bonding distance of TRAF2. The two sequences share the Glu in position four, and this residue makes similar interactions with TRAF2 Arg-393 and Tyr-395. Virtually every other side-chain contact is different. The serine tongs that recognize CD40 Gln-252 are 3.7–3.8 Å from the glutamate in position three of the TNF-R2 site. In the β3–β4 loop, the crucial hydrogen bond between TRAF2 Asp-399 and the CD40 Thr-254 hydroxyl has no analog in the TNF-R2 complex. Conversely, the TRAF2–CD40-p1 complex lacks an important specificity determinant in TNF-R2 recognition, a buried hydrogen bond between TNF-R2 Ser-424 and TRAF2 Ser-467 (12). Instead, CD40 Pro-250 at this position sequesters TRAF2 Ser-467 from solvent without making compensating hydrogen bonds.
These two different sets of contacts suggest that distinct, overlapping specificity determinants mediate recognition of two classes of receptor sequences. Consistent with this conclusion, TRAF2 binding to immobilized peptides is abolished by each single substitution in CD40 that makes the sequence more like the TNF-R2 recognition site (11). Covariation of the first and third residues (PXQ vs. XXE) suggest that positioning of CD40 Pro-250 and the interaction of Gln-252 with the serine tongs help distinguish the binding modes of the two receptor sequence families. The distinct receptor contacts predict that amino acid substitutions in TRAF2 may selectively influence recognition of different receptor classes.
Signaling Mechanism.
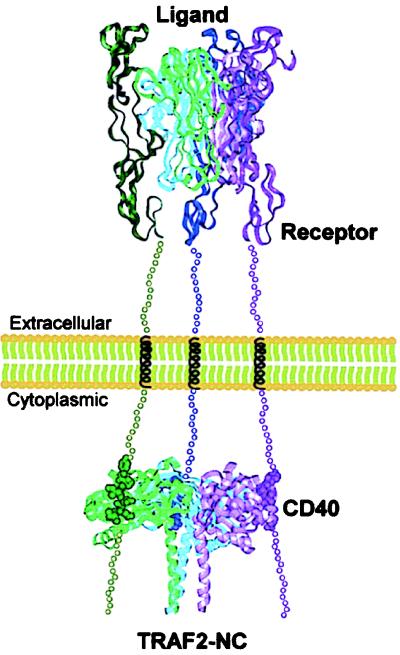
TNF receptor superfamily monomers contain an extracellular ligand-binding domain followed by a single transmembrane helix and the cytoplasmic domain. How does extracellular ligand binding lead to TRAF recognition of the intracellular domain? Binding of the TNF-R1 extracellular domain to the trimeric TNF positions three receptor molecules ≈33 Å apart on the corners of an equilateral triangle (27). Homologous TNF superfamily receptors, like CD40, are expected to be trimerized similarly (1, 28, 29). The spacing and triangular geometry of ligand-bound, extracellular receptor domains resembles the ≈54-Å distance between the three CD40-p1 peptides in the TRAF2-311 complex. This geometric match supports a simple signaling mechanism (12) in which increased avidity selectively promotes TRAF binding to ligand-bound, trimeric receptors (Fig. 4). This idea predicts that activated receptors bind simultaneously to all three TRAF subunits. In the TRAF2-310–TNF-R2 complex, however, only two of six TRAF subunits were complexed with TNF-R2 (12). As a consequence, the triply liganded TRAF2-311–CD40-p1 complex provides crucial experimental support for the signaling model.
Figure 4.
Signaling by TNF superfamily receptors. Ligand-promoted trimerization (top) positions the receptor intracellular domains to match the spacing between the receptor binding sites on the TRAF2 trimer (bottom). The trimeric receptor (top) is represented by the structure of the TNF–TNF-R1–extracellular-domain complex (27). Simultaneous TRAF binding to three receptor cytoplasmic domains (bottom) increases avidity and leads to selective recognition of activated, trimeric receptors. The region between the complexes is shown at approximately half scale. Assuming unfolded segments of ≈30 aa flanking the transmembrane helix, the intracellular and extracellular complexes are expected to be ≈150 Å apart.
The TNF receptor-signaling mechanism differs from previously described systems. In G protein-coupled receptors and bacterial chemotaxis receptors, for example, long-range conformational changes that cross the membrane are critical for signaling (30, 31). For the growth hormone and erythropoietin receptors, signaling depends on ligand-promoted, geometrically specific, receptor dimerization (32, 33). In contrast, the TNF receptors make no contacts, and orientational effects are most likely limited by unstructured connections to the transmembrane helix (27). Compared with dimeric systems or systems that feature receptor–receptor contacts, the trimeric TNF superfamily affords weaker intrinsic interactions, larger discrimination of liganded and unliganded receptors, and reduced spurious activation.
Because the cytoplasmic domains occur near the receptor C termini, the arrangements of CD40 and TNF-R2 (12) peptides on the TRAF domain imply that receptor binding orients TRAF2 with the top of the mushroom facing the membrane (Fig. 4). In this alignment, the coiled coil and the predicted N-terminal zinc-binding domains face the cytoplasm. The extended conformation of CD40-p1 suggests that TRAF2 binding can position neighboring receptor sequences to interact with other proteins in the signaling complex.
TRAF2 mediates signaling by associating with a variety of other factors, including NIK, TRADD, RIP, and cIAPs (1). The TRAF2-311–CD40-p1 structure provides insights into the locations of additional binding sites. For example, recruitment of NIK requires a TrpLysIle motif conserved in nearly all of the TRAFs (23). This sequence is located under the cap of the mushroom (Fig. 1). Trp-357 makes contact with the partially exposed Pro-379, which is conserved in TRAFs 1, 2, 4, and 6 and replaced by Gln in TRAFs 3 and 5. Interestingly, Trp-357 underlies the C terminus of CD40-p1. This juxtaposition suggests that receptor sequences C-terminal to the TRAF-binding site are positioned to interact with NIK (or other associated factors). Consistent with this idea, mutations in the TRAF-binding site of CD40 produce more severe signaling defects on deletion of the C-terminal region of the receptor cytoplasmic domain (34). Simultaneous receptor binding to TRAFs and to downstream components could help couple assembly of signaling complexes to receptor cross-linking.
The adapter protein TRADD promotes TRAF recruitment to TNF-R1, and the C terminus of TRAF2 is required for TRADD binding (8). The TRAF2 C terminus forms strand β8, which forms the highest point on the cap of the mushroom. Although deletion of the C terminus may produce pleiotropic structural defects, the C terminus abuts a potential binding site formed by the large bowl in the surface of TRAF2 C domain. This surface faces the membrane in the other TNF receptor superfamily complexes (Fig. 4), consistent with a location for TRADD directly between TRAFs and TNF-R1.
The surface of the TRAF domain (Fig. 2) displays many additional canyons and protrusions that may mediate protein–protein interactions. These features can guide mutagenesis experiments targeted to dissect TRAF functions. More directly, the crystal structure of trimeric TRAF2-311–CD40-p1 complex provides models for oligomerization specificity, receptor recognition and signaling through TNF receptor superfamily members.
Acknowledgments
We thank D. Reardon, S. Crowder, J. J. Plecs,and C. Day for helpful discussions and H. Bellamy, T. Earnest, and members of the Alber group for help with x-ray data collection. X-ray data were collected at Stanford Synchrotron Radiation Laboratory and the Advanced Light Source, which are operated by the Department of Energy and supported by the National Institutes of Health. We also thank H. Estes White for cloning; D. King, L. Frego, and W. Davidson for mass spectrometry; Dr. M. Labadia for Kd determinations; and A. Shrutkowski and Dr. J. Miglietta for DNA sequence analysis. This work was supported by National Institutes of Health Grant GM48958 to T.A.
ABBREVIATIONS
- TNF
tumor necrosis factor
- TRAF
TNF receptor-associated factor
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1QSC).
References
- 1.Arch R H, Gedrich R W, Thompson C B. Genes Dev. 1998;12:2821–2830. doi: 10.1101/gad.12.18.2821. [DOI] [PubMed] [Google Scholar]
- 2.VanArsdale T L, VanArsdale S L, Force W R, Walter B N, Mosialos G, Kieff E, Reed J C, Ware C F. Proc Natl Acad Sci USA. 1997;94:2460–2465. doi: 10.1073/pnas.94.6.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuhne M R, Robbins M, Hambor J E, Mackey M F, Kosaka Y, Nishimura T, Gigley J P, Noelle R J, Calderhead D M. J Exp Med. 1997;186:337–342. doi: 10.1084/jem.186.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rothe M, Wong S C, Henzel W J, Goeddel D V. Cell. 1994;78:681–692. doi: 10.1016/0092-8674(94)90532-0. [DOI] [PubMed] [Google Scholar]
- 5.Cheng G, Cleary A M, Ye Z S, Hong D I, Lederman S, Baltimore D. Science. 1995;267:1494–1498. doi: 10.1126/science.7533327. [DOI] [PubMed] [Google Scholar]
- 6.Force W R, Cheung T C, Ware C F. J Biol Chem. 1997;272:30835–30840. doi: 10.1074/jbc.272.49.30835. [DOI] [PubMed] [Google Scholar]
- 7.Pullen S S, Miller H G, Everdeen D S, Dang T T A, Crute J J, Kehry M R. Biochemistry. 1998;37:11836–11845. doi: 10.1021/bi981067q. [DOI] [PubMed] [Google Scholar]
- 8.Takeuchi M, Rothe M, Goeddel D V. J Biol Chem. 1996;271:19935–19942. doi: 10.1074/jbc.271.33.19935. [DOI] [PubMed] [Google Scholar]
- 9.Liu Z G, Hsu H, Goeddel D V, Karin M. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 10.Kawabe T, Naka T, Yoshida K, Tanaka T, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T, Kikutani H. Immunity. 1994;1:167–178. doi: 10.1016/1074-7613(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 11.Pullen S S, Dang T T A, Crute J J, Kehry M R. J Biol Chem. 1999;274:14246–14254. doi: 10.1074/jbc.274.20.14246. [DOI] [PubMed] [Google Scholar]
- 12.Park Y C, Burkitt v, Villa A R, Tong L, Wu H. Nature (London) 1999;398:533–538. doi: 10.1038/19110. [DOI] [PubMed] [Google Scholar]
- 13.Pullen, S. S., Labadia, M. E., Ingraham, R. H., McWhirter, S. M., Everdeen, D. S., Alber, T., Crute, J. J. & Kehry, M. R. (1999) Biochemistry, in press. [DOI] [PubMed]
- 14.Nautiyal S, Alber T. Protein Sci. 1999;8:84–90. doi: 10.1110/ps.8.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waksman G, Kominos D, Robertson S, Pant N, Baltimore D, Birge R B, Cowburn D, Hanafusa H, Mayer B J, Overduin, et al. Nature (London) 1992;358:646–653. doi: 10.1038/358646a0. [DOI] [PubMed] [Google Scholar]
- 16.Leslie A G W, Brick P, Wonacott A. Daresbury Laboratory Information Quarterly for Protein Crystallography. 1986;18:33–39. [Google Scholar]
- 17.Collaborative Computational Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 18.Terwilliger T C, Kim S-H, Eisenberg D. Acta Cryst A. 1987;43:1–5. [Google Scholar]
- 19.Cowtan K. Joint CCP 4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 20.Jones T A, Zou J-W, Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 21.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszuwski J, Nilges N, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 23.Song H Y, Regnier C H, Kirschning C J, Goeddel D V, Rothe M. Proc Natl Acad Sci USA. 1997;94:9792–9796. doi: 10.1073/pnas.94.18.9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholls A, Sharp K A, Honig B. Proteins Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 25.Harbury P B, Kim P S, Alber T. Nature (London) 1994;371:80–84. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- 26.Wolf E, Kim P S, Berger B. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banner D W, D’Arcy A, Janes W, Gentz R, Schoenfeld H J, Broger C, Loetscher H, Lesslauer W. Cell. 1993;73:431–445. doi: 10.1016/0092-8674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- 28.Karpusas M, Hsu Y M, Wang J H, Thompson J, Lederman S, Chess L, Thomas D. Structure. 1995;3:1031–1039. doi: 10.1016/s0969-2126(01)00239-8. [DOI] [PubMed] [Google Scholar]
- 29.Bajorath J, Aruffo A. Proteins. 1997;27:59–70. [PubMed] [Google Scholar]
- 30.Chervitz S A, Falke J J. Proc Natl Acad Sci USA. 1996;93:2545–2550. doi: 10.1073/pnas.93.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bourne H R. Curr Opin Cell Biol. 1997;9:134–142. doi: 10.1016/s0955-0674(97)80054-3. [DOI] [PubMed] [Google Scholar]
- 32.Wells J A, de Vos A M. Annu Rev Biophys Biomol Struct. 1993;22:329–351. doi: 10.1146/annurev.bb.22.060193.001553. [DOI] [PubMed] [Google Scholar]
- 33.Syed R S, Reid S W, Li C, Cheetham J C, Aoki K H, Liu B, Zhan H, Osslund T D, Chirino A J, Zhang J, et al. Nature (London) 1998;395:511–516. doi: 10.1038/26773. [DOI] [PubMed] [Google Scholar]
- 34.Hsing Y, Hostager B S, Bishop G A. J Immunol. 1997;159:4898–4906. [PubMed] [Google Scholar]