Abstract
In the budding yeast, Saccharomyces cerevisiae, protein kinases Ste20p (p21Cdc42p/Rac-activated kinase), Ste11p [mitogen-activated protein kinase (MAPK) kinase kinase], Ste7p (MAPK kinase), Fus3p, and Kss1p (MAPKs) are utilized for haploid mating, invasive growth, and diploid filamentous growth. Members of the highly conserved Ste20p/p65PAK protein kinase family regulate MAPK signal transduction pathways from yeast to man. We describe here a potent negative regulator of Ste20p in the yeast filamentous growth-signaling pathway. We identified a mutant, hsl7, that exhibits filamentous growth on rich medium. Hsl7p belongs to a highly conserved protein family in eukaryotes. Hsl7p associates with the noncatalytic region within the amino-terminal half of Ste20p as well as Cdc42p. Deletions of HSL7 in haploid and diploid strains led to cell elongation and enhancement of both haploid invasive growth and diploid pseudohyphal growth. However, deletions of STE20 in haploid and diploid greatly diminished these hsl7-associated phenotypes. In addition, overexpression of HSL7 inhibited pseudohyphal growth. Thus, Hsl7p may inhibit the activity of Ste20p in the S. cerevisiae filamentous growth-signaling pathway. Our genetic analyses suggest the possibility that Cdc42p and Hsl7p compete for binding to Ste20p for pseudohyphal development when starved for nitrogen.
The mitogen-activated protein kinase (MAPK) cascade is critical for various developmental events in eukaryotes (1–4). Elements of yeast single-MAPK pathway, Ste11p [MAPK kinase (MAPKK)], Ste7p [MAPK kinase kinase (MAPKK)], Kss1p, and Fus3p (MAPKs), are utilized for haploid mating, invasive growth, and diploid filamentous growth (5–9). Ste20p from Saccharomyces cerevisiae is a member of Ste20p/p65PAK family of proteins kinases, which regulate conserved MAPK pathways (10–13). Ste20p also functions in several signal transduction pathways, including those involved in haploid mating (14), invasive growth (8), and diploid filamentous growth (5, 15).
In response to nitrogen limitation, diploid yeast cells undergo a dimorphic transition known as pseudohyphal differentiation (15). A dominant-activated form of Ras2p, the yeast homologue of mammalian Ras, causes enhanced pseudohyphal growth (15), and this effect is blocked by the dominant-negative form of Cdc42p, placing Cdc42p downstream of Ras2p (18). Cdc42p binds to the noncatalytic region within the amino-terminal half of Ste20p (19, 20). Interestingly, the interaction of Cdc42p with Ste20p, which is essential for proper localization of Ste20p, is required for haploid invasive growth and diploid pseudohyphal development, but not for pheromone response (19, 20).
Here we describe the identification of Hsl7p, a potent negative regulator of Ste20p in yeast filamentous growth-signaling pathway. Hsl7p is a member of highly conserved protein family ranging from yeast to man. Biochemical and two-hybrid analyses revealed that Hsl7p interacts with the amino-terminal half of Ste20p. Using a genetic approach, we show that Hsl7p may negatively regulate pseudohyphal development by modulating the binding of Cdc42p to Ste20p.
MATERIALS AND METHODS
Media, Growth, and Yeast Strains.
Standard growth media and procedures for genetic manipulation of yeast have been described in detail (21). Synthetic low-ammonia medium (SLAD) for pseudohyphal development contains 50 μM ammonium sulfate, 2% glucose and 2% Bacto-agar (15). S. cerevisiae strains used in this study were YAF1151 (MATα his3 leu2 trp1 ura3 sir4Δ∷HIS3 ace2), YAF1108 (MATa leu2 ura3), YAF1110 (MATα leu2 ura3), YAF1111 (MATa/MATα leu2/leu2 ura3/ura3), YAF1191 (MATa hsl7Δ∷LEU2 leu2 ura3), YAF1192 (MATa ste20Δ∷kanr leu2 ura3), YAF1193 (MATa hsl7Δ∷LEU2 ste20Δ∷kanr leu2 ura3), YAF1194 (MATa/MATα hsl7Δ∷LEU2/hsl7Δ∷LEU2 leu2/leu2 ura3/ura3), YAF1195 (MATa/MATα ste20Δ∷kanr/ste20Δ∷kanr leu2/leu2 ura3/ura3), YAF1196 (MATa/MATα hsl7Δ∷LEU2/hsl7Δ∷LEU2 ste20Δ∷kanr/ste20Δ∷kanr leu2/leu2 ura3/ura3), and YAF2009L (MATα his3 leu2 ura3). All strains are derived from the Σ1278b genetic background except YAF1151. HSL7and STE20-disrupted strains were constructed by using a one-step gene-disruption procedure (22). To construct the HSL7Δ∷LEU2 disruption plasmid, a 4.5-kb fragment encompassing HSL7 was amplified by PCR and was cloned into pUC13 (pUC-HSL7). pUC-HSL7 was cleaved with EcoRV and Eco47III to remove the HSL7 coding region. A LEU2 fragment was ligated into the cleaved pUC-HSL7 to yield pUC-HSL7DL. To construct the STE20Δ∷kanr disruption plasmid, a 3.0-kb fragment encompassing STE20 was amplified by PCR and was cloned into pUC13 (pUC-STE20). pUC-STE20 was cleaved with HpaI to remove the STE20 coding region. A kanr PvuII-EcoRV fragment from pUG6 (23) was ligated into the cleaved pUC-STE20 to yield pUC-STE20DK. pUC-HSL7DL and pUC-STE20DK were treated with EcoRI and were used for yeast transformations.
Screen for Mutants Exhibiting Filamentous Growth.
The Tn3-LEU2 transposon mutagenized yeast genomic library (24) was transformed into the strain YAF1151 (MATα his3 leu2 trp1 ura3 sir4Δ∷HIS3 ace2). Fifty thousand transformants were plated on synthetic complete medium lacking leucine and were screened for filamentous growth by the observation of colony morphology and microscopic analysis. The yeast sequences adjacent to the Tn3-LEU2 insertion were determined by using plasmid-rescue procedures (24). The yeast sequences were compared with those in the GenBank database by using the blast program (25).
Plasmids.
To construct a fusion of glutathione S-transferase (GST) with full-length Ste20p, the STE20 coding region was amplified by PCR and ligated into pGEX-2T (Pharmacia) to yield pGEX-STE20. pGEX-STE201–495 and pGEX-STE20Δ258–842 carrying fusions of GST with the Ste20p1–496 and Ste20pΔ258–842 were constructed by removing an EcoRI fragment and a HindIII fragment of pGEX-STE20, respectively. pGEX-STE20258–495 carrying a fusion of GST with Ste20p258–495 was created by cloning an EcoRI-HindIII fragment of pUC-STE20 into pGEX-2T. To construct pMAL-P2-HSL7 carrying a fusion of maltose-binding protein (MBP) with full-length Hsl7p, a fragment including the HSL7 coding region was amplified by PCR and ligated into pMAL-P2 (New England Biolabs).
Plasmid YEUp-HSL7 was constructed by cloning of HSL7 into the high-copy vector YEUp3 (pUC13 + URA3 + 2 μm ori). Plasmid YEUp-HSL7Lp was constructed by cloning a 1.0 kb fragment containing the promoter of LEU2 into the NheI site of YEUp-HSL7 which corresponds to just upstream of HSL7 ORF. Plasmid YCLp-CDC42 was constructed by cloning of a BglII-HindIII DNA fragment containing CDC42 into the low-copy vector, pRS315 (pBluescript + LEU2 + CEN/ARS) (26). YELp-STE20 was constructed by cloning of the DNA fragment containing STE20 into the high-copy vector, pL1 (pUC19 + LEU2 + 2 μm ori).
Resin-Binding Assay.
GST-Ste20p and its deletion mutants expressed in the Escherichia coli strain MC1061 were purified on glutathione-Sepharose (Pharmacia) as described previously (27). Hsl7p tagged with MBP was purified by using the protease-deficient E. coli strain BL21. Cells of BL21 harboring pMAL-P2-HSL7 were grown at 37°C in LB. Expression of MBP-Hsl7p was induced by the addition of isopropyl β-d-thiogalactoside to a final concentration of 0.1 mM at OD600 = 0.4, followed by incubation at 37°C for 3 hr. Cells were harvested by centrifugation, resuspended in sonication buffer (20 mM Tris⋅HCl, pH 7.5/20 mM NaCl/50 mM EDTA/1 mM 2-mercaptoethanol/1% Triton X-100/1 mM PMSF/2 μg/ml leupeptin/2 μg/ml aprotinin), and disrupted by gentle sonication at 4°C. MBP-Hsl7p was purified by using amylose resin according to the manufacturer’s protocol (New England Biolabs) except for elution; MBP-Hsl7p was eluted from the beads by elution buffer (50 mM Tris⋅HCl, pH 7.5/150 mM NaCl/1.5 mM MgCl2/1 mM 2-mercaptoethanol/10 mM maltose). Ten micrograms of GST-Ste20p fusions were bound to 10 μl of glutathione-Sepharose beads in 200 μl of binding buffer (50 mM Tris⋅HCl/100 mM NaCl/5 mM MgCl2/1 mM DTT/0.5 mg/ml BSA/0.07% Triton X-100) by incubating at 4°C for 1 hr. The beads were washed once with binding buffer, resuspended in binding buffer containing 10 μg of MBP-Hsl7p to a total volume of 200 μl, and then incubated at 4°C for 1 hr with gentle shaking. After incubation, the beads were washed five times with binding buffer and resuspended in 30 μl of Laemmli buffer (39). Proteins were separated by SDS/PAGE and transferred to poly(vinylidene difluoride) membranes. Blots were probed with antiserum against MBP (New England Biolabs), and then bound antibodies were detected by using alkaline phosphatase-coupled secondary antibody (Bio-Rad).
Assays for Invasive Growth, Filament Formation, and β-Galactosidase Activity.
Assays for haploid invasive growth (8) and diploid pseudohyphal growth (5, 15) were performed as described. Plasmid YEpU-FTyZ (6), encoding the lacZ gene under the transcriptional control of filamentous response element (FRE) from Ty1 (28), was used for reporter assays. β-Galactosidase assays were performed with extracts from exponentially growing cells in synthetic dextrose (SD) medium at 30°C. For assays under nitrogen-starved conditions, extracts were prepared from the cells spread on SLAD medium plates for 3 days at 30°C. Cells always were washed with water before lysis. β-Galactosidase activities were normalized to the total protein in each extract by using a protein assay kit (Bio-Rad).
Two-Hybrid Analysis.
Plasmids for two-hybrid analysis were constructed for this study. We constructed a vector (pAD1) for the expression of activation domain (AD) fusion by introducing the HIS3 marker into the SwaI site of pYESTRP (Invitrogen) and a vector (pLBD1) for the expression of LexA-binding domain (LBD) fusion by introducing the LEU2 marker into the Ecl136II site of pHybLex/Zeo (Invitrogen). pAD-Ste20pNH contains the N-terminal region of STE20 (amino acids 1–495) fused to AD of pAD1. pLBD-Cdc42pL and pLBD-Hsl7pL contain full-length CDC42 and HSL7 fused to LBD of pLBD1, respectively. LBD-Cdc42p fusion incorporates the C188S substitution, which prevents prenylation. pLexAop-lucU is a luciferase reporter plasmid containing 8 LexA operator sites upstream of the luciferase gene on YEUp3 (pUC13 + URA3 + 2 μm ori). pLexAop-LucU-Cdc42p contains CDC42 on pLexAop-LucU. pLexAop-LucU-Hsl7p contains the LEU2-promoter-HSL7 ORF fusion on pLexAop-LucU. Two-hybrid experiments were performed with strain YAF2009L (MATα his3 leu2 ura3). The strain was transformed with an LBD plasmid, an AD plasmid, and a LexAop-luciferase reporter plasmid. Cells were grown to midlog phase in SD medium (21) and were assayed for luciferase activity with PicaGene assaying kit (Nippon Gene, Toyama, Japan). Each value represents the average for three independent cultures.
Microscopy.
Yeast cells were photographed with a Leitz DMRD microscope with a ×40 objective lens. Colonies on plates were photographed with a Leica MZ APO microscope.
RESULTS
Identification of Mutants Exhibiting Filamentous Growth on Nitrogen-Rich Medium.
To identify negative regulators of filamentous growth, we attempted to isolate mutants that exhibit filamentous growth on rich medium (not under the nitrogen-starved conditions). Pseudohyphal development under nitrogen-starved condition occurs only in MATa/MATα diploid cells (15). However, recessive mutations are difficult to isolate by using MATa/MATα diploid strains. Therefore, we used a haploid sir4−strain, YAF1151 (MATa his3 leu2 trp1 ura3 sir4∷HIS3 ace2). The sir4 mutation allows transcription of the silent mating-type loci HMRa and HMLα. Thus, some phenotypes of haploid sir4 strains (bipolar budding pattern, sterility, and pseudohyphal growth under nitrogen-starved conditions) are similar to those of MATa/MATα diploid strains because of simultaneous expression of both a and α information. Furthermore, because the ace2 mutation causes a defect in cell separation (29), we can distinguish the transformants exhibiting filamentous growth by their colony morphologies: colonies exhibiting filamentous growth are highly rugged (30). Mutagenesis for isolating mutants exhibiting filamentous growth was performed by using the Tn3-LEU2 transposon mutagenized yeast genomic library (24). Fifty thousand independent Leu+ colonies were visually screened for filamentous growth, and two mutants were obtained. Genomic DNAs immediately adjacent to the Tn3-LEU2 insertions were cloned and sequenced. In a blast search, these two mutants were found to have transposon insertions at 502389 and 503762 on chromosome II. Both insertion sites occurred within in the ORF YBR133c encoding Hsl7p (Fig. 1A), which had been identified as a negative regulator of Swe1p, an inhibitor of cyclin-dependent kinase known as Cdc28p (31). Hsl7p encodes a predicted protein of 828 aa (95,148 Da). As reported by other workers (31–33), the amino acid sequence of Hsl7p has sequence similarities with the amino acid sequences of Schizosaccharomyces pombe Skb1p, human Skb1p, and the Caenorhabditis elegans ORF C34E10.5 (Fig. 1B). Our blast search revealed the existence of a Drosophila homologue encoded by the capsuleen gene (Fig. 1B), thereby supporting the idea that Hsl7p/Skb1p/capsuleen protein family is conserved in eukaryotes.
Figure 1.
(A) Restriction map of HSL7. Positions of HSL7 ORF (open arrow) and the transposon insertion sites (triangles) are indicated. Structures of the fragment used to disrupt HSL7 are noted. (B) Sequence alignment of S. cerevisiae Hsl7p with S. pombe Skb1p, C. elegans C34E10.5, Drosophila capsuleen, and human Skb1p proteins. Numbers indicate amino acid residue position. Identical and conserved residues are shown in white letters against black and shaded letters, respectively. GenBank accession nos.: Sc-Hsl7p, U65920; Sp-Skb1p, U59684; Ce-C34E10.5, U10402; Dm-capsuleen, AJ002740; Hs-Skb1p, AF015913.
Interaction of Hsl7p with Ste20p.
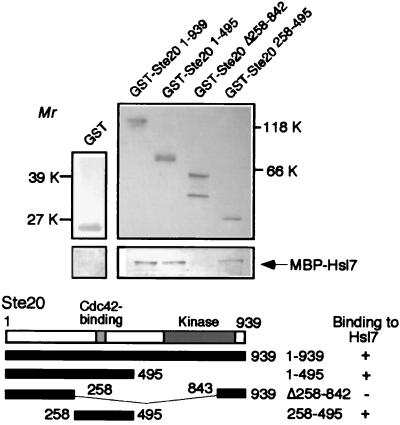
S. pombe Skb1p is a protein that interacts with Shk1p [S. pombe Ste20p/p21Cdc42p/Rac-activated kinase (PAK) homologue], as determined by using two-hybrid analyses (33, 34). S. pombe Shk1p is an essential component of the Ras- and Cdc42p-dependent signal transduction pathway required for cell viability, normal morphology, and sexual response. Skb1p may modulate Shk1p function in a signaling cascade to control morphology in S. pombe (33). Although biochemical interactions between Skb1p and Shk1p have not been detected (33), these results do suggest the possibility that Hsl7p interacts with Ste20p to regulate its function in S. cerevisiae. We used a resin-binding assay to examine the interaction of Hsl7p with Ste20p in vitro. We purified full-length Hsl7p as a fusion protein with MBP and full-length Ste20p as a fusion protein with GST. As shown in Fig. 2, MBP-Hsl7p bound to the full-length GST-Ste20p fusion, but not to GST alone. We next produced partial GST-Ste20p fusion proteins (Ste20p1–495, Ste20p258–495, and Ste20pΔ258–842) (Fig. 2). Both GST-Ste20p1–495 and GST-Ste20p258–495 fusions bound to MBP-Hsl7p, but GST-Ste20pΔ 258–842 did not (Fig. 2). These results demonstrate that Hsl7p can bind to the noncatalytic region of Ste20p (amino acid 258–495) in vitro. Previous studies showed that the Rho-family GTPase Cdc42p binds to a conserved region (amino acid 335–370) in the noncatalytic N-terminal region of Ste20p (19, 20, 35) and that binding of Cdc42p with this region of Ste20p is essential for filamentous growth (19, 20). The identified Hsl7p-binding domain of Ste20p (amino acid 258–495) includes the Cdc42p-binding domain (amino acid 335–370).
Figure 2.
Interaction of Hsl7p and Ste20p. Resin-binding assay was done to determine interactions between Hsl7p and Ste20p. GST fusion proteins with full-length Ste20p, Ste20p1–495, Ste20p258–495, and Ste20pΔ258–842. GST were bound to glutathione-Sepharose beads and incubated with MBP-Hsl7p. After washing, proteins eluted with SDS-sample buffer were subjected to SDS/PAGE in 12.5% (upper left blot), 7.5% (upper right blot), and 5% (lower blots) gels and immunoblotted for the presence of GST, GST-Ste20p fusion proteins (upper blots), and MBP-Hsl7p (lower blots) with antibodies to GST (upper blots) and MBP (lower blots), respectively. Interactions between Ste20p fragments and Hsl7p are summarized at the bottom.
We also used the two-hybrid system to confirm that Hsl7p can interact with the noncatalytic N-terminal region of Ste20p in vivo. A HIS3-based plasmid (pAD-Ste20pNH) was constructed to express the N-terminal region of Ste20p (amino acid 1–495) as a fusion to the C terminus of the B42 AD. LEU2-based plasmids (pLBD-Cdc42pL and pLBD-Hsl7pL) were constructed for expressing the full-length Cdc42p and Hsl7p as fusions to the C terminus of the LBD. An S. cerevisiae strain YAF2009L, which carries a plasmid (pLexAop-lucU) containing LexA-operator-luciferase reporter gene, was cotransformed with pLBD-Ste20pNH and pLBD-Cdc42pL or pLBD-Hsl7pL. Consistent with the previous observations, the interaction of the N-terminal region of Ste20p with Cdc42p was detected (refs. 19 and 20; shown in Table 1). By two-hybrid analysis, we found weak interaction of Hsl7p with Ste20p (Table 1). Thus, biochemical and two-hybrid analyses demonstrated that Hsl7p can associate with the N-terminal region of Ste20p.
Table 1.
Two-hybrid interactions among Ste20p, Cdc42p, and Hsl7p
| Activating domain fusion | LexA DNA-binding domain fusion | Relative luciferase activity* |
|---|---|---|
| Ste20p (1–495) | Vector | 1.0 |
| Ste20p (1–495) | Cdc42p | 18.6 |
| Ste20p (1–495) | Hsl7p | 7.4 |
| Vector | Cdc42p | 1.4 |
| Vector | Hsl7p | 1.5 |
| Ste20p (1–495) | Hsl7p (with overexpression of CDC42) | 1.7 |
| Ste20p (1–495) | Cdc42p (with overexpression of HSL7) | 3.1 |
The relative luciferase activities are normalized to that for Ste20p-vector.
Genetic Evidence for Functional Interaction Between Hsl7p and Ste20p in Yeast Filamentous Growth-Signaling Pathway.
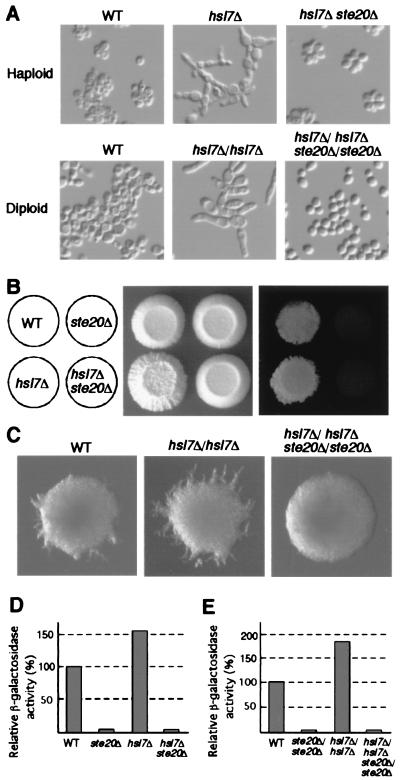
To determine whether Hsl7p regulates the signaling cascade that controls haploid invasive growth and diploid pseudohyphal development by affecting the activity of Ste20p, strains deleted for HSL7 (and/or STE20) were constructed. Because HSL7 is not essential for growth (31), the hsl7Δ∷LEU2 fragment (Fig. 1A) was used to transform the wild-type MATa and MATα haploid strains. The resultant MATa and MATα hsl7 disruptants were mated for the construction of MATa/MATα hsl7Δ/hsl7Δ homozygous diploid strains. Microscopic analysis revealed that the cells of both haploid and diploid HSL7 disruptants are highly elongated (Fig. 3A). An elongated cell morphology of pseudohyphae is generated by hyperpolarized cell growth and delay of G2/M cell cycle transition (36). Genetic study showed that Ste20p is essential for cell elongation (37). Disruption of STE20 in both haploid hsl7Δ strains and diploid hsl7Δ/hsl7Δ strains exhibited normal, round shapes (Fig. 3A). Haploid strains grown on rich-medium plates exhibit invasive growth (8). Ste20p and elements of the MAPK cascade (Ste11p, Ste7p, Kss1p, and Fus3p) are also required for invasive growth (6–8). Deletion of HSL7 resulted in greatly enhanced agar penetration (Fig. 3B), whereas the strain lacking both HSL7 and STE20 did not invade the agar (Fig. 3B). When starved for nitrogen, diploid strains enter a pseudohyphal growth mode that depends on elements of the same MAPK cascade (5, 15). In comparison with wild-type diploid strain, hsl7Δ/hsl7Δ diploid strain exhibited enhanced pseudohyphal growth (Fig. 3C), but the hsl7Δ/hsl7Δ ste20Δ/ste20Δ strain did not form pseudohyphae (Fig. 3C). These findings strongly suggest that Hsl7p negatively regulates the MAPK-signaling pathway essential for filamentous growth by interacting with Ste20p. We then asked whether Hsl7p is involved in haploid mating, a process that requires components of the same MAPK cascade. We assayed the mating efficiencies and the activities of mating-specific reporter (FUS1-promoter-luciferase fusion) of wild-type and hsl7Δ haploid cells. We did not find any difference either in the mating efficiencies or in the luciferase activities between wild-type cells and hsl7Δ cells (data not shown). Therefore, Hsl7p does not appear to be involved in the MAPK-dependent signaling pathway during mating.
Figure 3.
Disruption of HSL7 stimulates haploid invasive growth and diploid pseudohyphal development. (A) Elongated cell morphology of the haploid hsl7Δ and diploid hsl7Δ/hsl7Δ strains. Deletion of STE20 abolished their elongated cell morphology. Yeast strains were cultured at 30°C in yeast extract/peptone/dextrose (YPD) medium for 3 days. Cells were visualized under the microscope with Nomarski optics. (B) Disruption of HSL7-stimulated invasive growth in haploid cells. (Left) Genotypes of the strains examined. (Center) Total growth. (Right) Haploid invasive growth. Σ1278b-derived strains: YAF1108 (MATa HSL7+ STE20+ leu2 ura3) and its otherwise isogenic derivatives YAF1191 (MATa hsl7Δ∷LEU2), YAF1192 (MATa ste20Δ), and YAF1193 (MATa hsl7Δ∷LEU2 ste20Δ). Strains were streaked onto YPD plates and grown for 3 days at 30°C. The plate shown in Center was rinsed under a stream of running water to distinguish surface growth from invasive growth by the degree of agar penetration (8). (C) Disruption of HSL7 enhanced pseudohyphal growth in diploid cells. Diploid strain YAF1111 (MATa/MATα HSL7/HSL7 STE20/STE20 leu2/leu2 ura3/ura3) was constructed from YAF1108 and YAF1110 (MATα HSL7+ STE20+ leu2 ura3) in the Σ1278b background. Diploid strains YAF1194 (MATa/MATα hsl7Δ∷LEU2/hsl7Δ∷LEU2), YAF1195 (MATa/MATα ste20Δ/ste20Δ) and YAF1196 (MATa/MATα hsl7Δ∷LEU2/hsl7Δ∷LEU2 ste20Δ/ste20Δ) are YAF1111 derivatives. Every diploid strain contained vectors carrying URA3 and/or LEU2 to complement the ura3 and/or leu2 auxotrophies. These transformants were streaked on SLAD medium (15) and allowed to grow for 4 days. Resulting colonies were photographed. (D and E) Expression of the FRE-lacZ reporter gene. The eight strains described in B and C were transformed with a plasmid YEpU-FTyZ (6) (and a LEU2-containing vector to complement leu2), and β-galactosidase activity was measured. (D) Haploid strains on SD medium. (E) Diploid strains on nitrogen-starved medium. Activities were normalized to protein concentration and are shown as relative to the activities of wild-type strain. Values are the averages of measurements made in triplicate.
To confirm the possibility that Hsl7p negatively regulates MAPK cascade in filamentous growth-signaling pathway, we used the STE12-dependent transcriptional reporter (FRE-lacZ) (6). Because transcription from FREs, which contain the binding sites for Ste12 and Tec1p transcription factors (5, 38) (both factors were required for filamentous growth), is induced upon nitrogen starvation but not in response to mating pheromone (18, 28), FRE-lacZ reflects the activity of MAPK cascade during invasive growth and pseudohyphal development. Compared with the wild-type haploid strain, FRE-dependent transcription was induced about 1.5-fold in the haploid hsl7Δ strain (Fig. 3D). Deletion of STE20 in the hsl7Δ background eliminated FRE-dependent transcription (Fig. 3D). In the diploid hsl7Δ/hsl7Δ strain, FRE-lacZ expression was induced 1.8-fold on nitrogen-starved media (Fig. 3E) as compared with that of wild-type strain. Deletion of STE20 also eliminated the FRE-lacZ activity in the hsl7Δ/hsl7Δ background (Fig. 3E). These data support the model that Hsl7p functions as a negative regulator of Ste20p in filamentous growth-signaling pathway.
Diploid hsl7Δ/hsl7Δ Strain Forms Pseudohyphae on Nitrogen-Rich Medium.
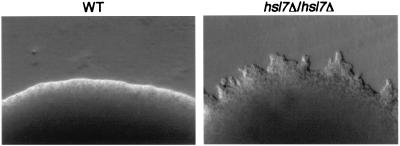
We determined whether or not hsl7Δ/hsl7Δ diploid strain would form pseudohyphae on nitrogen-rich medium. In contrast to the wild-type diploid strain, hsl7Δ/hsl7Δ diploid strain showed pseudohyphal development after 2 weeks (Fig. 4). In addition, FRE-lacZ expression of disruptants also was induced 1.3-fold on nitrogen-rich medium relative to that of wild-type strain, indicating that the MAPK-dependent cascade is constitutively activated in hsl7Δ/hsl7Δ diploid cells.
Figure 4.
Pseudohyphal growth of the hsl7Δ/hsl7Δ diploid strain on nitrogen-rich medium. Wild-type diploid (YAF1111) and hsl7Δ/hsl7Δ (YAF1194) strains containing a URA3-containing vector and/or a LEU2-containing vector to complement ura3 and/or leu2 were streaked on SLAD medium containing 5 mM ammonium sulfate and were incubated for 2 weeks at 30°C.
Overexpression of Hsl7p Inhibits Diploid Pseudohyphal Growth.
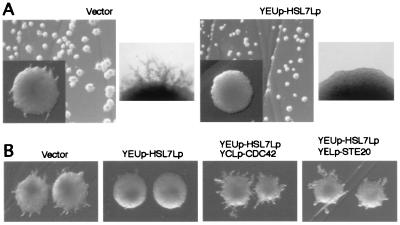
If binding of Hsl7p to Ste20p is essential for negative regulation of its activity, overexpression of HSL7 may inhibit diploid pseudohyphal growth. To overproduce Hsl7p, we fused the LEU2 promoter to the HSL7 ORF on a multicopy vector (YEUp-HSL7Lp). Indeed, overexpression of HSL7 in diploid strains inhibited pseudohyphal development (Fig. 5A).
Figure 5.
(A) Overproduction of HSL7 inhibits diploid pseudohyphal growth. Wild-type diploid strain YAF1111 containing a LEU2-vector (pRS315) was transformed with plasmids described at the top of the images. Cells of those strains were streaked on SLAD medium and incubated for 4 days at 30°C. (Left) Wild-type diploid cells containing vector, YEUp3. (Right) Wild-type diploid cells containing the plasmid for HSL7 overproduction, YEUp-HSL7Lp. (B) Simultaneous expression of CDC42 or STE20 reversed the inhibitory effect to pseudohyphal development of HSL7-overexpressed cells. Strain YAF1111 was transformed with plasmids described at the top of the images. Cells of those strains were streaked on SLAD medium and incubated for 4 days at 30°C. YCLp-CDC42 is the LEU2 plasmid for slight overproduction of CDC42; YELp-STE20 is the LEU2 plasmid for overexpression of STE20.
Slight Overexpression of CDC42 Reverses the Inhibitory Effect of HSL7 Overexpression on Diploid Pseudohyphal Growth.
The interaction between Cdc42p and Ste20p is essential for pseudohyphal development (19, 20). If both Hsl7p and Cdc42p bind to the same noncatalytic region of Ste20p, inhibition of pseudohyphal growth by HSL7 overexpression will be reversed by the simultaneous overexpression of CDC42. Thus, we examined the effect of overexpression of CDC42 on the inhibition of pseudohyphal development caused by the overexpression of HSL7. We found that slight overexpression of CDC42 (CDC42 on low-copy vector) eliminated the inhibitory effect of HSL7 overexpression (Fig. 5B). We also found that STE20 overexpression (STE20 on high-copy vector) reverses the inhibitory effect of HSL7 overexpression (Fig. 5B). These results suggested the possibility that Cdc42p competes with Hsl7p for Ste20p binding. We used the two-hybrid system to test this model. We cloned CDC42 into the reporter plasmid, pLexAop-LucU, for overexpression from its native promoter (pLexAop-LucU-Cdc42p) and used this construct for two-hybrid analysis. Overexpression of Cdc42p resulted in a decrease in luciferase activity associated with the Ste20p-Hsl7p two-hybrid interaction (Table 1). Moreover, We cloned HSL7 into pLexAop-LucU for HSL7 overexpression (pLexAop-LucU-Hsl7p). Overexpression of Hsl7p also resulted in a decrease in luciferase activity associated with the Ste20p-Cdc42p two-hybrid interaction (Table 1). These results suggested that Cdc42p competes with Hsl7p for Ste20p binding to allow diploids to form pseudohyphae in response to nitrogen starvation.
DISCUSSION
We have described the characterization of yeast HSL7, which we identified from a screen for mutants exhibiting filamentous growth on rich medium. HSL7 encodes a protein that is a member of a highly conserved protein family. One family member, the S. pombe Skb1p protein, had been identified from a two-hybrid screen for proteins that interact with S. pombe Shk1p (Ste20p/p65PAK homologue) (33, 34). Based on binding assays and two-hybrid analyses, Hsl7p interacts with Ste20p, as suggested by the interaction S. pombe of Skb1p with Shk1p. HSL7 disruption in haploid and diploid strains resulted in morphological changes (cell elongation) in both haploids and diploids, enhancement of invasive growth in haploids, enhancement of pseudohyphal growth on nitrogen starvation medium, and induction of pseudohyphal growth on nitrogen-rich medium in diploids. Moreover, all of these hsl7 phenotypes were blocked by disruption of STE20. These facts can be explained by positing that Hsl7p inhibits Ste20p through their interaction (Fig. 6). This assumption was supported by the fact that overexpression of HSL7 could inhibit pseudohyphal growth.
Figure 6.
Model for the role of Hsl7p during filamentous growth. Hsl7p binds to Ste20p to repress its activity. In response to nitrogen starvation, Ras2p activates Cdc42p and activated Cdc42p interferes with binding of Hsl7p to Ste20p. Two 14–3-3 proteins, Bmh1p and Bmh2p, which are essential for the pathway leading to pseudohyphal growth, also interact with the C terminus of Ste20p (37).
How is the activity of Ste20p regulated by Hsl7p in the filamentous growth-signaling pathway? One possible explanation is that the transcription of HSL7 is regulated in response to nitrogen starvation. To test this possibility, we made a HSL7 promoter–luciferase gene fusion to monitor the levels of HSL7 expression in response to nitrogen starvation. However, we could not find any decrease in the measured luciferase activities (data not shown). Moreover, the transcript level of HSL7 remained constant (data not shown), indicating that HSL7 was constitutively expressed. We imagined another explanation based on the observation that both Cdc42p and Hsl7p bind to the N-terminal region of Ste20p. The identified Hsl7p-binding domain of Ste20p (amino acid 258–495) includes the Cdc42p-binding domain (amino acid 335–370), suggesting that Cdc42p and Hsl7p may bind to the same or neighboring regions of Ste20p. Although overexpression of HSL7 inhibits pseudohyphal growth, slight overexpression of CDC42 with overexpression of HSL7 reverses pseudohyphal growth (Fig. 5B). Therefore, these findings suggest the possibility that Cdc42p may compete for binding to Ste20p with Hsl7p in response to nitrogen starvation (Fig. 6). Two-hybrid analysis of their interactions supported this idea. Recent work indicated that the interaction of Cdc42p to Ste20p is essential for pseudohyphal development (19, 20), but not for mating. Ste20p lacking the Cdc42p-binding region cannot localize properly (19, 20), causing a defect in pseudohyphal development. Overexpression of HSL7, which leads to inhibition of pseudohyphal growth, may interfere with the binding of Cdc42p to Ste20p. In this context there is a possibility that HSL7 overexpression causes mislocalization of Ste20p. A detailed analysis of localization of Ste20p in HSL7-overexpressing cells may be necessary. In conclusion, Hsl7p may be a potent negative regulator of Ste20p in S. cerevisiae filamentous growth-signaling pathway. The identification of a novel Ste20p regulator will provide additional insights into the regulation of PAKs in both yeasts and higher organisms.
Acknowledgments
We thank J. Thorner, M. Lorentz, J. Heitman, J. Chant, K. Tanaka, C. Oka, Y. Arikawa, Y. Koyama, T. Murakami, W. E. Timberlake, C. J. Gimeno, C. A. Styles, J. R. Pringle, Y. Kikuchi, and G. R. Fink for generously providing yeast strains and plasmids. We also thank J. Chant and M. Ohara for the critical reading of the manuscript.
ABBREVIATIONS
- MAPK
mitogen-activated protein kinase
- MAPKK
MAPK kinase
- MAPKKK
MAPKK kinase
- PAK
p21Cdc42p/Rac-activated kinase
- GST
glutathione S-transferase
- MBP
maltose-binding protein
- SLAD
synthetic low-ammonia medium
- AD
activation domain
- LBD
LexA-binding domain
- ori
origin of replication
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Bardwell L, Cook J G, Inouye C J, Thorner J. Dev Biol. 1994;166:363–379. doi: 10.1006/dbio.1994.1323. [DOI] [PubMed] [Google Scholar]
- 2.Cobb M H, Goldsmith E J. J Biol Chem. 1995;270:14843–14846. doi: 10.1074/jbc.270.25.14843. [DOI] [PubMed] [Google Scholar]
- 3.Denhardt D T. Biochem J. 1996;318:729–747. doi: 10.1042/bj3180729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herskowitz I. Cell. 1995;80:187–195. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 5.Liu H, Styles C A, Fink R G. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 6.Cook J G, Bardwell L, Thorner J. Nature (London) 1997;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- 7.Madhani H D, Styles C A, Fink G R. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 8.Roberts R L, Fink G R. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Z, Gartner A, Cade R, Ammerer G, Errede B. Mol Cell Biol. 1993;13:2069–2080. doi: 10.1128/mcb.13.4.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polverino A, Frost J, Yang P, Hutchison M, Neiman A M, Cobb M H, Marcus S. J Biol Chem. 1995;270:26067–26070. doi: 10.1074/jbc.270.44.26067. [DOI] [PubMed] [Google Scholar]
- 11.Brown J L, Stowers L, Baer M, Trejo J, Coughlin S, Chant J. Curr Biol. 1996;6:598–605. doi: 10.1016/s0960-9822(02)00546-8. [DOI] [PubMed] [Google Scholar]
- 12.Frost J A, Xu S, Hutchison M R, Marcus S, Cobb M H. Mol Cell Biol. 1996;16:3707–3713. doi: 10.1128/mcb.16.7.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bagrodia S, Derijard B, Davis R J, Celione R A. J Biol Chem. 1995;270:27995–27998. doi: 10.1074/jbc.270.47.27995. [DOI] [PubMed] [Google Scholar]
- 14.Leberer E, Dignard D, Harcus D, Thomas D Y, Whiteway M. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 16.Simon M N, De Virgilio C, Souza B, Pringle J R, Abo A, Reed S I. Nature (London) 1995;376:702–705. doi: 10.1038/376702a0. [DOI] [PubMed] [Google Scholar]
- 17.Zao Z S, Leung T, Manser T, Lim L. Mol Cell Biol. 1995;15:5246–5257. doi: 10.1128/mcb.15.10.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosch H-U, Roberts R L, Fink G R. Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peter M, Neiman A M, Park H-O, Lohuizen M, Herskowitz I. EMBO J. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- 20.Leberer E, Wu C, Leeuw T, Fourest-Lieuvin A, Segal J E, Thomas D Y. EMBO J. 1997;16:83–97. doi: 10.1093/emboj/16.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherman F, Fink G R, Hicks J B. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 22.Rothstein R J. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 23.Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann J H. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burns N, Grimwade B, Ross-Macdonald P B, Choi E-Y, Finberg K, Roederm G S, Snyder M. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- 25.Altschul S F, Gish W, Miller W, Meyers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 26.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu C, Whiteway M, Thomas D Y, Leberer E. J Biol Chem. 1995;270:15984–15992. doi: 10.1074/jbc.270.27.15984. [DOI] [PubMed] [Google Scholar]
- 28.Madhani H D, Fink G R. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 29.Butler G, Thiele D J. Mol Cell Biol. 1991;11:476–485. doi: 10.1128/mcb.11.1.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujita A, Oka C, Arikawa Y, Katagai T, Tonouchi A, Kuhara S, Misumi Y. Nature (London) 1994;372:567–570. doi: 10.1038/372567a0. [DOI] [PubMed] [Google Scholar]
- 31.Ma X, Lu Q, Grunstein M. Genes Dev. 1996;10:1327–1340. doi: 10.1101/gad.10.11.1327. [DOI] [PubMed] [Google Scholar]
- 32.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonifield J, Burton J, Connell M, Copsey T, Cooper J, et al. Nature (London) 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 33.Gilbreth M, Yang P, Wang D, Frost J, Polverino A, Cobb M H, Marcus S. Proc Natl Acad Sci USA. 1996;93:13802–13807. doi: 10.1073/pnas.93.24.13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcus S, Polverino A, Chang E, Robbins D, Cobb M H, Wigler M H. Proc Natl Acad Sci USA. 1995;92:6180–6184. doi: 10.1073/pnas.92.13.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manser E, Leung T, Salihuddin H, Zao Z S, Lim L. Nature (London) 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 36.Kron S J, Styles C A, Fink G R. Mol Biol Cell. 1994;5:1003–1022. doi: 10.1091/mbc.5.9.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts R L, Mosch H-U, Fink G R. Cell. 1997;89:1055–1065. doi: 10.1016/s0092-8674(00)80293-7. [DOI] [PubMed] [Google Scholar]
- 38.Gavrias V, Andrianopoulos A, Gimeno C J, Timberlake W E. Mol Microbiol. 1996;19:1255–1263. doi: 10.1111/j.1365-2958.1996.tb02470.x. [DOI] [PubMed] [Google Scholar]
- 39.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]