Abstract
We discuss here the problems in identifying sequence motifs of peptides that bind to I-Ag7, the class II histocompatibility molecule of NOD diabetic mice. We present studies that indicate a minor contribution of amino acid side chains for binding. A peptide from the Eα chain binds to I-Ag7 molecules and is recognized by CD4 T cells. By producing single-residue mutations we identified four residues that were considered to contact the T cell receptor. No residue was found to be essential for binding to I-Ag7: a peptide that contained the T cell contact residues, on a backbone of alanines, bound to I-Ag7 and stimulated the T cells. We conclude that peptides can bind to I-Ag7 without the requirement for residues with prominent side chains to anchor them.
The sequence motif of peptides responsible for their interaction with the diabetogenic class II MHC molecule, I-Ag7, has not been convincingly defined. Defining peptide sequences that favor interaction with I-Ag7 is important in the context of identifying the immunogens that trigger autoimmune diabetes. However, reports in the literature differ markedly in establishing a particular sequence responsible for binding (1–6). For example, in Reich et al.’s detailed analysis of a mouse albumin peptide (1), four residues were found important for binding in that their replacement by alanine markedly affected the interaction; two natural alanine residues could not be replaced with tyrosine; and positive charged residues at the amino terminus and an acidic one at the carboxyl terminus influenced binding. However, no clear sequence motif was identified common to many other peptides studied by them and others (1–6). Amor et al. (2), on the other hand, examining encephalitogenic peptides, paid attention to a central core sequence of four residues made up of a hydrophobic residue, a basic residue, a small residue, and another large hydrophobic residue. But again, this core tetramer was not present in all peptides that stimulated I-Ag7 T cells. Reizis et al. (3) identified some residues that hindered or favored binding in what was assumed to be the P4, P6, and P9 pockets. Alignment of some I-Ag7 binding peptides favored a hydrophobic sequence at P4 with an acidic residue at P9, akin to the sequence of an albumin peptide identified in Reich et al. (1). Finally, Harrison et al. (4) had evidence for two anchor residues in a lysozyme peptide without much specificity (i.e., claiming interaction in a P6 and P9 position).
These variations could be attributed to technical differences with the assays used. After all, I-Ag7 is a weak peptide binding molecule, which forms loose complexes having a fast off-rate and no SDS/PAGE stability (7). We have discussed these features in the context of the autoimmune trait of NOD mice. Another difference could be that peptides bind in several different positions or registers to I-Ag7 and that with each there may be particular residues important for the interaction. Precedence for this alternative was documented in the recent study of peptide binding to I-Ad (8). We report here the detailed findings with a peptide, which bear no major amino acid side chains responsible for anchoring to I-Ag7. However, a number of residues have a marked negative influence on peptide binding. These results could help explain the discrepancies in the literature, by realizing that no major side chain anchors the peptide to I-Ag7.
MATERIALS AND METHODS
Peptides.
Peptides were synthesized by fluorenylmethoxycarbonyl procedures, using an Applied Biosynthesizer (model 432A) (Foster City, CA). Peptides were purified by RP-HPLC on a C18 column (600E, Waters) and analyzed by MS for purity. Stock peptides at 1 mM concentration were dissolved in culture media and stored until its use at −70°C.
Cell Lines.
The Eα12.25 T cell hybridoma was obtained by immunization of NOD mice with a peptide, residues 52–68, of the I-Eα chain (1) in complete Freund’s adjuvant. C3.G7 is a B cell lymphoma cell line that expresses I-Ag7 by transfection with the βg7 chain into M12C3 cell line (9, 10). The OVA-1 T cell is a T cell clone obtained by immunization of NOD mice with residues 323–339 of the protein ovalbumin (OVA) (9, 10). The IL-2-dependent CTLL-2 cell line, the IL-3-dependent cell line, and FDCP3 (11) were maintained in our laboratory.
T Cell Assays.
Eα12.25 T cell hybridoma cells (5 × 104) and 5 × 104 C3.G7 cells were incubated in 96-well plates with DMEM containing 5% FCS and the peptide to test at various concentrations; 24 hr later, 100 μl of supernatant was assayed for IL-2 production by using 4 × 103 CTLL-2 cells as indicator. After 24 hr, cells were pulsed with 1 μCi/ml of [3H]thymidine for 16 hr. The cultures then were harvested, and the incorporation of [3H]thymidine into dividing cells was estimated by liquid scintillation on a β counter. All experiments were done 3–6 times. Results indicated in the tables and figures are mean of three samples. Variations never exceeded 15% of the mean.
Competition Assay.
Competitor peptides in a concentration range of 100 μM to 2 μM were incubated at 37°C with 5 × 103 C3.G7 cells irradiated at 3,000 rad, in a 100 μl final volume of DMEM plus 5% FCS. After 1 hr, 5 × 103 OVA-1 T cells plus 1 μM of 323–339 OVA peptide were added in 100 μl of DMEM with 5% FCS. After 24 hr, 25 μl of the supernatant was used to quantify IL-3 production by using 4 × 103 FDCP3 cells per well as indicator. All competition experiments were done 5–6 times.
RESULTS
Effect of Alanine Substitutions of Eα 52–68 Peptide on T Cell Activation.
The Eα peptide from residues 52–68 was shown to stimulate the T cell hybridoma H12.25. In Table 1 we examined the effects of testing peptides truncated at either the amino or carboxyl terminus. Each peptide was tested in different concentrations, and the amounts required to stimulate the T cell hybridoma by 50% were estimated (and given a value of 1 for the wild-type sequence). Usually the 50% point was reached at the 0.1–1 μM concentration. Examining the amino terminal truncations, we concluded that the optimal stimulatory peptides required the Phe-54 and Glu-55 residues. Ala-64 and Val-65 were required on the carboxyl terminus. A peptide from residues 54–65 was as stimulatory as the wild-type peptide 52–68 (Table 1).
Table 1.
Minimum size of Eα-peptide capable of stimulating the H12.25 T cell hybridoma
| Amino acid residues of the Eα peptide | T cell stimulation | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | 64 | 65 | 66 | 67 | 68 | |
| A | S | F | E | A | Q | G | A | L | A | N | I | A | V | D | K | A | 1 |
| A | S | F | E | A | Q | G | A | L | A | N | I | A | V | D | K | 2 | |
| A | S | F | E | A | Q | G | A | L | A | N | I | A | V | D | 4 | ||
| A | S | F | E | A | Q | G | A | L | A | N | I | A | V | 8 | |||
| A | S | F | E | A | Q | G | A | L | A | N | I | A | 10 | ||||
| A | S | F | E | A | Q | G | A | L | A | N | I | 175 | |||||
| S | F | E | A | Q | G | A | L | A | N | I | A | V | D | K | A | 1 | |
| F | E | A | Q | G | A | L | A | N | I | A | V | D | K | A | 1 | ||
| E | A | Q | G | A | L | A | N | I | A | V | D | K | A | 10 | |||
| E | A | Q | G | A | L | A | N | I | A | >250 | |||||||
| A | Q | G | A | L | A | N | I | A | V | >250 | |||||||
| A | Q | G | A | L | A | N | I | A | (−) | ||||||||
| F | E | A | Q | G | A | L | A | N | I | A | V | 2 | |||||
Truncated peptides of the 52-68 Eα-wild-type peptide were tested for their capacity to stimulate the production of IL-2 by Eα 12.25 T cell hybridoma. T cell stimulation refers to the concentration of peptide that gave 50% of the maximum T cell response compared to the wild-type peptide Eα 52-68 (stimulation equal to 1). The 50% of the maximum T cell response was reached with 0.4 μM of wild-type peptide.
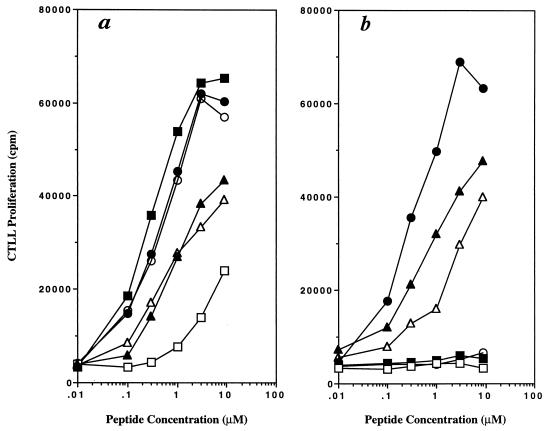
Amino acids of the Eα peptide 52–68 from residue 53–66 then were substituted with alanine, and the modified peptides were assayed for their ability to stimulate the T cell hybridoma H12.25 through a concentration range from 0.1 to 10 μM. Peptides with single substitution of residues 60 (Leu), 62 (Asn), or 63 (Ile) to alanine did not stimulate the T cell hybridoma (Fig. 1). The substitution of residue 55 (Glu) with alanine significantly reduced the T cell stimulatory activity (Fig. 1). Residues 53 and 54 had no effect, and those with substitutions of residues 57, 58, 65, and 66 had a slight reduction in stimulation (≈ 10-fold).
Figure 1.
Alanine scanning of 52–68 residues from Eα peptide. Different concentrations of alanine-substituted peptides were incubated in the presence of 5 × 104 H12.25 T cell hybridoma and 5 × 104 C3.G7 cells for 24 hr. Later, 100 μl of the supernatants was removed and assayed for IL-2 production by using 5 × 103 CTLL-2 cells and incubated for 24 hr. Cells then were pulsed with 1 μCi/ml of [3H]thymidine for 16 hr. Two representative experiments are shown. (a) The T cell response of H12.25 hybridoma in the presence of the following alanine-substituted peptides: (●) wild-type 52–68, (○) Ala-53, (■) Ala-54, (□) Ala-55, (▴) Ala-57, and (▵) Ala-58. (b) The T cell response of H12.25 hybridoma in the presence of the following alanine-substituted peptides: (●) wild-type 52–68, (○) Ala-60, (■) Ala-62, (□) Ala-63, (▴) Ala-65, and (▵) Ala-66.
The Alanine-Substituted Peptides That Did Not Stimulate T Cells Bind to I-Ag7.
The lack of T cell stimulation with an alanine-substituted peptide could be the result of either a lack of MHC binding of the peptide or the lack of interaction of the MHC peptide complex with T cells (12).
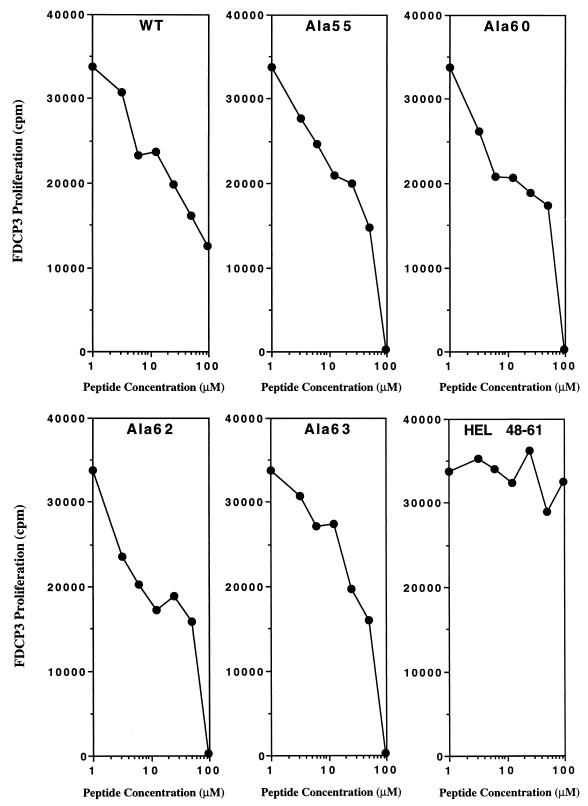
To evaluate whether the alanine substitution of the four residues that failed to stimulate was affecting either binding to MHC or the interaction with T cells, we measured their binding to I-Ag7 MHC molecules. In this assay, each peptide was tested for the inhibition of the IL-3 production by the I-Ag7 restricted OVA-specific T cell clone (OVA-1) stimulated with OVA peptide 326–339. As shown in Fig. 2, alanine-substituted Eα 52–68 peptide, at residues 55, 60, 62, or 63, as well as the wild-type Eα peptide, inhibited the OVA-specific T cell response in a dose-dependent fashion. An unrelated peptide, from HEL residues 48–61, did not inhibit the response of the OVA-T cell clone even at 100 μM.
Figure 2.
Alanine-substituted peptides: Ala-55, Ala-60, Ala-62, and Ala-63 can bind to I-Ag7 specifically. Different concentrations of alanine-substituted Eα 52–68 peptide (Ala-55, Ala-60, Ala-62, and Ala-63) or an unrelated nonbinding control peptide (HEL 48–61) were assayed for its ability to compete with OVA 323–339 peptide and block the T cell response of OVA-1 T cell clone. Competitor peptides plus 1 μM of 323–339 OVA peptide were incubated in the presence of 5 × 103 OVA T cells and 5 × 103 C3.G7 irradiated cells for 24 hr. Later, 25 μl of culture supernatant was removed and tested for IL-3 production by using FDCP3 cells as indicator cells.
These results strongly suggest that alanine substitution at positions 55, 60, 62, and 63 affected the interaction between the I-Ag7-bound peptide and the T cells. Thus Glu-55, Leu-60, Asn-62, and Ile-63 are likely T cell receptor (TCR) contact residues in the Eα peptide 52–68. It should be noted that a high dose of Ala-55-substituted peptide stimulated the Eα-specific T cell hybridoma (Fig. 1). This finding suggests that Glu at position 55 plays a lesser role in the interaction between I-Ag7-Eα peptide complex and the H12.25 T cell hybridoma.
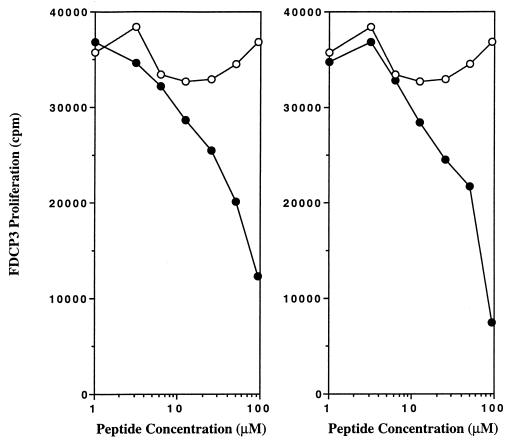
The other alanine-substituted Eα peptides stimulated the Eα-specific hybridoma, indicating their binding to the I-Ag7 MHC molecule. This result indicated that no single residue in the Eα peptide was critical for MHC binding. To address more closely the nature of the peptide-I-Ag7 binding, we tested a synthetic polyalanine peptide for I-Ag7 binding. A 15-mer polyalanine peptide with an arginine at the carboxyl terminus (to improve the solubility of the peptide) was used in a competition assay as described in the previous section. As shown in Fig. 3 (Right), the polyalanine 15-mer peptide binds to I-Ag7 in a dose-dependent fashion. (In experiments not shown, we found that the terminal arginine was not required for binding. Polyalanine peptides containing eight or 10 residues bound considerably less than the 15-residue peptide.) Moreover, a polyalanine peptide containing the TCR contact residues Glu, Leu, Asp, and Ile with the same spacing as in the Eα peptide 52–68 (AAAEAAAALANIAAA) instead of ASFEAQGALANIAVD also bound to I-Ag7 MHC (Fig. 3).
Figure 3.
A 15-mer polyalanine peptide is able to compete for binding to I-Ag7 with the 323–339 OVA peptide. (Left) A competition assay using different concentrations of a polyalanine peptide containing residues Glu-55, Leu-60, Asn-62, Ile-63 (●) or with a nonbinder peptide (○) 48–61 HEL. (Right) A competition assay using different concentrations of a 15-mer polyalanine peptide with an arginine at the carboxyl terminus to improve solubility (●). The results with the nonbinder peptide (○) 48–61 HEL from the left panel are included. Competitor peptides were tested as detailed in the legend of Fig. 2.
Polyalanine Peptides Bearing the TCR Contact Residues Stimulate T Cells.
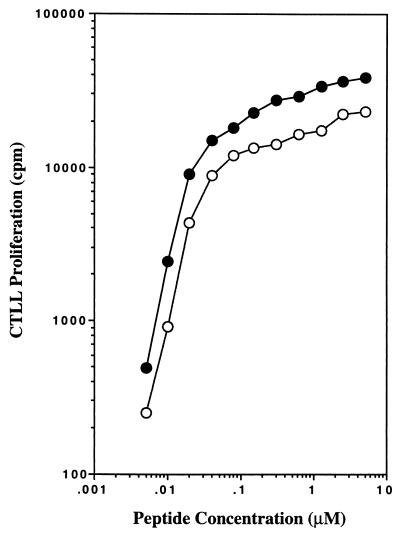
The polyalanine peptide containing the four TCR contact residues was tested for their capacity to stimulate the T cell hybridoma H12.25 (Fig. 4). We obtained the surprising result that this peptide exhibited stimulatory activity identical to the wild-type Eα peptide. Thus, Glu-55, Leu-60, Asn-62, and Ile-63 represent T cell contact residues of the Eα peptide that can be displayed on a polypeptide backbone lacking large amino acid side chains.
Figure 4.
A polyalanine peptide containing T cell contact residues: Glu-55, Leu-60, Asn-62, and Ile-63 can stimulate H12.25 T cell hybridoma. Different concentrations of a polyalanine peptide containing residues Glu-55, Leu-60, Asn-62, and Ile-63 (○) or wild-type 52–68 Ea-peptide (●) were incubated in the presence of T cell hybridoma H12.25, and production of IL-2 was measured 24 hr later.
Not All Amino Acid Substitutions Are Well Tolerated.
We examined the influence of different amino acids on six of the peptide residues of peptide 54–65 thought to be involved in contacting I-Ag7, rather than in providing TCR contacts (Table 2). Except for some amino acid substitutions at the Gly-58 and Ala-61 positions, most of the changes were well tolerated, as judged by both stimulation of the H12.25 hybridoma and inhibition of the OVA-reactive T cell. Thus, at residue 56 we could substitute different amino acids with some effect on binding but no major alteration in the T cell stimulation. The same happened with substitution of Gln-57, Ala-59, or Ala-64. (A peptide with glutamic acid at residue 59, however, was not tolerated.) As noted, residue Gln-58 could not be substituted with negatively charged or some hydrophobic or polar residues. Either basic or acidic charged residues inhibited binding at the 61 residue.
Table 2.
Effect of amino acid substitutions of MHC contact residues
| Amino acid position at Ea peptide | T cell stimulation | Binding | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | 64 | ||
| E | A | Q | G | A | L | A | N | I | A | ||
| − | Y | − | − | − | − | − | − | − | − | + | ++ |
| − | K | − | − | − | − | − | − | − | − | ++ | + |
| − | E | − | − | − | − | − | − | − | − | ++ | ++ |
| − | I | − | − | − | − | − | − | − | − | ++ | + |
| − | L | − | − | − | − | − | − | − | − | ++ | + |
| − | V | − | − | − | − | − | − | − | − | ++ | + |
| − | T | − | − | − | − | − | − | − | − | +++ | + |
| − | S | − | − | − | − | − | − | − | − | +++ | + |
| − | − | Y | − | − | − | − | − | − | − | + | ++ |
| − | − | K | − | − | − | − | − | − | − | +++ | ++ |
| − | − | E | − | − | − | − | − | − | − | + | + |
| − | − | I | − | − | − | − | − | − | − | +++ | + |
| − | − | − | Y | − | − | − | − | − | − | + | ++ |
| − | − | − | K | − | − | − | − | − | − | Negative | ++ |
| − | − | − | E | − | − | − | − | − | − | +/− | Negative |
| − | − | − | I | − | − | − | − | − | − | Negative | Negative |
| − | − | − | L | − | − | − | − | − | − | + | + |
| − | − | − | V | − | − | − | − | − | − | +/− | Negative |
| − | − | − | A | − | − | − | − | − | − | +++ | + |
| − | − | − | T | − | − | − | − | − | − | +/− | Negative |
| − | − | − | S | − | − | − | − | − | − | +++ | + |
| − | − | − | H | − | − | − | − | − | − | Negative | Negative |
| − | − | − | − | Y | − | − | − | − | − | + | + |
| − | − | − | − | K | − | − | − | − | − | Negative | + |
| − | − | − | − | E | − | − | − | − | − | +/− | Negative |
| − | − | − | − | I | − | − | − | − | − | +++ | ++ |
| − | − | − | − | L | − | − | − | − | − | +++ | ++ |
| − | − | − | − | V | − | − | − | − | − | ++ | ++ |
| − | − | − | − | T | − | − | − | − | − | +++ | + |
| − | − | − | − | S | − | − | − | − | − | +++ | + |
| − | − | − | − | H | − | − | − | − | − | ++ | + |
| − | − | − | − | − | − | Y | − | − | − | Negative | Negative |
| − | − | − | − | − | − | K | − | − | − | Negative | Negative |
| − | − | − | − | − | − | E | − | − | − | Negative | Negative |
| − | − | − | − | − | − | I | − | − | − | +/− | Negative |
| − | − | − | − | − | − | L | − | − | − | + | + |
| − | − | − | − | − | − | V | − | − | − | +/− | + |
| − | − | − | − | − | − | G | − | − | − | +/− | + |
| − | − | − | − | − | − | T | − | − | − | + | + |
| − | − | − | − | − | − | S | − | − | − | +++ | + |
| − | − | − | − | − | − | F | − | − | − | +/− | + |
| − | − | − | − | − | − | − | − | − | Y | +/− | + |
| − | − | − | − | − | − | − | − | − | K | +/− | + |
| − | − | − | − | − | − | − | − | − | E | +++ | ++ |
| − | − | − | − | − | − | − | − | − | I | + | + |
| − | − | − | − | − | − | − | − | − | L | + | + |
| − | − | − | − | − | − | − | − | − | V | ++ | + |
| − | − | − | − | − | − | − | − | − | T | ++ | + |
| − | − | − | − | − | − | − | − | − | S | +++ | + |
This table summarizes all experiments done testing peptides with different residues at key positions thought to be MHC contact. T cell stimulation was done by titrating various amounts of each peptide: a +++ indicates a peptide that required less than 10 μM concentration for 50% response; a ++ response required up to 40 μM; a + response was elicited with 40–100 μM; a +/− response is one where at 40 or 100 μM the extent of the response never reached a 50% level. Binding was carried out by a functional assay, testing for blocking of the response by an OVA reactive peptide: a +++ binding indicates that 10 μM peptide or less was required to inhibit by 50% the response to 1 μM OVA peptide. A ++ binding required up to 25 μM; a + binding required from 25 to 100 μM for 50% inhibition. No binding refers to a lack of inhibition with peptides at 100 μM. All assays with each peptide were done 4–5 times.
Comments.
The main result to come from our analysis of the Eα peptide is the lack of a defined residue that anchors the peptide to I-Ag7 and fixes its position in the binding groove. Moreover, we defined four residues that influenced the interaction with the TCRs and that operationally were defined as TCR contact residues. These four amino acids placed on a backbone of polyalanines were sufficient to stimulate the T cell as effectively as the wild-type peptide. Other observations were consistent with the above findings: polyalanine peptide was able to interact with I-Ag7 (see also ref. 4); and furthermore, no clear anchor residue could be identified in the Eα peptide, among the residues thought to be involved in the interactions with I-Ag7.
The position that the Eα peptide (or its alanine-substituted form) occupies in the binding groove could be influenced by some of the residues assumed to be TCR contact. Some TCR contact residues could have an influence in binding and positioning the peptide, and indirectly influencing the display of the set of solvent exposed residues (13–16). Alternatively, the Eα peptide may interact in diverse registers: it is our T cell that reads out the register to which it has complementarity. (A second T cell hybridoma that we have examined in limited experiments responded very similarly to the Eα 12.25 clone reported here, except for a lack of requirement of Asn-62.) Thus, the relatively poor influence of its component amino acid side chains may lead to different alignments of the peptide backbone. This possible difference in registers may be one explanation for the results obtained by various groups and the frustration in defining a binding motif. Finally, the binding reactions with I-Ag7 have limitations and are not precisely quantitative (see Table 2). The issue of residues inhibiting the interaction of the peptide with the MHC binding site was apparent for the Eα peptide and is an important consideration in determining whether a peptide binds.
The I-Ag7 molecule binds to many peptides with broad specificity (1–7) and with low binding strength (7) as expected by the limited influence imparted by side-chain interactions. This highly promiscuous and weak interaction may be the feature that sets the I-Ag7 molecule in a unique situation to favor autoreactivity, as we (7) and others have discussed (ref. 17, and also ref. 18, regarding autoimmune encephalomyelitis). I-Ag7 could tilt toward autoimmunity by not favoring thymic negative selection combined with the facile interactions with self peptides in lymphoid organs. We recently have shown the high level of autoreactive T cells in mice bearing I-Ag7 proteins (9). A situation similar to the polyalanine peptide containing the critical Eα residues was reported for the also weak interaction of a myelin basic protein peptide and I-Au molecules (19, 20).
Acknowledgments
This research was supported by the National Institutes of Health and the Kilo Research Foundation.
ABBREVIATIONS
- OVA
ovalbumin
- TCR
T cell receptor
References
- 1.Reich E-P, von Grafenstein H, Barlow A, Swenson K E, Williams K, Janeway C A., Jr J Immunol. 1994;152:2279–2288. [PubMed] [Google Scholar]
- 2.Amor S, O’Neill J K, Morris M M, Smith R M, Wraith D C, Groome N, Travers P J, Baker D. J Immunol. 1996;156:3000–3008. [PubMed] [Google Scholar]
- 3.Reizis B, Eisenstein M, Bocková J, Könen-Waisman S, Mor F, Elias D, Cohen I R. Int Immunol. 1997;9:43–51. doi: 10.1093/intimm/9.1.43. [DOI] [PubMed] [Google Scholar]
- 4.Harrison L C, Honeyman M C, Trembleau S, Gregori S, Gallazzi F, Augstein P, Brusic V, Hammer J, Adorini L. J Exp Med. 1997;6:1013–1021. doi: 10.1084/jem.185.6.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao C C, McDevitt H O. Immunogenetics. 1997;46:29–34. doi: 10.1007/s002510050238. [DOI] [PubMed] [Google Scholar]
- 6.Oiso M, Matsushita S, Nishi T, Ishikawa T, Nakano N, Yoshida K, Kikutani H, Nishimura Y. Immunogenetics. 1998;47:411–417. doi: 10.1007/s002510050377. [DOI] [PubMed] [Google Scholar]
- 7.Carrasco-Marin E, Shimizu J, Kanagawa O, Unanue E R. J Immunol. 1996;56:450–458. [PubMed] [Google Scholar]
- 8.Scott A C, Peterson P A, Teyton L, Wilson I A. Immunity. 1998;8:319–320. doi: 10.1016/s1074-7613(00)80537-3. [DOI] [PubMed] [Google Scholar]
- 9.Kanagawa O, Martin S M, Vaupel B A, Carrasco-Marin E, Unanue E R. Proc Natl Acad Sci USA. 1998;95:1721–1724. doi: 10.1073/pnas.95.4.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanagawa O, Vaupel B A, Xu G, Unanue E R, Katz J. J Immunol. 1998;161:4489–4492. [PubMed] [Google Scholar]
- 11.Kelso A. J Immunol. 1986;136:2930–2938. [PubMed] [Google Scholar]
- 12.Allen P M, Matsueda G R, Evans R J, Dunbar J B, Jr, Marshall G, Unanue E R. Nature (London) 1987;327:713–715. doi: 10.1038/327713a0. [DOI] [PubMed] [Google Scholar]
- 13.Evavold B D, Williams S G, Hsu B L, Buus S, Allen P M. J Immunol. 1992;148:347–353. [PubMed] [Google Scholar]
- 14.Fremont D H, Hendrickson W A, Marrack P, Kappler J. Science. 1996;272:1001–1004. doi: 10.1126/science.272.5264.1001. [DOI] [PubMed] [Google Scholar]
- 15.Kersch G J, Allen P M. J Exp Med. 1996;184:1259–1268. doi: 10.1084/jem.184.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dadaglio G, Nelson C A, Deck M B, Petzold S J, Unanue E R. Immunity. 1997;6:727–738. doi: 10.1016/s1074-7613(00)80448-3. [DOI] [PubMed] [Google Scholar]
- 17.Ridgway W M, Ito H, Fasso M, Yu C, Fathman C G. J Exp Med. 1998;188:2267–2275. doi: 10.1084/jem.188.12.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fairchild P J, Wildgoose R, Atherton E, Webb S, Wraith D C. Int Immunol. 1993;5:1151–1156. doi: 10.1093/intimm/5.9.1151. [DOI] [PubMed] [Google Scholar]
- 19.Gautam A M, Pearson C I, Smilek D E, Steinman L, McDevitt H O. J Exp Med. 1992;176:605–609. doi: 10.1084/jem.176.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gautam A M, Lock C B, Smilek D E, Pearson C I, Steinman L, McDevitt H O. Proc Natl Acad Sci USA. 1994;91:767–771. doi: 10.1073/pnas.91.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]