Abstract
The proteins bound in vivo at the human lamin B2 DNA replication origin and their precise sites of binding were investigated along the cell cycle utilizing two novel procedures based on immunoprecipitation following UV irradiation with a pulsed laser light source. In G1, the pre-replicative complex contains CDC6, MCM3, ORC1 and ORC2 proteins; of these, the post-replicative complex in S phase contains only ORC2; in M phase none of them are bound. The precise nucleotide of binding was identified for the two ORC and the CDC6 proteins near the start sites for leading-strand synthesis; the transition from the pre- to the post-replicative complex is accompanied by a 17 bp displacement of the ORC2 protein towards the start site.
Keywords: lamin B2 origin/ORC proteins/origin of replication/UV cross-linking
Introduction
DNA replication is regulated in all living organisms by modulating the activation of the replication origins, herein abbreviated as ori(s). The study of relatively simple eukaryotic organisms (Saccharomyces cerevisiae and Schizosaccharomyces pombe) has shown that during the G1 phase of the cell cycle a multiprotein complex assembles on the ori sequence to form a structure called the pre-replicative complex. This occurs when a six protein origin recognition complex (ORC) binds in a sequence-specific fashion to the ori (Bell and Stillman, 1992; Aparicio et al., 1997; Ogawa et al., 1999). The selection of the sequence may be influenced by another specific protein, Cdc6p (Mizushima et al., 2000; Sun et al., 2002). Once ori-bound this protein complex drives the recruitment of other ori activation-specific proteins, Ctd1p (Maiorano et al., 2000; Nishitani et al., 2000; Tanaka and Diffley, 2002), another six protein complex called the MCM (Tanaka et al., 1997) and other yet unidentified proteins. This pre-replicative complex triggers the start of bi-directional replication. This event is followed by the destruction or inactivation of Cdc6p, the removal of the MCM proteins and the reduction of the extension of the protected sequence down to a shorter post-replicative complex (Diffley et al., 1994; Bielinsky and Gerbi, 2001; Diffley and Labib, 2002). When the cell enters, after mitosis and cell division, a new cell cycle, the pre-replicative complex is again constituted.
Homologues of the yeast pre-replicative complex proteins have been identified and characterized in Drosophila, Xenopus and Homo sapiens (Kelly and Brown, 2000; Blow, 2001); the lack of well defined oris has not yet allowed a comparably detailed description of the ori activation and inactivation cycle in these organisms, but some similarities and differences are already apparent. Contrary to what happens in yeast, where all the ORC subunits remain bound to chromatin throughout the cell cycle, in Drosophila, Xenopus, as well as in hamster and human cells, one or more ORC subunits leave the chromatin at different times (Coleman et al., 1996; Romanowski et al., 1996; Hua and Newport, 1998; Findeisen et al., 1999; Rowles et al., 1999; Natale et al., 2000; Kreitz et al., 2001; Ladenburger et al., 2002; Li and DePamphilis, 2002; Mendez et al., 2002); indeed, the programmed release of Xenopus ORC from somatic cell chromatin has recently been demonstrated in Xenopus egg extracts and found to be dependent on pre-replication complex assembly (Sun et al., 2002). Furthermore, the putative pre-replicative and post-replicative complexes assembled around the human lamin B2 ori are much more extended (110 and 70 bp, respectively) than the corresponding yeast complexes (Dimitrova et al., 1996; Abdurashidova et al., 1998). It has to be expected therefore that, besides the human homologues of the ORC, MCM, etc. proteins, other components of the replicative complexes could be identified, and that the dynamics of the ori activation and inactivation processes may reveal more elaborate modulations than in yeast.
Previously we have shown that the human lamin B2 ori, that is activated immediately after the onset of the S phase (Biamonti et al., 1992), is the site of the assembly of the large pre- and post-replicative complexes mentioned above, and that these disassemble in mitosis. We have also identified the start sites of leading-strand synthesis which correspond to single nucleotides overlapping by four nucleotides on the complementary strands (Abdurashidova et al., 2000). The initiation event occurs within the area protected in both the G1 and S phases. The lamin B2 ori thus offers the possibility of investigating the composition and the structure (particularly as concerns the precise protein–DNA interactions involved) of the replication complexes as well as their functionally relevant modifications occurring along the cell cycle.
To this purpose, we have developed two novel procedures that allow identification of (i) proteins bound in vivo at the portion of the lamin B2 ori protected at different time points of the cell cycle and (ii) the precise nucleotides to which they are bound. The procedures are based on the principle of in vivo cross-linking coupled to chromatin immunoprecipitation. We were thus able to demonstrate the presence of some of these proteins in the area adjacent to the start sites of leading-strand synthesis, to identify for some of them the precise nucleotide to which they are bound, and to follow the variations of these events in the G1 and S phases. The results are reported in detail and discussed below.
Results and discussion
Cross-linking of specific proteins to ori DNA in UV-irradiated cells
Formaldehyde-induced chemical cross-linking is widely used for the study of protein–DNA interactions occurring in vivo (Solomon et al., 1988). However this method has the disadvantage of also inducing extensive protein–protein cross-links and of forming, by subsequent steps, chemical bridges of significant length, with the risk of creating artifacts. These drawbacks can be overcome, in principle, by the use of UV irradiation as the cross-linking agent (Gilmour and Lis, 1986), but in this case, if conventional lamps are used, the process is relatively inefficient, generating heat, which may perturb the existing DNA–protein interactions. Also both methods suffer from the fact that the times of the cross-linking treatments are rather long (of the order of minutes), therefore not suitable for the study of rapid kinetics and are also likely to create covalent links among structures that only casually come in fleeting contact. The use instead of a pulsed UV-laser light source avoids both disadvantages: the excitation times are of the order of nanoseconds or shorter, hence much faster than the standard times of microconformational transitions of macromolecules that are of the order of 100 µs (Careri et al., 1975); thus laser irradiation ‘freezes’ only the prevalent protein–DNA interactions present at any one time and allows snapshots to be taken of the subsequent steps during the assembly of a large protein–DNA complex (Moss et al., 1997; Mutskov et al., 1997; Russman et al., 1998). Furthermore, the DNA base excitation process is a two-step process, involving the consecutive absorption of two photons; hence, a short pulse excitation reduces the persistence of the excited molecule in the intermediate states where competing processes may initiate and thus increases the probability of effective cross-linking. The efficiency of the process can be further enhanced by exploiting the fact that the second excitation step (S1–Sn or T1–Tn) can be induced by a longer wavelength photon in the blue range; thus, a simultaneous two-wavelength excitation with harmonics of a solid-state laser (Russman et al., 1998) allows a significant increase in the cross-linking efficiency while keeping DNA damage low.
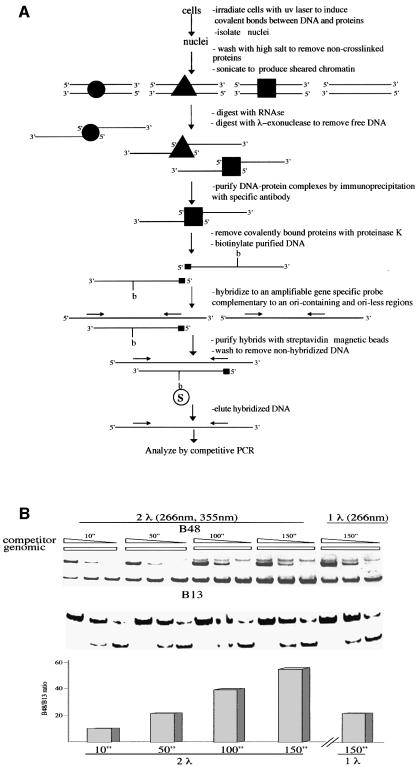
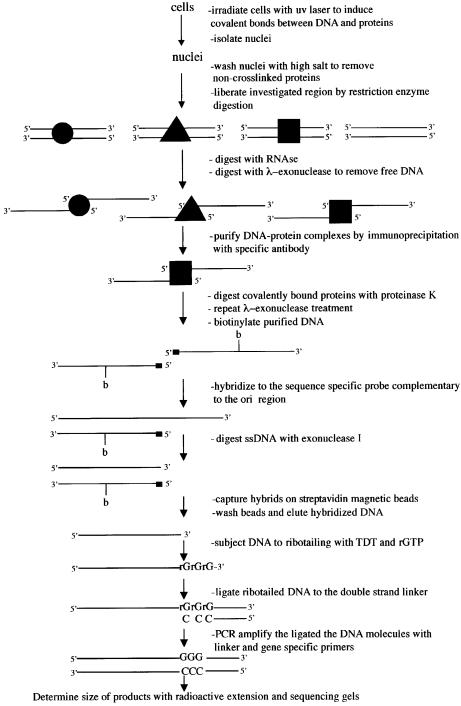
We have designed a procedure based on the simultaneous pulse irradiation of HeLa cells with laser light at 266 and 355 nm, with the purpose of determining the presence of specific proteins on the lamin B2 ori sequence. The procedure is depicted in Figure 1 and can be thus summarized: the cells are irradiated, nuclei are isolated and depleted of non-cross-linked proteins by salt wash; the chromatin is sheared by sonication, freed of RNA and treated with λ-exonuclease, which hydrolyzes the single strands of a duplex from the 5′ end and also progresses across UV photoproducts (Mattes, 1990) until it reaches a protein-bound nucleotide; the residual DNA fragments bearing a protein bound to their 5′ end are precipitated with specific antibodies, isolated and digested with proteinase K, leaving only short peptides bound to DNA. The material must now be analyzed to see whether a significant enrichment for the lamin B2 ori was obtained. In view of the limited amount of material available quantitative PCR procedures are imperative to this purpose, but the presence of DNA polymerase blocking photoproducts imposes a detour as follows: the DNA fragments are biotinylated, annealed to defined single-strand probes complementary to the lower or upper strand of the ori sequence or to a non-ori, 5 kb removed DNA sequence; the duplexes are captured from the solution with streptavidin-coated magnetic beads and the annealed probes are eluted and assayed for the abundance of ori and non-ori DNA with competitive PCR (Giacca et al., 1997). This entails the simultaneous amplification in the same sample of the DNA fragment of unknown abundance and of a known number of molecules of a competitor having the same sequence as the probe but with an extra short nucleotide sequence allowing the separation from the probe in electrophoresis but assuring an identical amplification.
Fig. 1. Procedure for the identification of proteins bound to the lamin B2 ori sequence and validation with the USF binding. (A) Flow-chart of the procedure; for a detailed description see Materials and methods. (B) Cross-linking of USF to the ori sequence: the cells were exposed to the treatment described in (A) and the isolated DNA was analyzed for the abundance of ori (B48 probe) and non-ori (B13 probe) sequences. From left to right, the results of five sets of competitive PCR experiments (Giacca et al., 1997) are shown: the first four performed on DNA isolated after treatment of the cells with two-wavelength pulsed laser light for the indicated times and the fifth one with one wavelength light and one exposure time only. The immunoprecipitation was performed with anti-USF antibody. Each set of PCR reactions contained the same amount of isolated DNA and concentrations of competitor decreasing by a factor of 10. The upper portion of the figure shows the results of the analysis with a competitor for the ori sequence, the lower portion with a competitor for the non-ori sequence. The amounts of the corresponding sequences in genomic DNA are interpolated by comparison with the known abundance of competitor molecules present. The quantitation of the relative abundance of ori versus non-ori sequence is shown at the bottom. The two-wavelength treatment appears significantly more efficient.
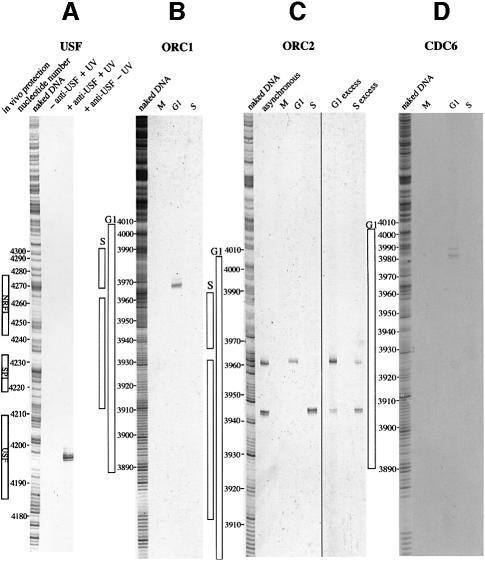
The procedure was validated by testing for the presence on the lamin B2 ori region of the USF transcription factor, that is known to reside on the promoter of the ppv gene, located ∼250 bp to the right of the replicative complex (Dimitrova et al., 1996). The results are reported in Figure 1; the B48 probe corresponds to the core of the ori region, whereas the B13 probe represents an area 5 kb removed to the right of the ori. Four different irradiation times were tested and for each of them three competitive PCR reactions were performed with primers appropriate to amplify the ori sequence and three with primers for the non-ori one. The three reactions contained concentrations of the appropriate competitor decreasing each by an order of magnitude. The abundance of the probe DNA was estimated by comparing the intensity of the corresponding electrophoretic band with that of the known amounts of competitor present in each reaction. The samples deriving from non-irradiated cells contain the same abundance of B48 and B13 molecules (not shown). Even at the shortest time of cumulative simultaneous pulse irradiation with the two laser wavelengths a significant excess of the ori DNA versus non-ori is visible; the abundance of ori DNA keeps increasing up to 150 s, whereas the non-ori DNA remains constant. During this time the laser source emits ∼1000 pulses of 8 ns each, so that the total time of exposure to the cross-linking agent is of the order of 10 µs. The simultaneous irradiation with two wavelengths appears significantly more efficient than with UV light only, since at 150 s we can estimate in the latter conditions an ori versus non-ori excess of ∼25-fold compared with a value of 50-fold for the two-wavelength irradiation, corresponding to an efficiency of cross-linking close to 5% for USF (data not shown).
We conclude that the chromatin immunoprecipitation procedure we have devised, based on UV laser-induced in vivo cross-linking, is adequate to provide evidence for the presence of specific proteins bound to specific regions of human DNA. The procedure was thus applied to proteins homologous to the key components of the yeast replicative complexes.
Binding of hOrc1p and hOcr2p to the lamin B2 ori in different cell cycle phases
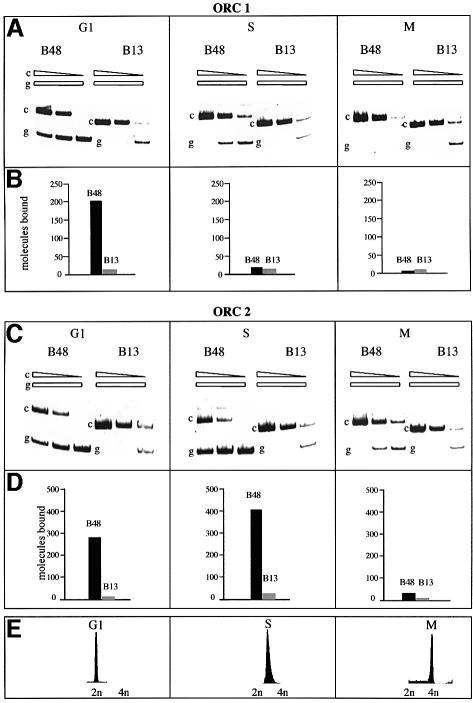
We investigated first the presence of the human homologues of Orc1p and Orc2p in the ori region in cells accumulated in G1, S and mitosis and irradiated with the two wavelengths for 150 s. These two proteins remain stably bound to the yeast chromatin throughout the cell cycle (Diffley et al., 1994; Aparicio et al., 1997; Liang and Stillman, 1997) and undoubtedly play an important role in ori activation and in the prevention of reactivation. However in human cells it was recently shown, by formaldehyde cross-linking studies, that whereas both hOrc1p and hOrc2p are DNA-bound in G1, hOrc1p (but not hOrc2p) leaves the replicative complex in S (Ladenburger et al., 2002). With our procedure we confirm and extend these observations to the lamin B2 ori. The experiments performed using precipitating antibodies specific for hOrc1p and hOrc2p show a clear enrichment of the ori region over the non-ori control in G1 (Figure 2), demonstrating that both proteins are bound in the region of the lamin B2 ori in this phase and are most probably part of the pre-replicative complex, by analogy to that reported in yeast. But as we move into the S phase and the ori activation is followed by the start of bi-directional synthesis and the passage from the pre- to the more compact post-replicative complex, hOrc1p leaves the ori region; this is in contrast to what happens in yeast and in agreement with what has been observed in metazoan chromatin (Hua and Newport, 1998; Rowles et al., 1999; Natale et al., 2000; Kreitz et al., 2001; Li and DePamphilis, 2002; Mendez et al., 2002; Sun et al., 2002), as well as for the human ori located in the region between the two divergently transcribed genes MCM4 and PRKD (Ladenburger et al., 2002). When we extended our analysis to the M phase we could detect, with our procedure, no enrichment of the ori DNA when using the antibody precipitating hOrc1p, and a very modest enrichment (in comparison with the data in G1 and S) with the anti-hOrc2p antibody. This observation is in agreement with our previous report that the human replicative complex is not present in packed mitotic chromosomes, and with the data showing that mitotic Xenopus and Drosophila chromatin does not contain ORC proteins (Coleman et al., 1996; Romanowski et al., 1996; Hua and Newport, 1998; Rowles et al., 1999; Sun et al., 2002); alternatively, it is conceivable that the chemical details of the interaction of, at least, hOrc2p with chromatin in M are different than in G1 and S, resulting in a different efficiency of cross-linking.
Fig. 2. Presence of hOrc1p and hOrc2p on the ori sequence along the cell cycle. The cells arrested in the G1, S or M phase were irradiated with two-wavelength light for 150 s and treated with the procedure described in Figure 1A. The isolated DNA was analyzed for the abundance of ori and non-ori sequences as described in Figure 1B. (A and C) PCR analysis following precipitation with antibodies versus hOrcp1 and hOrcp2, respectively. (B and D) Quantitation of the abundance of ori and non-ori sequences interpolated from the PCR analysis. (E) Flow-cytometric analysis of the treated cell cultures.
Binding of hCdc6p and hMcm3p to the lamin B2 ori in different cell cycle phases
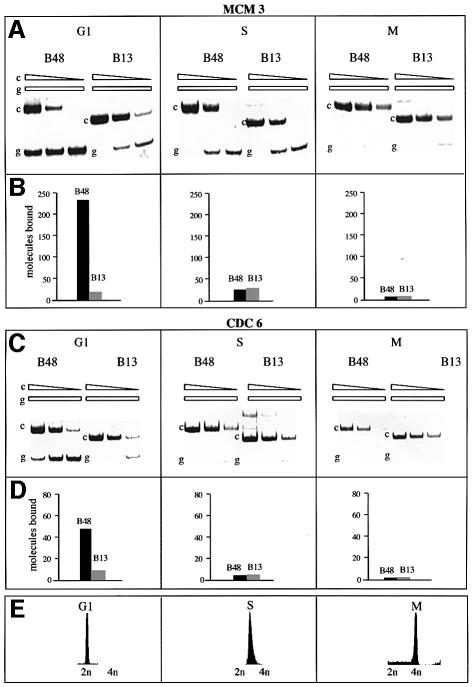
We extended our studies to other key components of the human replicative complexes, namely the CDC6 and MCM3 proteins. Cells in the G1, S or M phase were irradiated as indicated previously for 150 s and the chromatin was analysed with the procedure illustrated in Figure 1 using antibodies raised against hCdc6p and hMcm3p (Figure 3). We were able to demonstrate that indeed these two molecules assemble on the DNA of the ori region in G1, completing, conceivably, the assembly of the pre-replicative complex. Once the ori has been activated, replication has started and the cells have entered the S phase, both proteins depart from the ori region whereas Mcmp is likely to move along the replicon in concert with the replisome (Schaarschmidt et al., 2002).
Fig. 3. Presence of hMcm3p and hCdc6p on the ori sequence along the cell cycle. As Figure 2, except that the antibodies were those raised versus the indicated proteins.
The absence from chromatin of both these proteins in the mitotic chromosomes is in agreement with our previously reported inability to detect the presence of replication or transcription cofactors on the ori region following chromosome condensation in mitosis. However, as indicated above, we cannot rule out the possibility that some factor may be bound in a fashion not detectable with our procedure.
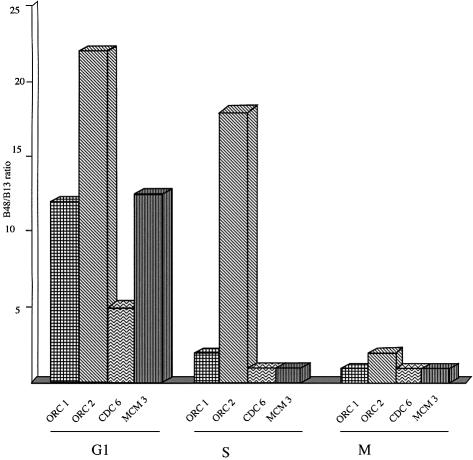
Figure 4 reports a summary of the variations described above of the occupation of the ori DNA by the different proteins along the cell cycle. These observations fit with our previous description of the cell cycle modulation of the overall protein–DNA interactions occurring in vivo at the lamin B2 ori that can be thus summarized: in G1 a large 110 bp complex builds up around the ori on both strands, that corresponds in all probability to the pre-replicative complex; when the cells enter the S phase the area covered by proteins shrinks to a length of 70 bp, in agreement with the contention that we are dealing now with a different structure, namely the post-replicative complex; in mitosis we cannot detect any significant protein contact in the ori area.
Fig. 4. Summary of the presence of DNA replication proteins on the ori sequence along the cell cycle. The data of the experiments reported in Figures 2 and 3 are reported for the four investigated proteins as the relative abundance of ori versus non-ori sequences in the DNA isolated after treatment with the appropriate antibody at different time points of the cell cycle.
A procedure for mapping with single nucleotide resolution proteins bound to DNA
In order to describe the modifications of the specific protein–DNA contacts occurring as the replicative complex passes from the pre- to the post-replicative state, we tried to identify the precise nucleotides at which the replicative complex proteins are bound and the modifications correlated with their functional transition.
To this purpose we devised a novel procedure based also on in vivo protein–DNA cross-linking and immunoprecipitation. The procedure entails initially the same steps as previously, namely in vivo UV irradiation with 266 plus 355 nm laser light followed by isolation of nuclei and elimination of the non-DNA cross-linked protein (Figure 5). The chromatin is then digested with appropriate restriction enzymes in order to obtain fragments with well defined ends. These are treated with λ-exonuclease, which again, besides destroying all non-cross-linked fragments, will produce single-strand stretches having a well defined 3′ end and an unknown 5′ end blocked by a covalently bound specific protein. These complexes are precipitated, isolated with appropriate antibodies and the bound proteins hydrolyzed with proteinase K. The task of identifying, for instance, the precise nucleotide bound to the residual peptide on the lower strand, as found previously, cannot be approached by amplification of the isolated fragments, since the 5′ end is blocked and there may be photoproducts in the fragment forbidding DNA polymerase action. Consequently, the peptide–DNA fragments are labeled with biotin and later annealed to a specific fragment complementary to the lower strand but bearing a 3′ end overhanging the unknown 5′ end of the isolated fragment. This overhang is hydrolyzed with Escherichia coli exonuclease I that is strictly specific for single-stranded DNA. The digested duplexes are isolated with streptavidin-coated magnetic beads and the partially hydrolyzed, upper strand probe fragment obtained now bears a 3′ end complementary to the bound 5′ end of the lower strand; hence, we can now identify the 3′ end of the probe. To this purpose, the partially hydrolyzed probe is subjected to the action of terminal deoxynucleotidyl transferase (TdT) in the presence of rGTP, conditions in which the enzyme adds only three GMP residues to the 3′ end of the probe (Schmidt and Mueller, 1996; Komura and Riggs, 1998). The resulting tailed probe is then annealed to a short duplex linker bearing a 3′ tail of three CMP residues which is covalently ligated to the probe. The resulting probe–linker construct is amplified with appropriate primers and analyzed for its size and this will give precise information on the position of the protein-bound nucleotide. The relatively low efficiency of DNA–protein cross-link (∼5% for USF, as indicated above, and somewhat lower for all other investigated proteins) ensures that it is extremely unlikely that more than one of the in vivo bound proteins will be cross-linked to the same restriction fragment, so that precipitation with the appropriate antibody will eventually reveal the position of only the specifically probed protein molecule.
Fig. 5. Procedure for the identification of the nucleotide(s) bound in vivo by a particular protein. For a detailed description see Materials and methods.
Also in this case we validated the procedure by checking whether we could identify the precise nucleotide at which the USF transcription factor is bound on the PPV1 promoter; the canonical binding sequence of USF is found 247 nucleotides upstream of the transcription start (Giacca et al., 1994), and the area protected in vivo and in vitro by this factor corresponds to nucleotides 4194–4211 of the humlambbb file of GenBank (Dimitrova et al., 1996). Terminal transferase-dependent (TD)-PCR analysis shows clearly a nucleotide in position 4201 as the site of binding, whereas non-irradiated or non-immunoprecipitated controls did not give any evidence of exonuclease arrest (Figure 6). The procedure appears to be able to identify the positions of binding to DNA of defined, immunoprecipitable proteins; we proceeded to investigate this aspect for the two studied ORC proteins and for hCdc6p at different time points of the cell cycle.
Fig. 6. Validation of the procedure with USF and identification of the binding sites of hOrc1p, hOrc2p and hCdc6p on the ori region at different time points of the cell cycle. (A) Asynchronously growing cells were treated with the procedure reported in Figure 5 utilizing anti-USF antibody. The results obtained with non-irradiated and non-immunoprecipitated controls are shown. On the left, the nucleotide position identified from the parallel treatment of naked DNA is shown. The protections observed in vivo (Dimitrova et al., 1996) on the PPV1 gene promoter are also shown. (B–D) Results of the application of the procedure described in Figure 5 to cells in different conditions of synchronization. (B) Anti-hOrc1p antibody; (C) anti-hOrc2p antibody; (D) anti-hCdc6 antibody. The cell cycle phase is indicated at the top of each lane. The nucleotide positions identified from the parallel treatment of naked DNA and the protection observed in vivo in G1 or S on the ori sequence (Abdurashidova et al., 1998) are reported.
Localization of hOrc1p, hOrc2p and hCdc6p on the ori at different time points of the cell cycle
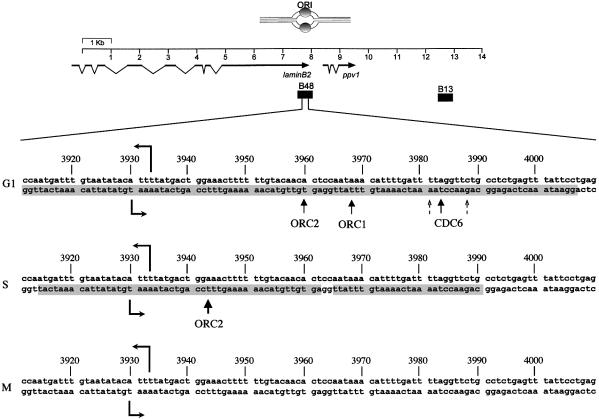
The results of the application of the procedure described above for determining the localizations of the hORC1, hORC2 and hCDC6 proteins on the lower strand of the ori region in the succeeding cell cycle phases are reported in Figure 6; on the left-hand side of the gels are shown the protections observed in vivo in this area on the lower strand in G1 and in S (in M no protection is visible, as mentioned above). Using anti-hOrc1p antibody a clear exonuclease stop is observed, but only in the G1 sample. This is not surprising, considering our previous observation that this protein leaves the replicative complex when synthesis starts. Interestingly, the position of the binding corresponds to nucleotides 3967–3969, that is a region which is protected in G1 but becomes relatively more exposed when the cell moves into S and the pre-replicative complex is restructured into the smaller post-replicative form. We might surmise that this exposition is due to the departing of hOrc1p from the G1 site as the ori is licensed and poised for the start of replication.
Conversely, as we look at the position of hOrc2p, we observe, in the first place, that in asynchronously growing cells two well defined exonuclease stops are identifiable, indicating the presence of two binding sites for hOrc2p in the lamin B2 ori. This observation is clarified by the data with the synchronized cultures, which show that the nucleotide involved in hOrc2p binding is nucleotide 3960 in G1 and 3943 in S (corresponding to the two locations observed in asynchronous cultures). As expected, no indication of bound protein is observed for either molecule in mitosis. This clarification is further strengthened by the results reported in the last two lanes of Figure 6C, obtained from only partially synchronized cell cultures. The cultures having an excess of G1-phase cells show a clear excess of fragments bound at nucleotide 3960, whereas those mainly containing S-phase cells have an excess of fragments bound at nucleotide 3943.
Finally, the hCDC6 protein shows one clear position of binding at nucleotide 3984, flanked by two minor ones at nucleotides 3982 and 3988, only in G1, in agreement with the results summarized in Figure 4. The identified position fits nicely with previous observations on the close association of Orc1p with Cdc6p in the pre-replication complexes of human and murine cells (Saha et al., 1998; Kneiss et al., 2003).
Figure 7 reports a summary of our observations. (i) In G1, a large pre-replicative complex builds around the future start sites of the lamin B2 ori, covering as much as 110 bp on both strands and containing, at some time point, hOrcp1, hOrcp2, hCdc6p and hMcm3. This is in agreement with what has been described in yeast and in other metazoa. (ii) As the cell moves into S and the lamin B2 ori (one of the first to be activated in human cells) is licensed for firing, hCdc6p and hMcm3p leave the pre-replicative complex which rearranges its structure into the smaller post-replicative one, covering ∼70 bp on the lower and 30 on the upper strand. (iii) This rearrangement entails the departure of hOrc1p from the ori DNA, leaving exposed a previously protected nucleotide. (iv) The rearrangement causes the displacement of hOrc2p by 17 bp towards the start sites of synthesis, that are now between 10 and 13 bp removed from the new hOrc2p location.
Fig. 7. Variations in the localization of hOrc1p, hOrc2p and hCdc6p at different time points of the cell cycle. Top, a map of the ori region, indicating the position of the surrounding genes and of the probes utilized for ori (B48) and non-ori DNA. Below, a 100 nucleotide portion of the ori region is shown. The two oppositely directed horizontal arrows indicate the start sites of leading-strand synthesis (Abdurashidova et al., 2000). The shadowed portions of the sequence show the protection observed in vivo on the lower strand in the indicated phase of the cell cycle (Abdurashidova et al., 1998). The upward pointing arrows indicate the nucleotide bound by the three proteins in each phase.
Considering the previous nucleotide-level identification of the start sites for DNA synthesis at the lamin B2 ori, the observations reported here allow us to conclude unambiguously that hOrc1p, hOrc2p and hCdc6 are elements of the human pre-replication complex, since they are located in G1 only a few tens of nucleotides away from the initiation site and well within the in vivo identified complex. We believe that the procedures we have developed here will offer useful tools for dissecting the composition, structure and functional dynamics of the ori activation process.
Materials and methods
Cell culture and synchronization
Available as Supplementary data at The EMBO Journal Online.
Sequence-specific probe preparation
Available as Supplementary data.
Laser light irradiation of cells
HeLa cells were harvested and washed twice in PBS. The cross-linking was induced by exposures to UV light generated by a nanosecond laser. The laser source was a Q-switched Nd:YAG laser (Quanttel, YG-511), delivering ∼30 mJ at 1064 nm (fundamental) and 20 mJ of energy in the second harmonic (532 nm) beam at 10 Hz repetition rate, pulse duration being 8 ns. By frequency doubling and mixing in two consecutive β-barium borate crystals, ∼2 mJ at the fourth harmonic (266 nm) and 3 mJ at the third (355 nm) harmonic were produced. They were separated from the fundamental and second harmonic light by a dichroic mirror and focused by a fused silica lens of 400 mm focal length. The standard rectangular fused silica cuvette containing 1 ml of cell suspension kept under constant stirring was placed a few centimeters in front of the focus where the beam spot size was ∼1 mm in diameter.
The optical density of the solution were kept at the range 0.1 < A260 < 0.2; the optical density of the cells can be determined in 8 M urea or 1% SDS.
Identification of the proteins directly interacting with the lamin B2 ori
Samples of ∼2 × 108 irradiated cells were resuspended in 10 ml of hypotonic buffer RSB (10 mM Tris–HCl pH 7.5, 10 mM NaCl, 3 mM MgCl2, protease inhibitors mix; Roche) and incubated in ice for 10 min. An equal volume of 0.2% NP-40 in RSB was added followed by incubation for 10 min at 4°C. The released nuclei were then collected and washed twice in RSB buffer. Unbound proteins were extracted twice in 5 ml of high-salt buffer (10 mM Tris–HCl, 1 M NaCl, 0.1% NP-40, 10 mM Na2S2O5 pH 8, protease inhibitors mix) and the nuclear pellet was resuspended in λ-exonuclease buffer (67 mM glycine pH 8.8, 2.5 mM MgCl2, 5% glycerol, 1 mM DTT). Chromatin was sonicated to yield an average DNA size of 700 bp, followed by incubation with RNase I (DNase free; Roche; 100 U) and λ-exonuclease (Boehringer Mannheim; 2 U/µg DNA) at 37°C for 2 h. For immunoprecipitation, samples of fixed chromatin were lysed in 1 ml of RIPA buffer (50 mM Tris–HCl, 150 mM NaCl, 0.1% deoxycholate, 0.1% SDS, 5 mM EDTA pH 8) and incubated with 10 µg of anti-USF mAb (Santa Cruz Biotechnology), 15 µg of anti-cdc6 mAb (Santa Cruz Biotechnology), 15 µg of anti-MCM3 mAb (MBL), 10 µg of anti-ORC2 mAb (Stress Gen Biotechnology) and 15 µg of anti-ORC1 mAb (kind gift of Drs Mauro Giacca and Ramiro Mendoza) on a rocker platform for 2 h at 4°C. A/G plus Agarose beads (100 µl; Santa Cruz Biotechnology) were added and incubation continued for another 2 h. The beads were washed five times, 15 min each, in RIPA buffer, five times in LiCl buffer (250 mM LiCl, 0.5% Triton X-100, 0.1% SDS, 0.1% deoxycholate, 1 mM EDTA, 10 mM Tris–HCl pH 8) and five times in TE buffer (10 mM Tris–HCl, 1 mM EDTA pH 8). The beads were finally resuspended in 100 µl of TE buffer, containing 1% SDS and incubated with proteinase K (1 mg/ml) for 1 h at 56°C. The DNA was purified by standard phenol/chloroform/isoamylalcohol extraction and ethanol precipitation. Purified DNA samples resuspended in 50 µl of TE were then biotinylated with Biotin Chem. Link (Boehringer Mannheim) in the presence of 1 µg of E.coli tRNA according to the manufacturer’s instructions. Biotinylated DNA was then purified by gel filtration, ethanol precipitated and resuspended in 10 µl of 6× SSC, 0.1% SDS, 1× Denhardt mix, 2 µl of E.coli tRNA (8 mg/ml), 10 mg/ml BSA and 50 ng/ml sequence-specific probe. The solutions were overlaid with mineral oil, the DNA was denatured for 5 min at 98°C and allowed to anneal overnight at 70°C. The final hybridization mixture was diluted in 200 µl of 3× SSC and genomic biotinylated DNA plus hybridized probe were captured using 100 µl of prewashed, preblocked, streptavidin-coated paramagnetic beads (Promega) for 30 min at room temperature. The beads were washed twice for 15 min each with 3× SSC, 0.1% SDS at 20°C, followed by four washes for 15 min each with 0.5× SSC, 0.1% SDS at 68°C. The hybridized DNA was then eluted with 50 µl of water at 75°C for 10 min and analyzed by competitive PCR. The quantitation of the abundance of two different sequences was performed as described by Diviacco et al. (1992). The sequences chosen for amplification are defined as B48 and B13, the B48 corresponding to the lamin B2 ori, whereas B13 is displaced by 5 kb. A single competitor molecule was used for both markers. The core of the molecule is a 110 bp region derived from the β-globin gene harboring a 20 bp insertion. The core competitor was built directly from the amplification products obtained by the overlap extension method (Higuchi et al., 1988; Ho et al., 1989). A set of four primers was synthesized for a region of the β-globin gene. Two external primers (PCO3 and PCO4) were synthesized together with two internal primers (PCO/+1 and PCO/+2) consisting of a common 5′ tail of 20 nucleotides linked to the specific sequences complementary to genomic targets on the 3′ end. Pairs of internal and external primers were used in two separate PCR reactions for the construction of two intermediate products. These intermediate products were eluted from polyacrylamide gel, mixed, denatured and annealed. Subsequently, after one round of extension, the hybrid product was amplified using the external primers to obtain the core competitor molecule, which has the same sequence as the β-globin genomic target, except for the addition of 20 nucleotides in the middle. The forward and reverse primers spanning the lamin B2 origin area (SB12, SE10, B13, B48 see Giacca et al., 1994, 1997), arranged in a head-to-tail fashion, were joined to the core molecule by PCR amplifications, using chimerical primers. This approach allowed us to use a single competitor for the quantification of the relative abundance of different PCR markers. The competitor was quantified in competitive PCR experiments against a known amount of plasmid molecules harboring the lamin B2 origin area. The PCR cycle profile was as follows: denaturation at 95°C, annealing at 56°C and extension at 72°C, time for each step was 30 s; 35 cycles were performed with 1 U of Taq polymerase (Boehringer Mannheim) in the conditions recommended by the manufacturer.
Localization of the proteins bound to DNA
For TD-PCR analysis, nuclei isolated from cross-linked cells were washed with high-salt buffer (as described before), resuspended in restriction enzyme digestion buffer (10 mM Tris–HCl pH 7.9, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, protease inhibitors mix) and incubated with BstNI (New England Biolabs; 10 U/µg DNA) at 60°C. After overnight digestion, another 10 U/µg DNA of enzyme were added and the digestion was allowed to continue for a further 2 h. The nuclei were then resuspended in λ-exonuclease buffer and incubated overnight with λ-exonuclease (2 U/µg DNA) and RNase I (100 U) at 37°C. Samples of fixed chromatin were lysed in RIPA; immunoprecipitation and DNA isolation were carried out as described above. DNA samples were then treated with polynucleotide kinase (New England Biolabs) to phosphorylate 5′ hydroxyl ends exposed by non-specific DNA shearing and make all the molecules available for λ-exonuclease digestion. The reaction was carried out in a total volume of 50 µl with 50 U of PNK (10 U/µl) in the presence of 1 mM ATP, 1× PNC buffer at 37°C for 30 min. The phosphorylated DNA was resuspended in λ-exonuclease buffer and subjected to λ-exonuclease treatment with 1 µl of enzyme (5 U/µl) in a final volume of 20 µl at 37°C for 3 h. The enzyme was heat inactivated for 10 min at 75°C, digested by proteinase K (1 mg/ml) for 1 h at 56°C. The DNA was extracted once with phenol/chloroform/isoamylalcohol, ethanol precipitated in the presence of 0.3 M sodium acetate pH 7.2, 2 µg of glycogen as carrier, and resuspended in 50 µl of TE buffer. The DNA was then biotinylated and purified as described above. Purified DNA was resuspended in 10 µl of 50 mM HEPES pH 8.3, 0.5 M NaCl, 0.5 mM EDTA, 10% w/v PEG 8000, 1% BSA, 50 ng/ml sequence probe. The solution was overlaid with mineral oil, the DNA was denatured for 5 min at 98°C and allowed to anneal overnight at 68°C. The hybridization mixture was then diluted in 200 µl of 50 mM HEPES pH 8.3, 5 mM MgCl2, 10 mM β-mercaptoethanol and incubated with 20 U of exonuclease I (New England Biolabs) for 30 min at 37°C, followed by addition of SDS (2% final concentration). The hybrids were captured using 100 µl of prewashed, preblocked, streptavidin-coated paramagnetic beads. The beads were washed four times with 2× SSC, 1% SDS at room temperature, four times with 0.5× SSC, 0.1% SDS at 65°C. The hybridized DNA was then eluted with 50 µl of water at 75°C for 10 min, ethanol precipitated, resuspended in 10 µl of 1/10 TE buffer (10 mM Tris–HCl, 1 mM EDTA pH 8). After the addition of 10 µl of TdT mix containing 10 U of TdT (Gibco-BRL), 2× buffer supplied by the manufacturer, 4 mM rGTP, the samples were incubated at 37°C for 15 min; the DNA was precipitated in the presence of 80 µl of 2.5 M ammonium acetate, 2.5 mM EDTA followed by addition of 300 µl of ethanol. The precipitates were dissolved in 15 µl of 1/10 TE. After the addition of 10.5 µl of ligation solution (100 mM Tris–HCl pH 7.5, 20 mM MgCl2, 20 mM DTT, 2 mM ATP), 3 µl of 20 µM TD-PCR linker (gamma linker) described by Komura and Riggs (1998) and 2 µl of T4 DNA ligase (Promega; 3 U/µl), the mixtures were incubated at 17°C overnight. After the addition of 70 µl of Vent exo– mix (5 U of Vent exo–), 1.43× ThermoPol buffer, 2.9 mM MgSO4, 0.4 mM each dNTP, 10 pmol primer 2: 5′-GTAAACAGGACCCAGGCGATGCATG-3′ (ppv1) or 5′-CAA AAACGGAGCTGGGCTGCAGCTG-3′ (ori), 10 pmol of linker primer, the samples were subjected to PCR using 18 cycles of 1 min at 95°C (5 min at 95°C for the first cycle), 2 min at 70°C, 3 min at 76°C, plus 3 s added per cycle, and 5 min at 76°C. Forty microliters of the amplification reactions were subjected to radioactive extension in the presence of 1 pmol of γ-33P-labeled primer 3: 5′-CAGGACCCAGGCGATGCATGG GACCCT-3′ (ppv1) or 5′-GGGCTGCACTGGGGCTGGCATGGAC-3′ (ori) in 1× ThermoPol buffer, 2.6 mM MgSO4, 0.2 mM dNTP, 1 U of Vent exo– in the final volume of 60 µl. The conditions of the radioactive extension were as follows: 1 min at 95°C (5 min at 95°C for the first cycle), 2 min at 72°C, 7 min at 76°C. Five cycles of extension were performed. The DNA was purified by phenol/chloroform extraction, precipitated with ethanol in the presence of 0.3 M sodium acetate, washed with 70% ethanol, dried briefly and resuspended in loading buffer for sequencing gel (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol). The samples were denatured for 3 min at 95°C and loaded on 8% polyacrylamide sequencing gel. The control sequencing ladder was obtained by treating the TD-PCR reaction genomic DNA in vitro with dimethyl sulfoxide (DMS) as follows: 175 µg of total genomic DNA were incubated with 0.2% DMS for 2 min at room temperature. The reaction was blocked by the addition of 200 mM (final concentration) β-mercaptoethanol and the DNA was immediately precipitated with ethanol in the presence of 0.3 M sodium acetate, resuspended in TE, heated 30 min at 95°C. Two micrograms of DMS-treated genomic DNA were linearly amplified in 30 µl of reaction containing 1 U of Vent exo– (New England Biolab), 1× ThermoPol buffer (New England Biolab), 1 µl of 100 mM MgSO4, 1 µl of 100 mM Tris–HCl pH 7.5, 250 µm each dNTP, 0.6 pmol of primer 1 biotinylated at the 5′ end: 5′-biotin–GGCTAGTGTAGCTAGTGTAAACAGGACC-3′ (ppv1) and 5′-biotin–GTCACAGCACAACCTGCAAAAACGG-3′ (ori). The temperature cycles were 1 min at 95°C (5 min at 95°C for the first cycle), 3 min at 60°C and 2 min at 72°C. After thermal cycling, the samples were denatured at 95°C for 2 min and then cooled to 4°C. Streptavidin-coated paramagnetic beads were prepared immediately before use by washing twice in 1× WBB (washing and binding buffer, 10 mM Tris–HCl pH 7.5, 1 mM EDTA, 2.5 M NaCl). Washed beads (20 µl) were added to 30 µl of extension product from the previous step and the mixture was rotated at room temperature for 30 min. The beads were washed twice with WBB, 100 µl of 0.15 M NaOH were added and the suspension was incubated for 5–10 min; the beads were washed once more with 100 µl of 0.15 M NaOH. The beads were neutralized by washing 3× with 200 µl TE buffer (10 mM Tris–HCl, 1 mM EDTA pH 8) and resuspended in 20 µl of 0.1× TE. After the addition to the neutralized beads of 20 U of TdT (Gibco-BRL), 2× buffer supplied by manufacturer and 4 mM rGTP, the samples were incubated at 37°C for 15 min. The supernatant was removed and the beads were washed twice with 200 µl of TE buffer and resuspended in 30 µl of 0.1× TE pH 7.5. Thirty microliters of 2× ligation mixture, containing 100 mM Tris–HCl pH 7.5, 20 mM MgCl2, 20 mM DTT, 2 mM ATP, 100 µg/ml BSA, plus 6 µl of 20 µM TD-PCR linker (gamma linker) and 3 µl of T4 DNA ligase were added to the beads suspension. The reaction was incubated overnight at 17°C. The beads were washed twice with 200 µl of TE pH 7.5 and resuspended in 60 µl of 0.1× TE. The washed beads (20 µl) from the ligation step were transferred to a new tube and 30 µl of PCR mix, 5 µl of 10× ThermoPol, 1 µl of 10 mM each dNTP, 1 µl of 100 mM MgSO4, 10 pmol of primer 2: 5′-GTAAACAGGACCCAGGCGATGCATG-3′ (ppv1) or 5′-CAAAAA CGGAGCTGGGCTGCAGCTG-3′ (ori), 10 pmol of linker primer, 20.5 µl of H2O, 5 U of Vent exo– were added, the samples were subjected to 18 cycles of exponential amplification: 1 min at 95°C (5 min at 95°C for the first cycle), 2 min at 70°C, 3 min at 76°C plus 3 s extension per cycle. For complete extension of all product, 5 min at 76°C. Twenty microliters of the amplification reaction were subjected to radioactive extension in the presence of 1 pmol of γ-33P-labeled primer 3: CAGGACCCAGGCGA TGCATGGGACCCT (ppv1) and GGGCTGCAGCTGGGGCTGGCAT GGAC (ori), in 1× ThermoPol buffer, 2.6 mM MgSO4, 0.2 mM of each dNTP, 1 U of Vent exo– in a final volume of 30 µl. The conditions of the radioactive extension were as follows: 1 min at 95°C (5 min at 95°C for the first cycle), 2 min at 72°C, 7 min at 76°C. Five cycles of extension were performed. PCR tubes were centrifuged, the supernatant (not the beads) was transferred to a new tube and the DNA was purified by standard phenol/chloroform extraction and ethanol precipitation. Purified DNA was resuspended in 10 µl of sequencing gel labeling buffer, denatured for 3 min at 95°C and loaded onto a 8% polyacrylamide sequencing gel.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Professor Gallieno Denardo, head of the Laser Laboratory of the Sincrotrone Trieste, for invaluable support and advice. The assistance of Ms Maria Elena Lopez for the preparation of HeLa cells is gratefully acknowledged. The work was partially supported by grants from the Consiglio Nazionale delle Ricerche of Italy (CNR01/007), from the Associazione Italiana Ricerche per il Cancro and from the European Commission (HPRN-CT-2000–00089).
References
- Abdurashidova G., Riva,S., Biamonti,G., Giacca,M. and Falaschi,A. (1998) Cell cycle modulation of protein–DNA interactions at a human replication origin. EMBO J., 17, 2961–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdurashidova G., Deganuto,M., Klima,R., Riva,S., Biamonti,G., Giacca,M. and Falaschi,A. (2000) Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science, 287, 2023–2026. [DOI] [PubMed] [Google Scholar]
- Aparicio O.M., Weinstein,D.M. and Bell,S.P. (1997) Components and dynamics of DNA replication complexes in S.cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell, 91, 59–69. [DOI] [PubMed] [Google Scholar]
- Bell S.P. and Stillman,B. (1992) ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature, 357, 128–134. [DOI] [PubMed] [Google Scholar]
- Biamonti G. et al. (1992) The gene for a novel human lamin maps at a highly transcribed locus of chromosome 19 which replicates at the onset of S-phase. Mol. Cell. Biol., 12, 3499–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielinsky A.K. and Gerbi,S.A. (2001) Where it all starts: eukaryotic origin of DNA relication. J. Cell Sci., 114, 643–651. [DOI] [PubMed] [Google Scholar]
- Blow J.J. (2001) Control of chromosomal DNA replication in the early Xenopus embryo. EMBO J., 20, 3293–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Careri G., Fasella,P. and Gratton,E. (1975) Statistical time events in enzymes: a physical assessment. CRC Crit. Rev. Biochem., 3, 141–164. [DOI] [PubMed] [Google Scholar]
- Coleman T.R., Carpenter,P.B. and Dunphy,W.G. (1996) The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell, 87, 53–63. [DOI] [PubMed] [Google Scholar]
- Diffley J.F.X. and Labib,K. (2002) The chromosome replication cycle. J. Cell Sci., 115, 869–872. [DOI] [PubMed] [Google Scholar]
- Diffley J.F., Cocker,J.H., Dowell,S.J. and Rowley,A. (1994) Two steps in the assembly of complexes at yeast replication origins in vivo. Cell, 78, 303–316. [DOI] [PubMed] [Google Scholar]
- Dimitrova D., Giacca,M., Demarchi,F., Biamonti,G., Riva,S. and Falaschi A. (1996) In vivo protein–DNA interactions at human DNA replication origin. Proc. Natl Acad. Sci. USA, 93, 1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diviacco S., Norio,P., Zentilin,L. Menzo,S., Clementi,M., Biamonti,G., Riva,S., Falaschi,A. and Giacca,M. (1992) A novel procedure for quantitative polymerase chain reaction by coamplification of competitive templates. Gene, 122, 313–320. [DOI] [PubMed] [Google Scholar]
- Findeisen N., El-Denary,M., Kapitza,T., Graf,R. and Strausfeld,U. (1999) Cyclin A-dependent kinase activity affects chromatin binding of ORC, Cdc6 and MCM in egg extracst of Xenopus laevis. Eur. J. Biochem., 264, 415–426. [DOI] [PubMed] [Google Scholar]
- Giacca M. et al. (1994) Fine mapping of a replication origin of human DNA. Proc. Natl Acad. Sci. USA, 91, 7119–7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacca M., Pelizon,C. and Falaschi,A. (1997) Mapping replication origin by quantifying relative abundance of nascent DNA strands using competitive polymerase chain reaction. Methods, 13, 301–312. [DOI] [PubMed] [Google Scholar]
- Gilmour D.S. and Lis,J.T. (1986) RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol. Cell. Biol., 6, 3984–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R., Krummel,B. and Saiki,R.K. (1988) A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res., 16, 7351–7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S.N., Hunt,N.D., Horton,R.M., Pullen,J.K. and Pease,L.R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Hua X.H. and Newport,J. (1998) Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J. Cell Biol., 140, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T.J. and Brown,G.W. (2000) Regulation of chromosome replication. Annu. Rev. Biochem., 69, 829–880. [DOI] [PubMed] [Google Scholar]
- Kneiss M., Pütter,V., Szalay,A.A. and Grummt,F. (2003) Interaction and assembly of murine pre-replicative complexes proteins in yeast and mouse cells. J. Mol. Biol., 327, 111–128. [DOI] [PubMed] [Google Scholar]
- Komura J.I. and Riggs,A. (1998) Terminal transferase-dependent PCR: a versatile and sensitive method for in vivo footprinting and detection of DNA adducts. Nucleic Acids Res., 26, 1807–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitz S., Ritzi,M., Baack,M. and Knippers,R. (2001) The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. J. Biol. Chem., 276, 6337–6342. [DOI] [PubMed] [Google Scholar]
- Ladenburger E.M., Keller,C. and Knippers,R. (2002) Identification of a binding region for human origin recognition complex protein 1 and 2 that coincides with an origin of DNA replication. Mol. Cell. Biol., 22, 1036–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.J. and DePamphilis,M.L. (2002) Mammalian Orc1 protein is selectively released from chromatin and ubiquitinated during the S-to-M transition in the cell division cycle. Mol. Cell. Biol., 23, 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C. and Stillman,B. (1997) Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev., 11, 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorano D., Moreau,J. and Mechali,M. (2000) XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature, 404, 622–625. [DOI] [PubMed] [Google Scholar]
- Mattes W.B. (1990) Lesion selectivity in blockage of λ exonuclease by DNA damage. Nucleic Acids Res., 18, 3723–3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J., Zou-Yang,X.H., Kim,S.X., Hidaka,M., Tansey,W.P. and Stillman,B. (2002) Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell, 9, 481–491. [DOI] [PubMed] [Google Scholar]
- Mizushima T., Takahashi,N. and Stillman,B. (2000) Cdc6 modulates the structure and DNA binding activity of the origin recognition complex in vitro. Genes Dev., 14, 1631–1641. [PMC free article] [PubMed] [Google Scholar]
- Moss T., Dimitrov,S.I. and Houde,D. (1997) UV-laser crosslinking of proteins to DNA. Methods, 11, 225–234. [DOI] [PubMed] [Google Scholar]
- Mutskov V., Angelov,D. and Pashev,I. (1997) A preparative method for crosslinking proteins to DNA in nuclei by single-pulse UV laser irradiation. Photochem. Photobiol., 66, 42–45. [DOI] [PubMed] [Google Scholar]
- Natale D.A., Li,C.J., Sun,W.H. and DePamphilis,M.L. (2000) Selective instability of Orc1 protein accounts for the absence of functional origin recognition complexes during the M–G1 transition in mammals. EMBO J., 19, 2728–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H., Lygerou,Z., Nishimoto,T. and Nurse,P. (2000) The Cdt1 protein is required to license DNA for replication in fission yeast. Nature, 404, 625–628. [DOI] [PubMed] [Google Scholar]
- Ogawa Y., Takahashi,T. and Masukata,H. (1999) Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol. Cell. Biol., 19, 7228–7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski P., Madine,M.A., Rowles,A., Blow,J.J. and Laskey,R.A. (1996) The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr. Biol., 6, 1416–1425. [DOI] [PubMed] [Google Scholar]
- Rowles A., Tada,S. and Blow,J.J. (1999) Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J. Cell Sci., 112, 2011–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russman C., Stallhof,J., Weiss,C., Beigang,R. and Beato,M. (1998) Two wavelength femtosecond laser induced DNA–protein crosslinking. Nucleic Acids Res., 26, 3967–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha P., Chen,J., Thome,K.,C., Lawlis,S.J., Hou,Z.,Hendricks,M., Parvin,D.J. and Dutta,A. (1998) Human CDC6/Cdc18 associates with Orc1 and cyclin–cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol. Cell. Biol., 18, 2758–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaarschmidt D., Ladenburger,E., Keller,C. and Knippers,R. (2002) Human Mcm proteins at a replication origin during the G1 to S phase transition. Nucleic Acids Res., 30, 4176–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt W.M. and Mueller,M.W. (1996) Controlled ribonucleotide tailing of cDNA ends (CRTC) by terminal deoxynucleotidyl transferase: a new approach in PCR-mediated analysis of mRNA sequences. Nucleic Acids Res., 24, 1789–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon M.J., Larsen,P.L. and Varshavsky,A. (1988) Mapping protein–DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell, 53, 937–947. [DOI] [PubMed] [Google Scholar]
- Sun W.H., Coleman,T.R. and DePamphilis,M.L. (2002) Cell cycle-dependent regulation of the association between origin recognition proteins and somatic cell chromatin. EMBO J., 21, 1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S. and Diffley,J.F.X. (2002) Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2-7 during G1 phase. Nat. Cell Biol., 4, 198–206. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Knapp,D. and Nasmyth,K. (1997) Loading of an Mcm protein onto DNA replication origin is regulated by Cdc6p and CDKs. Cell, 90, 649–660. [DOI] [PubMed] [Google Scholar]