Abstract
Genome-wide synthetic genetic interaction screens with mutants in the mus81 and mms4 replication fork-processing genes identified a novel replication factor C (RFC) homolog, Elg1, which forms an alternative RFC complex with Rfc2–5. This complex is distinct from the DNA replication RFC, the DNA damage checkpoint RFC and the sister chromatid cohesion RFC. As expected from its genetic interactions, elg1 mutants are sensitive to DNA damage. Elg1 is redundant with Rad24 in the DNA damage response and contributes to activation of the checkpoint kinase Rad53. We find that elg1 mutants display DNA replication defects and genome instability, including increased recombination and mutation frequencies, and minichromosome maintenance defects. Mutants in elg1 show genetic interactions with pathways required for processing of stalled replication forks, and are defective in recovery from DNA damage during S phase. We propose that Elg1-RFC functions both in normal DNA replication and in the DNA damage response.
Keywords: cell cycle checkpoints/DNA damage/DNA replication/genome stability/replication factor C
Introduction
DNA replication is typically highly processive. Replication fork stalling or arrest can result when replication forks encounter damage in the DNA, and at naturally occurring sequences such as replication fork barriers and replication slow zones (Rothstein et al., 2000; Cha and Kleckner, 2002). Several mechanisms by which replication forks can be restarted following arrest have been described in bacteria (reviewed in Michel, 2000; Michel et al., 2001). Stalled forks are susceptible to breakage and to replication fork reversal, both of which generate a double-stranded DNA end. A replication fork can then be re-established by homologous recombination followed by Holliday junction resolution or by branch migration. By analogy, some of these same processes are believed to occur in eukaryotes. In addition to these pathways, which are principally involved in restarting what are presumably collapsed replication forks, recent work has demonstrated that the S phase checkpoint pathway is responsible for stabilizing replication forks and preventing fork collapse and formation of DNA structures that are substrates for replication restart pathways (Lopes et al., 2001; Tercero and Diffley, 2001; Sogo et al., 2002).
Replication factor C (RFC) was first identified as a protein complex required for SV40 DNA replication in vitro (Tsurimoto and Stillman, 1989; Virshup and Kelly, 1989). RFC is a five-subunit complex that recognizes the primer terminus and catalyzes the loading of the sliding clamp proliferating cell nuclear antigen (PCNA; Hubscher, 1996; Mossi and Hubscher, 1998). PCNA acts as a processivity factor for DNA polymerase δ on the leading and lagging strands. At least two alternative forms of RFC have been identified recently in yeast and humans. In the first of these, the large subunit of RFC, Rfc1, is replaced by the Rad24 protein (Green et al., 2000). Rad24-RFC loads a PCNA-like clamp consisting of Rad17, Ddc1 and Mec3 (Bermudez et al., 2003; Majka and Burgers, 2003), is required for DNA damage checkpoint responses in G1 and G2, and contributes to S phase checkpoint responses. In the second alternative RFC, Rfc1 is replaced by Ctf18 (Hanna et al., 2001; Mayer et al., 2001; Naiki et al., 2001). Mutants in ctf18 have defects in sister chromatid cohesion (Hanna et al., 2001; Mayer et al., 2001), the process by which newly replicated chromatids remain physically associated until entry into anaphase. Ctf18-RFC also contains the Ctf8 and Dcc1 proteins, and mutations in ctf8 or dcc1 recapitulate the cohesion defects observed in ctf18 mutants (Mayer et al., 2001). There is evidence that like Rad24, Ctf18 also contributes to the S phase checkpoint response. ctf18 rad24 mutants have a mild S–M checkpoint defect that is not evident in the single mutants, and have a Rad53 activation defect in S phase (Naiki et al., 2001).
Here we describe a functional genomics approach to identify previously uncharacterized factors required for the DNA damage response, particularly those involved in replication fork progression. To this end, we conducted genome-wide synthetic lethality screens with deletion mutants in mus81 and mms4. Mus81 and Mms4 are subunits of an endonuclease with a preference for branched DNA structures (Boddy et al., 2001; Chen et al., 2001; Kaliraman et al., 2001). In addition to this substrate preference, several lines of evidence connect this enzyme to the processing of stalled DNA replication forks (Haber and Heyer, 2001; Kaliraman et al., 2001; Mullen et al., 2001).
We identified a previously uncharacterized RFC1 homolog, ELG1. We find that Elg1 forms an RFC-like complex with the Rfc2–5 proteins, but not with Rfc1 or its homologs Rad24 and Ctf18. ELG1 is functionally redundant with RAD24 in the DNA damage response, yet does not share its primary role in checkpoint activation. Of particular significance, elg1 mutants display defects in DNA replication in both the presence and absence of DNA damage, suggesting that Elg1 functions directly in DNA replication. Cells lacking elg1 require the intra-S phase checkpoint, homologous recombination proteins and pathways involved in replication restart following replication fork stalling for wild-type growth, and are defective in recovery from DNA damage in S phase. We propose that Elg1-RFC functions in lagging strand DNA synthesis to prevent replication fork stalling and to facilitate re-start of stalled replication forks.
Results
Genome-wide synthetic lethal screens with mus81Δ and mms4Δ identify ELG1
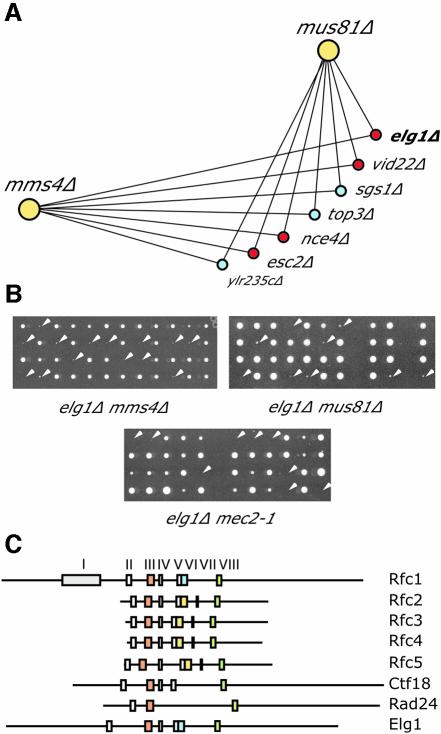
In order to identify novel genes that function to stabilize replication forks in vivo, we conducted genome-wide synthetic genetic interaction screens (Tong et al., 2001) with strains carrying deletions of MUS81 or MMS4. Several lines of evidence suggest that Mus81 and Mms4 are involved in the processing of stalled replication forks (Haber and Heyer, 2001; Kaliraman et al., 2001; Mullen et al., 2001). In these screens, the query mutant was crossed with the ∼4600 strains that make up the complete set of Saccharomyces cerevisiae viable haploid deletion mutants. Double mutants that show reduced fitness compared with single mutants, as evidenced by lack of growth or slow growth, were scored as positive. Each screen was performed three times and interactions that were scored at least twice were confirmed by tetrad analysis. The result of these screens is presented as a genetic interaction network in Figure 1A, with tetrad analysis of the elg1Δ interactions shown in Figure 1B. Each line on the network represents a synthetic lethal or synthetic sick (slow growth) interaction between the linked genes. Consistent with models in which Mus81 and Mms4 function exclusively as a heterodimer (Boddy et al., 2001; Kaliraman et al., 2001), the screen with mus81Δ identified the same set of seven genes as the screen with mms4Δ. We found strong genetic interactions between mus81Δ and mms4Δ and deletions of either sgs1 or top3. These interactions have been described previously and are believed to reflect the redundant roles of the Sgs1/Top3 and Mus81/Mms4 pathways in the repair of stalled replication forks and/or the resolution of recombination intermediates (Mullen et al., 2001). YLR235C overlaps the TOP3 gene and so was probably identified due to its effect on Top3 function. In addition to these known interactions, we identified four novel interactions, with elg1Δ, esc2Δ, nce4Δ and vid22Δ. We also found that elg1Δ is synthetic lethal with mec2-1, a checkpoint-defective allele of RAD53 (Figure 1B), which indicates that elg1Δ mutant cells require a Rad53-dependent checkpoint for viability. Together, these data suggest that Elg1 is important for the integrity of DNA replication forks in vivo.
Fig. 1. Genome-wide synthetic lethal screens with mus81Δ and mms4Δ identify the RFC homolog Elg1. (A) The results from synthetic genetic array analysis with mus81Δ and mms4Δ presented as a genetic interaction map. Lines connecting genes represent synthetic lethality or synthetic slow growth. Red circles indicate novel genetic interactions. (B) Tetrad confirmation of the elg1Δ crosses. Each column represents the four spores from a single ascus. Double mutant colonies, as detected by selection for the dominant selectable marker linked to each gene, are indicated by white arrowheads. (C) Schematic representation of the conserved sequence blocks in the S.cerevisiae RFC family genes. Elg1 contains six of the seven RFC boxes found in Rfc1.
ELG1 is a member of the replication factor C family
ELG1 (enhanced level of genome instability) was first identified in a screen for increased Ty transposon mobility, and elg1 mutants have been reported to confer an increase in direct repeat recombination (Scholes et al., 2001). Using a LEU2 direct repeat assay (Smith and Rothstein, 1999), we confirmed that deletion of elg1 causes a 7-fold increase in recombination rate (data not shown). Detailed examination of the hypothetical translation product of ELG1 revealed extensive similarity to the RFC-like protein family. Eight regions of sequence similarity have been defined in RFC proteins (Cullmann et al., 1995). As indicated in Figure 1C and in the Supplementary data (available at The EMBO Journal Online), Elg1contains all of the RFC boxes present in Rfc1 with the exception of the ligase homology region, RFC box I. The Rfc-specific boxes II, IV, VI and VIII are present in Elg1, as is the ATP-binding motif contained in boxes III and V. RFC box VII is present only in the small Rfc subunits, Rfc2–5, and is absent from Elg1. The RFC box VI in Elg1 bears greater similarity to box VIa, found in the large Rfc subunits, than it does to box VIb, found in the small subunits. Like the Rfc1 homologs Rad24 and Ctf18, Elg1 is conserved throughout Eucaryota. Homologs of Elg1 are readily identifiable in Schizosaccharomyces pombe (NP_595265), Drosophila melanogaster (AAF49530), mouse (BAC39389.1) and human (CAC44537). Together, these data indicate that Elg1 is an Rfc1 homolog and, by analogy with the Rfc1 homologs Rad24 and Ctf18, suggests that Elg1 forms an alternative RFC complex with the small RFC subunits Rfc2–5.
Elg1 forms a novel RFC complex
High-throughput protein–protein interaction screens have detected Elg1 in complexes with Rfc2, Rfc4 and Rfc5 (Gavin et al., 2002; Ho et al., 2002). In order to test the possibility that Elg1 forms an RFC-like complex distinct from the canonical replication RFC, we conducted co-immunoprecipitation experiments with Elg1 and the five subunits of RFC. Elg1 protein was not detectable in Rfc1 immunoprecipitates (Figure 2A, lane 4). In contrast, immunoprecipitates of Rfc2, Rfc3, Rfc4 or Rfc5 all contained Elg1 (Figure 2A, lanes 6, 8, 10 and 12). Elg1 was not immunoprecipitated in the absence of RFC, indicating that the immunoprecipitations were specific (Figure 2A, lane 2). Therefore, Elg1 forms a complex with Rfc2–5 but not with Rfc1. The simplest interpretation of these data is that Elg1 forms an alternative RFC complex in which it substitutes for Rfc1 in binding to Rfc2–5.
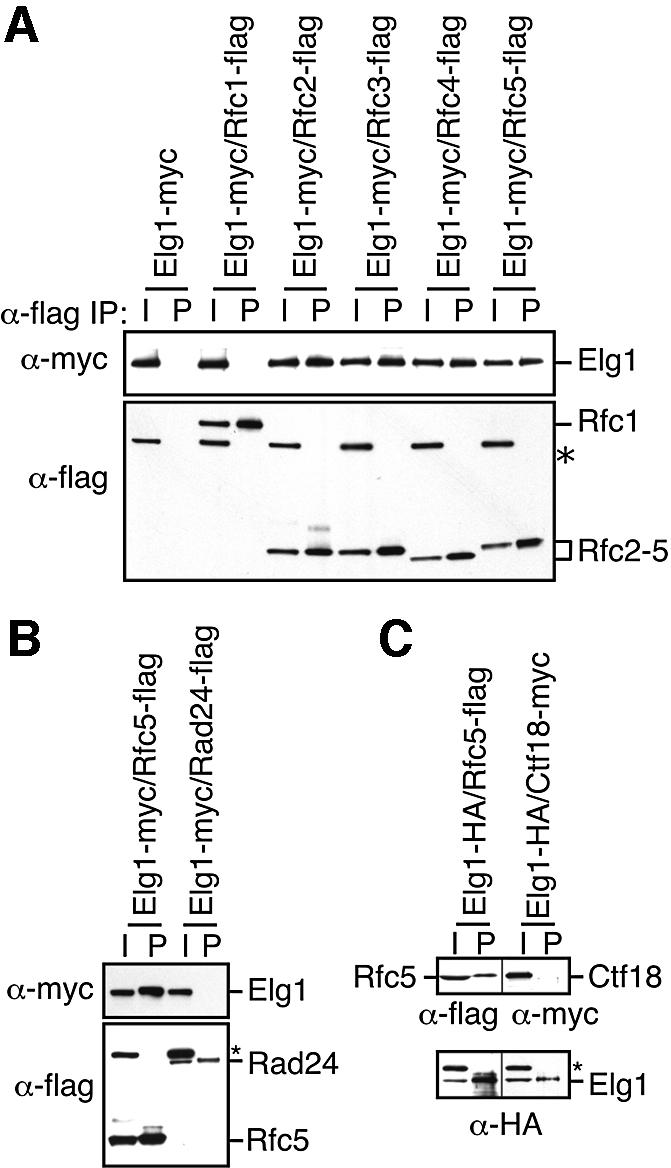
Fig. 2. Elg1 forms complexes with Rfc2, 3, 4 and 5, but not with Rfc1, Rad24 or Ctf18. (A) Extracts from yeast strains expressing the indicated epitope-tagged RFC proteins were immunoprecipitated with antibody against the flag epitope. Ten percent of the input extract (I) and the immunoprecipitate (P) were fractionated by SDS–PAGE. Immunoblots were probed with anti-flag antibody to detect Rfc1, Rfc2, Rfc3, Rfc4 and Rfc5, and with anti-myc antibody to detect Elg1. A non-specific cross-reacting polypeptide is indicated (*). (B) Extracts from strains expressing the indicated proteins were immunoprecipitated with anti-flag antibody to precipitate Rfc5 and Rad24. Immunoblots were probed with anti-flag antibody to detect Rfc5 and Rad24 and with anti-myc to detect Elg1. (C) Extracts were immunoprecipitated with anti-HA antibody to precipitate Elg1. Immunoblots were probed with anti-flag antibody to detect Rfc5, anti-myc to detect Ctf18 and anti-HA to detect Elg1.
Although genetic data suggest that Elg1 is functionally distinct from Ctf18 and Rad24 as neither ctf18Δ nor rad24Δ shows as strong a genetic interaction with mus81Δ, mms4Δ or rad53 (data not shown), it remained formally possible that Elg1 is a member of the Rad24- RFC or the Ctf18-RFC complexes. To exclude these possibilities, we first immunoprecipitated Rad24 and assayed for the presence of Elg1 (Figure 2B). Elg1 was detected in Rfc5 complexes, but not in Rad24 complexes. We next immunoprecipitated Elg1 and assayed for the presence of Ctf18 (Figure 2C). Ctf18 was not present in Elg1 complexes, although again Rfc5 was present. We conclude that Elg1 forms a novel RFC complex with Rfc2–5.
Elg1 function in the DNA damage response is redundant with that of Rad24
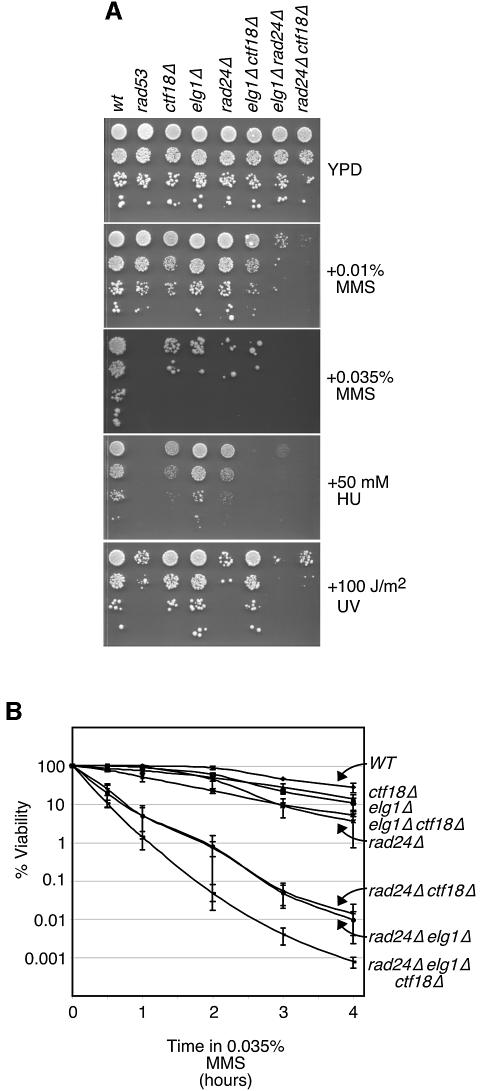
Several lines of evidence have implicated RFC complexes in the checkpoint response to DNA damage, particularly during S phase. We tested the sensitivity of elg1Δ, rad24Δ and ctf18Δ mutants, as well as all pairwise double mutants, and the triple mutant, to the DNA alkylating agent methyl methanesulfonate (MMS), the replication inhibitor hydroxyurea (HU) and UV radiation (Figure 3A). In addition, we measured mutant cell viability following exposure to 0.035% MMS in a 4 h time course (Figure 1B). First, we found that the elg1Δ mutant was only mildly sensitive to MMS, and was resistant to HU and UV. This contrasts with rad24Δ, which was significantly sensitive to all three agents, and with ctf18Δ, which displayed sensitivity to MMS and to HU, but was only modestly UV sensitive. These different sensitivities distinguish the Rfc1 homologs, and indicate that Elg1 is not a central player in DNA damage repair.
Fig. 3. Elg1 is required for the DNA damage response. (A) Ten-fold serial dilutions of cultures of the indicated mutants were spotted on YPD, YPD containing 0.01% (v/v) MMS, 0.035% (v/v) MMS, 50 mM HU, or on YPD that was subsequently exposed to 100 J/m2 UV. Plates were incubated at 30°C for 2–3 days. (B) Logarithmically growing cultures of the indicated mutants were incubated in YPD containing 0.035% (v/v) MMS at 30°C. At the indicated times, samples were withdrawn and plated on medium lacking MMS to determine the number of viable cells. The percentage of viable cells relative to the number of viable cells at t = 0 is shown. Plots represent the average of three experiments, and error bars span 1 SD.
We next examined the sensitivity of each possible double mutant combination and the triple mutant. Both double mutants with rad24Δ were significantly more sensitive to all agents than any of the single mutants, whereas the rad24Δ ctf18Δ elg1Δ triple mutant conferred the greatest sensitivity to MMS. These findings indicate that the Rfc1 homologs are in partially redundant pathways for DNA damage resistance.
Elg1 contributes to Rad53 activation
Rad24 has a well-documented role in activation of the checkpoint kinase Rad53 (Shimomura et al., 1998; Pellicioli et al., 1999; Naiki et al., 2000). Activation of Rad53 can be readily assessed by immunoblot detection of a phosphorylation-dependent shift in Rad53 mobility (Pellicioli et al., 1999). We first assayed Rad53 activation in HU-arrested cells to assess S–M checkpoint function in elg1Δ cells (Figure 4). ELG1 was not required for Rad53 activation following HU arrest, as we observed wild-type levels of Rad53 phosphorylation in the elg1Δ mutant (Figure 4A). The ctf18Δ rad24Δ mutant displayed a clear defect in Rad53 activation, as previously reported (Naiki et al., 2001) and consistent with the increased HU sensitivity of this double mutant relative to the ctf18Δ and rad24Δ single mutants (Figure 3). The elg1Δ rad24Δ and elg1Δ ctf18Δ double mutants displayed slight defects in Rad53 activation, as evidenced by the smaller fraction of Rad53 present in the most slowly migrating form when compared with wild-type (Figure 4A). As with ctf18Δ rad24Δ, this might account for the increased HU sensitivity of the double mutants compared with the single mutants (Figure 3). In the elg1Δ rad24Δ ctf18Δ triple mutant, phosphorylation of Rad53 was almost completely absent (Figure 4A). We conclude that ELG1 contributes to the S–M checkpoint in the context of a rad24 mutation. The near absence of Rad53 phosphorylation in the triple mutant indicates that all of the Rfc1 homologs can contribute to Rad53 activation in S phase.
Fig. 4. Rad53 activation defects in elg1Δ. (A) S–M checkpoint. Logarithmically growing cultures were arrested in G1 with α-factor and released into medium containing 200 mM HU. At the indicated times, samples were fixed and extracts fractionated by SDS–PAGE. Following transfer, the immunoblot was probed with anti-Rad53 antibody. Phosphorylation of Rad53 causes a shift in electrophoretic mobility (Rad53-P) and is a marker for checkpoint activation. (B) Intra-S phase checkpoint. Logarithmically growing cultures were treated with 0.035% (v/v) MMS. At the indicated times, samples were withdrawn and Rad53 activation was analyzed by immunoblotting. (C) Cell cycle progression in the presence of MMS was assessed by flow cytometry. Logarithmically growing cultures were treated with 0.035% (v/v) MMS. At the indicated times, samples were fixed and the DNA contents of cells in each sample were analyzed by flow cytometry. The positions of cells with 1C and 2C DNA contents are indicated on the histograms.
We next treated cells with MMS to assess the role of Elg1 in the intra-S phase checkpoint (Figure 4B). Again, elg1Δ cells displayed no defect in Rad53 activation. However, when combined with rad24Δ, both elg1Δ and ctf18Δ have significant Rad53 activation defects, consistent with roles in intra-S checkpoint response. It is worth noting, however, that the elg1Δ ctf18Δ double mutant had little, if any, Rad53 activation defect, indicating that Rad24 plays the more important role in the intra-S checkpoint.
Since we had hypothesized a role for Elg1 in replication fork integrity, we assessed intra-S phase checkpoint function directly. Cells in asynchronous culture were treated with MMS, and their accumulation in S phase was monitored by flow cytometry (Figure 4C). Wild-type cells, with an intact intra-S phase checkpoint, accumulate in S phase by 2 h, and remain blocked with an intermediate DNA content for the 4 h duration of the experiment. This S phase accumulation is due to checkpoint-independent slowing of DNA replication fork progression combined with a checkpoint-dependent inhibition of dormant and late origin firing (Tercero and Diffley, 2001). In the rad53 mutant, the checkpoint-dependent inhibition of origin firing is abrogated and cells appeared to move through S phase more rapidly, accumulating with a 2C DNA content by 3 h. In the rad24Δ mutant, which is partially defective in the intra-S checkpoint (Paulovich et al., 1997), cells moved through S phase more rapidly than wild-type but not as rapidly as rad53 mutant cells. The elg1Δ resembled wild-type, and deleting elg1 did not enhance the intra-S checkpoint defect in rad24Δ. In contrast, the ctf18Δ rad24Δ mutant was more defective in the intra-S checkpoint, accumulating with 2C DNA content with kinetics similar to those observed with the rad53 mutant.
Cells lacking Elg1 require homologous recombination and replication fork re-start pathways for optimum growth
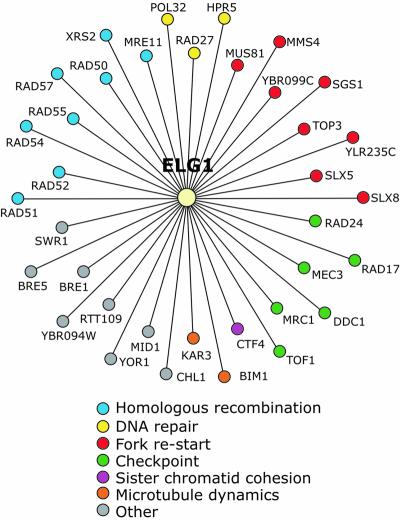
To gain further insight into Elg1 function in vivo, we performed a genome-wide synthetic lethal screen with elg1Δ. The synthetic genetic interactions confirmed by tetrad analysis are presented in Figure 5 and in the Supplementary data. Several functional clusters of genes are readily apparent. First, elg1Δ interacts with members of the RAD52 epistasis group (rad51Δ, rad52Δ, rad54Δ, rad55Δ and rad57Δ), which are required for homologous recombination. The genes rad50Δ, mre11Δ and xrs2Δ were also identified. These interactions are likely to reflect the role of these genes in homologous recombination rather than in non-homologous end joining, as elg1Δ is not synthetic lethal or sick with dnl4Δ or yku80 (data not shown). Of particular significance, genes thought to be in redundant pathways for re-starting stalled replication forks (Kaliraman et al., 2001; Mullen et al., 2001) have synthetic genetic interactions with elg1Δ. mus81Δ, mms4Δ, sgs1Δ, top3Δ, slx5Δ and slx8Δ all displayed a fitness defect in combination with elg1Δ. The last functional group of note comprises rad24Δ, rad17Δ, mec3Δ, ddc1Δ, mrc1Δ and tof1Δ. These genes are all linked to defects in S phase checkpoints (Alcasabas et al., 2001; Foss, 2001; Tanaka and Russell, 2001; Osborn et al., 2002), and their identification in the screen is consistent with lack of Elg1 causing DNA lesions or arrested replication forks during S phase. Our comprehensive genetic analysis indicates that cells lacking Elg1 and homologous recombination, replication fork re-start or S phase checkpoint pathways have a significant fitness defect, indicating that these pathways are required for optimal growth when ELG1 is deleted. Finally, the functional classes of genes identified in the elg1 synthetic genetic screen bear a striking similarity to those identified in screens with rad27Δ (Tong et al., 2001). Therefore, the genetic data suggest that Elg1 performs a function that is similar to that of Rad27, which plays an important role in Okazaki fragment maturation (Merrill and Holm, 1998; Parenteau and Wellinger, 1999).
Fig. 5. Genome-wide synthetic genetic screens with elg1Δ identify homologous recombination, fork re-start and S phase checkpoint pathways. The results of synthetic genetic array analysis with elg1Δ presented as a genetic interaction map. Lines connecting genes represent synthetic lethality or synthetic slow growth. Colored circles designate the cellular role of the interacting genes.
Elg1 is required for replication fidelity
Mutants defective in Okazaki fragment maturation, such as rfc1-1, pol30-52 and rad27Δ, all have increased forward mutation rates, but differ in the spectrum of mutants produced (Xie et al., 1999, 2001). Whereas rfc1-1 and pol30-52 cause predominantly point mutations and small insertions or deletions, rad27Δ causes large rearrangements. We assessed the forward mutation rate to canavanine resistance of elg1Δ, and found that it was almost 8-fold higher than the wild-type strain (24.2 × 10–7 versus 3.24 × 10–7), indicating that Elg1 is important for replication fidelity. To determine the spectrum of mutations caused by elg1Δ, we examined the CAN1 gene from 20 independent Canr mutants from both wild-type and elg1Δ for the presence of large insertion or deletion mutations. A difference in the size of a CAN1-derived fragment was observed in two out of 20 Canr wild-type strains, and in only one out of 20 Canr elg1Δ strains (data not shown). DNA sequencing revealed that the mutations in the elg1Δ strains were predominantly base substitution mutations, with some small (<5 bp) deletions and insertions. Therefore, the elg1Δ mutation spectrum resembles that caused by defects in Rfc1 or PCNA.
elg1Δ mutants are defective in S phase progression
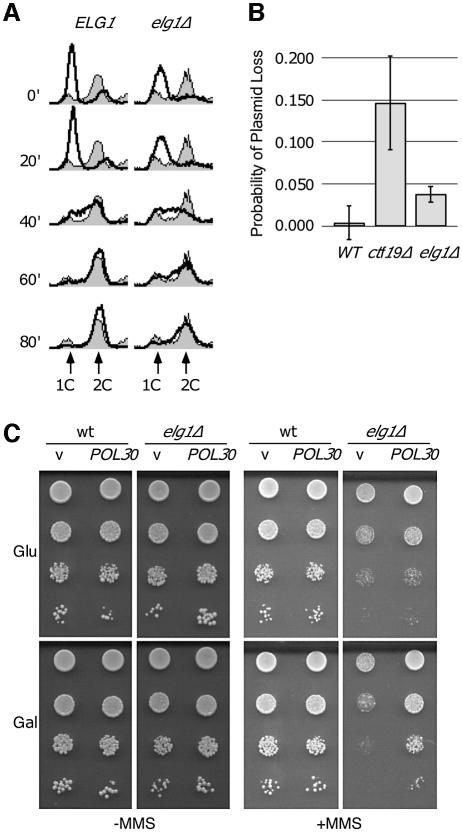
If elg1Δ cells are defective in fork re-start or have increased levels of fork stalling, we would expect to detect defects in S phase progression. We arrested cells in G1 phase and released them synchronously into the cell cycle. Progression through S phase was measured by flow cytometry. As shown in Figure 6A, the wild-type culture completed S phase by 60 min after release, as evidenced by the accumulation of cells with a 2C DNA content. In the elg1Δ culture, however, a significant fraction of cells still had a 1C or intermediate (<2C) DNA content at 60 min. Even at 80 min post-release, some elg1Δ cells had not completed S phase. Consistent with a role for Elg1 in S phase progression, we found that elg1Δ has a probability of plasmid loss that is 7.6 times higher than wild-type in a minichromosome maintenance assay (Figure 6B). This elevated plasmid loss was not suppressed significantly by the presence of additional origins of replication, and 2D gel analysis did not reveal any initiation defects in elg1Δ (data not shown). Furthermore, we found that elg1Δ did not have any detectable defect in sister chromatid cohesion (data not shown), suggesting that plasmid loss in elg1Δ was not due to a segregation defect.
Fig. 6. elg1Δ mutants display DNA replication defects. (A) Progression through S phase. Wild-type or elg1Δ cells were arrested in G1 (t = 0) and released synchronously into the cell cycle. Samples were removed at the indicated times and analyzed by flow cytometry. The shaded histograms represent the cell cycle distribution of the asynchronous cultures before the G1 arrest. Overlaid histograms represent the cell cycle distribution at the indicated times after release from the G1 arrest. The positions of cells with 1C and 2C DNA contents are indicated. (B) Plasmid loss in wild-type, elg1Δ and ctf19Δ. The probability of plasmid loss per generation is plotted, and error bars span 1 SD. (C) Suppression of elg1Δ MMS sensitivity by PCNA overexpression. Serial dilutions of wild-type or elg1Δ cells carrying empty vector (v) or GAL1-POL30 plasmid (POL30) were plated on synthetic medium with 2% glucose (Glu; uninduced) or 2% galactose + 2% raffinose (Gal; induced), plus or minus 0.01% MMS.
Since Elg1-RFC most probably functions as a clamp loader, we tested whether overexpression of PCNA could suppress the elg1Δ phenotype. As shown in Figure 6C, overexpression of PCNA rescued the MMS sensitivity of elg1Δ, as evidenced by improved growth on MMS plates when PCNA overexpression was induced by the presence of galactose. Taken together, these results suggest that Elg1 plays a direct role in DNA replication, most probably during the elongation phase of DNA synthesis.
Abnormal recovery from replication fork stalling in elg1Δ
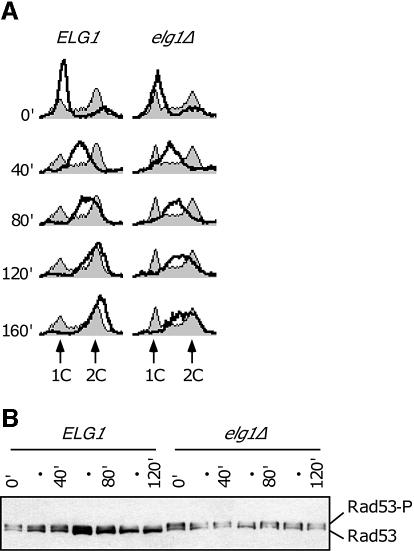
We measured S phase progression in medium containing 0.035% MMS, a concentration of MMS known to cause extensive fork stalling in wild-type cells (Tercero and Diffley, 2001) (Figure 7A). In the presence of MMS, S phase was prolonged in the wild-type cells, which take 120 min to accumulate with 2C DNA content. The elg1Δ mutant was clearly defective in S phase progression in the presence of MMS, with a significant fraction of cells containing <2C DNA at 160 min post-release. Thus the introduction of fork stalls causes further defects in DNA synthesis in elg1 mutants, consistent with Elg1 functioning in preventing fork stalling or in re-starting stalled replication forks.
Fig. 7. elg1Δ mutants are defective in recovery from MMS-induced replication fork stalling. (A) S phase progression in the presence of MMS. Wild-type or elg1Δ cells were arrested in G1 (t = 0) and released synchronously into medium containing 0.035% (v/v) MMS. Samples were removed at the indicated times and analyzed by flow cytometry. The shaded histograms represent the cell cycle distribution of the asynchronous cultures before the G1 arrest. Overlaid histograms represent the cell cycle distribution at the indicated times after release from the G1 arrest. (B) Checkpoint activation of Rad53 during recovery from MMS damage. Cells were arrested in G1, released into MMS for 1 h, and then transferred to medium lacking MMS (t = 0). At the indicated times, samples were withdrawn and Rad53 activation was analyzed by immunoblotting.
If Elg1 is required for replication fork integrity, we expected to see prolonged activation of Rad53 following MMS treatment during S phase. Wild-type and elg1Δ cells were arrested in G1 and released into medium containing MMS for 1 h to activate Rad53. The MMS was then washed out and cells were allowed to recover. Rad53 activation during recovery from MMS damage was assayed by immunoblot analysis (Figure 7B). In the wild-type strain, Rad53 is dephosphorylated by 80 min following removal of MMS. In contrast, activated Rad53 persisted in the elg1Δ strain for at least 120 min, indicating that Elg1 is required for downregulation of the intra-S checkpoint response. Since the elg1Δ strain is at least 10-fold less sensitive to MMS than mutants in MMS repair pathways (data not shown), these results suggest that Elg1 is required for efficient fork re-start rather than direct repair of DNA lesions.
Discussion
A novel RFC-like complex
Using a functional genomics approach, we have identified Elg1 as a novel DNA replication protein. Elg1 associates with Rfc2, Rfc3, Rfc4 and Rfc5, forming a fourth eukaryotic RFC-like complex that is functionally distinct from the canonical RFC, and from Rad24-RFC and Ctf18-RFC. Whether Elg1-RFC functions as a pentameric complex, like RFC and Rad24-RFC (Mossi and Hubscher, 1998; Green et al., 2000), or requires accessory factors as found in Ctf18-RFC (Mayer et al., 2001), awaits purification of Elg1-RFC. Eukaryotic cells have at least two sliding clamps that are loaded by RFC-like enzymes, PCNA and the 9-1-1 complex. RFC loads PCNA during leading and lagging strand DNA synthesis (reviewed in Mossi and Hubscher, 1998), whereas Rad24-RFC loads the 9-1-1 clamp in vitro but cannot load PCNA (Bermudez et al., 2003; Majka and Burgers, 2003). Ctf18-RFC binds to PCNA in vitro and in vivo (Ohta et al., 2002), and so may function as a PCNA clamp loader under specialized circumstances. Our data suggest a role for Elg1-RFC in DNA replication, and we therefore propose that it functions to load or unload PCNA. In support of this, we find that overexpression of PCNA suppresses the MMS sensitivity of elg1Δ. Furthermore, elg1Δ is synthetic sick when combined with rad17Δ, mec3Δ and ddc1Δ, which encode the 9-1-1 clamp, but has no genetic interaction with pol30-1, a mutant in the gene encoding PCNA (data not shown), suggesting that Elg1 functions in the same pathway as PCNA, but in a pathway that is parallel to that in which the 9-1-1 clamp functions.
Elg1-RFC and checkpoint activation
A number of RFC family proteins have been implicated in checkpoint activation. Rad24 is required for the G1 and G2 DNA damage checkpoints (Siede et al., 1994; Weinert et al., 1994), and is important (although not essential) for the intra-S damage checkpoint (Paulovich et al., 1997; Pellicioli et al., 1999). A role for Rad24 in the S–M checkpoint is revealed in the context of mutations in other RFC family genes (Shimomura et al., 1998; Naiki et al., 2001). Similarly, deletion of ctf18 has little effect on checkpoint activation unless rad24 is also deleted (Naiki et al., 2001). Consistent with formation of Rad24–Rfc2–5 complexes (Green et al., 2000), mutants in rfc2, rfc4 and rfc5 with checkpoint defects have also been described (Sugimoto et al., 1996, 1997; Noskov et al., 1998; Kim and Brill, 2001). Our data indicate that Elg1 does not play a primary role in checkpoint activation, as elg1Δ mutants show normal Rad53 activation in response to DNA damage in S phase and in response to replication fork arrest by HU. Double mutants in rad24 and either elg1 or ctf18 were more defective in Rad53 activation in S phase, and the triple mutant lacked detectable Rad53 activation. Thus it appears that Ctf18, and to a lesser extent Elg1, can partially substitute for the primary role of Rad24 in Rad53 activation in S phase. Alternatively, Elg1 could contribute to Rad53 activation in an indirect manner, for example through its effects on DNA replication.
When assessed directly, elg1Δ mutants display an intact intra-S phase checkpoint response as these cells exhibit the slow progression through S phase in the presence of MMS that is seen in wild-type cells. We did note, however, that elg1Δ rad24Δ double mutants do not display an intra-S phase checkpoint defect that is greater than that seen with rad24Δ alone. This was unexpected, as elg1Δ rad24Δ was more defective in Rad53 activation than rad24Δ. In contrast, in rad24Δ ctf18Δ, S phase progression in the presence of MMS was more rapid than wild-type or rad24Δ, and resembled the progression seen in the completely checkpoint-defective mec2-1 strain. Thus deletion of ctf18 in a rad24Δ strain exacerbates the rad24Δ checkpoint defect, whereas deletion of elg1Δ does not. The implications of these data are 2-fold. First, the slow progression through S phase in elg1Δ is at least partially checkpoint independent, which is consistent with the elg1Δ mutant having a defect in DNA replication fork integrity or re-start. Secondly, there is not a strict correlation between Rad53 activation and S phase progression in the presence of MMS in these mutants. This probably reflects that the slow progression through S phase that is the hallmark of the intra-S phase checkpoint (Paulovich and Hartwell, 1995; Paulovich et al., 1997) has both a checkpoint-dependent and a checkpoint-independent component (Tercero and Diffley, 2001). Our data suggest that Elg1 functions in the checkpoint-independent component of the slow S phase progression, again consistent with a role for Elg1 in preventing replication fork stalling or in re-starting stalled forks.
Elg1-RFC and genome integrity
The phenotypes of elg1Δ indicate that Elg1 has an important role in maintaining genome integrity. Elg1 suppresses recombination, protects against mutation and is important for recovery from DNA damage. Several aspects of the elg1Δ phenotype point to the presence of double-strand DNA breaks (DSBs), including the importance of homologous recombination for growth of the elg1Δ mutant and the hyper-recombination phenotype. Other aspects of the phenotype, while consistent with the presence of DSBs, point to a more fundamental role for Elg1 in DNA replication. These include the genetic requirement for the SGS1/TOP3 and MUS81/MMS4 replication fork re-start pathways (Kaliraman et al., 2001; Doe et al., 2002; Fabre et al., 2002) in the elg1Δ mutant cells, the slow S phase progression observed in both the presence and absence of DNA damage, the increase in plasmid loss, the increase in mutation rate and the failure to recover efficiently from increased replication stalling induced by MMS. We propose that deletion of ELG1 causes a decrease in replication fork processivity (an increase in fork stalling or a defect in re-start of stalled forks) perhaps due to defective Okazaki fragment maturation. Precedent for such a model comes from studies in both bacteria and yeast which have indicated that mutants in Okazaki fragment processing cause DSBs, stimulate recombination, confer DNA damage sensitivity, cause an increase in mutation frequency and render homologous recombination essential for viability (Rothstein et al., 2000; Michel et al., 2001), phenotypes similar to those observed in elg1Δ. Furthermore, defects in Okazaki fragment synthesis can cause replication fork stalling (Flores et al., 2001), and there is a strong correlation between replication stalling and genome instability (Aguilera et al., 2000; Michel, 2000; Rothstein et al., 2000; Michel et al., 2001).
Given the similarity of Elg1 to Rfc1 and its presence in RFC-like complexes, it is likely that Elg1-RFC functions as a clamp loader or unloader. Biochemical experiments in vitro indicate that a complex interplay between RFC, PCNA and DNA polymerase δ governs appropriate polymerase switching and Okazaki fragment sealing during lagging strand DNA synthesis. Although it is clear that RFC is responsible for limiting the length of the nascent strand synthesized by pol α, and for loading of PCNA to initiate the switch to synthesis by pol δ (Maga et al., 2001; Ayyagari et al., 2003), it is not clear how PCNA is recycled on the lagging strand. Eukaryotic cells in S phase have a great excess of Okazaki fragments over molecules of PCNA (Mossi and Hubscher, 1998), and the stability of PCNA–DNA complexes suggests that an unloading activity is required to recycle PCNA (Yao et al., 1996). A role for Elg1 in unloading and recycling PCNA on the lagging strand fits well with the available data. Additionally, this model predicts that PCNA should be limiting in elg1Δ mutants, and indeed we found that PCNA overexpression suppresses the MMS sensitivity of elg1Δ. Since Elg1 is not essential, its role in this process must be redundant, perhaps with RFC. Alternatively, Elg1 could be important for loading of PCNA during re-start of stalled replication forks, although in this model there must be a significant amount of fork stalling in an unperturbed S phase to account for the increased plasmid loss and the slow S phase progression observed in elg1Δ. It will be of great interest to determine if the human homolog of Elg1 plays a similar role in maintaining genome integrity.
Materials and methods
Yeast strains and media
Yeast strains used in this study are listed in table S1 in the Supplementary data. Non-essential haploid deletion strains were made by the Saccharomyces Gene Deletion Project (Winzeler et al., 1999) and can be obtained from Research Genetics (Huntsville, AL) or EUROSCARF (Frankfurt, Germany). Standard yeast media and growth conditions were used (Sherman, 1991).
Synthetic genetic array (SGA) analysis
SGA analysis was carried out as described (Tong et al., 2001). The MATα SGA starting strains containing mus81Δ::natR (Y3597), mms4Δ::natR (Y3561) and elg1Δ::natR (Y4521) were used to identify viable gene deletions that show synthetic genetic interactions with deletions in mus81Δ, mms4Δ and elg1Δ, respectively. Genetic interactions were confirmed by tetrad analysis on YPD (for mms4Δ and mus81Δ) or on synthetic medium supplemented with sodium glutamate as a nitrogen source (for elg1Δ). Confirmed interactions and extent of fitness defect are listed in table S2 in the Supplementary data.
Epitope tagging, immunoprecipitation and immunoblotting
The construction of strains carrying 3HA- or 13MYC-tagged Elg1 was performed as described (Longtine et al., 1998). Immunoprecipitation was performed essentially as described (Naiki et al., 2001). Proteins were resolved on 12% polyacrylamide–SDS gels, transferred to nitrocellulose membranes and subjected to immunoblot analysis with anti-HA (16B12; Covance), anti-myc (9E10; Santa Cruz) or anti-flag (M2; Sigma) antibodies. Immunoblots were developed using Supersignal ECL (Pierce). For detection of Rad53 phosphorylation, cells were fixed and extracts prepared essentially as described (Pellicioli et al., 1999). Proteins were separated on 8 or 4–12% polyacrylamide gels (Invitrogen), and immunoblots were probed with anti-RAD53 (yC-19; Santa Cruz).
MMS, HU and UV sensitivity measurements
Cells were grown in YPD, serially diluted, spotted onto plates and incubated at 30°C. MMS (Aldrich) plates contained 0.01 or 0.035% (v/v) MMS in YPD and were used within 24 h of preparation. HU plates contained 50 mM HU in YPD. For the UV radiation sensitivity assay, cells were serially diluted, spotted onto YPD plates, exposed to UV light at 100 J/m2 and incubated at 30°C. To determine viability after transient MMS treatment, mid-log phase cultures were incubated with 0.035% MMS in YPD liquid at 30°C. Samples were collected at the indicated time points, diluted, plated on YPD, and colonies were counted after incubation at 30°C for 3 days.
Synchronization and flow cytometry
Cells were arrested in G1 by culturing in the presence of 2 µg/ml α-factor for 2 h at 30°C in YPD pH 3.9. Cells were released into the cell cycle by harvesting, washing and resuspending in YPD. Flow cytometry was performed as described (Chang et al., 2002).
Plasmid loss, forward mutation rate and Canr mutation spectra
Plasmid loss rate was measured using the plasmid YCp1 (Tye, 1999). Transformants were streaked on YPD, and single colonies were inoculated into YPD and grown to saturation. Probabilities of plasmid loss represent the averages of 10 independent experiments for wild-type and elg1Δ, and seven independent experiments for ctf19Δ, and were calculated as described (Boe and Rasmussen, 1996). Mutation rates were determined by measuring the rate of forward mutation to canavanine resistance, as described previously (Huang et al., 2002). Fluctuation tests were performed with 10 parallel cultures, and the median value from each was used to calculate the spontaneous mutation rate by the method of the median (Lea and Coulson, 1949). Values represent the average of three experiments. To examine the spectrum of Canr mutations, the complete open reading frame of the CAN1 gene was amplified by PCR from independent Canr colonies. Amplified DNA was analyzed by HphI restriction digestion and by sequencing.
Supplementary data
Supplememtary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Brenda Andrews, George Brush, Phil Hieter, Rodney Rothstein and Bik Tye for yeast strains and plasmids, Yiqun Chen, Xin Cheng and Lizhu Ke for help with the SGA analysis, Tania Roberts and Amy Tong for helpful discussions, and Dan Durocher for communicating results prior to publication. This work was supported by grants from the National Cancer Institute of Canada (to G.W.B. and C.B.) and the Canadian Institutes of Health Research (to C.B.).
References
- Aguilera A., Chavez,S. and Malagon,F. (2000) Mitotic recombination in yeast: elements controlling its incidence. Yeast, 16, 731–754. [DOI] [PubMed] [Google Scholar]
- Alcasabas A.A. et al. (2001) Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol., 3, 958–965. [DOI] [PubMed] [Google Scholar]
- Ayyagari R., Gomes,X.V., Gordenin,D.A. and Burgers,P.M. (2003) Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 and DNA2. J. Biol. Chem., 278, 1618–1625. [DOI] [PubMed] [Google Scholar]
- Bermudez V.P., Lindsey-Boltz,L.A., Cesare,A.J., Maniwa,Y., Griffith,J.D., Hurwitz,J. and Sancar,A. (2003) Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17–replication factor C complex in vitro. Proc. Natl Acad. Sci. USA, 100, 1633–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M.N., Gaillard,P.H., McDonald,W.H., Shanahan,P., Yates,J.R.,3rd and Russell,P. (2001) Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell, 107, 537–548. [DOI] [PubMed] [Google Scholar]
- Boe L. and Rasmussen,K.V. (1996) Suggestions as to quantitative measurements of plasmid loss. Plasmid, 36, 153–159. [DOI] [PubMed] [Google Scholar]
- Brachmann C.B., Davies,A., Cost,G.J., Caputo,E., Li,J., Hieter,P. and Boeke,J.D. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast, 14, 115–132. [DOI] [PubMed] [Google Scholar]
- Cha R.S. and Kleckner,N. (2002) ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science, 297, 602–606. [DOI] [PubMed] [Google Scholar]
- Chang M., Bellaoui,M., Boone,C. and Brown,G.W. (2002) A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc. Natl Acad. Sci. USA, 99, 16934–16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.B. et al. (2001). Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell, 8, 1117–1127. [DOI] [PubMed] [Google Scholar]
- Cullmann G., Fien,K., Kobayashi,R. and Stillman,B. (1995) Characterization of the five replication factor C genes of Saccharomyces cerevisiae. Mol. Cell. Biol., 15, 4661–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C.L., Ahn,J.S., Dixon,J. and Whitby,M.C. (2002) Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem., 277, 32753–32759. [DOI] [PubMed] [Google Scholar]
- Fabre F., Chan,A., Heyer,W.D. and Gangloff,S. (2002) Alternate pathways involving Sgs1/Top3, Mus81/Mms4 and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl Acad. Sci. USA, 99, 16887–16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores M.J., Bierne,H., Ehrlich,S.D. and Michel,B. (2001) Impairment of lagging strand synthesis triggers the formation of a RuvABC substrate at replication forks. EMBO J., 20, 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss E.J. (2001) Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics, 157, 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A.C. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- Green C.M., Erdjument-Bromage,H., Tempst,P. and Lowndes,N.F. (2000) A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr. Biol., 10, 39–42. [DOI] [PubMed] [Google Scholar]
- Haber J.E. and Heyer,W.D. (2001) The fuss about Mus81. Cell, 107, 551–554. [DOI] [PubMed] [Google Scholar]
- Hanna J.S., Kroll,E.S., Lundblad,V. and Spencer,F.A. (2001) Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell. Biol., 21, 3144–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y. et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature, 415, 180–183. [DOI] [PubMed] [Google Scholar]
- Huang M.E., Rio,A.G., Galibert,M.D. and Galibert,F. (2002) Pol32, a subunit of Saccharomyces cerevisiae DNA polymerase δ, suppresses genomic deletions and is involved in the mutagenic bypass pathway. Genetics, 160, 1409–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubscher U., Maga,G. and Podust,V. (1996) DNA replication accessory proteins. In DePamphilis,M. (ed.), DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–543. [Google Scholar]
- Kaliraman V., Mullen,J.R., Fricke,W.M., Bastin-Shanower,S.A. and Brill,S.J. (2001) Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev., 15, 2730–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.S. and Brill,S.J. (2001) Rfc4 interacts with Rpa1 and is required for both DNA replication and DNA damage checkpoints in Saccharomyces cerevisiae. Mol. Cell. Biol., 21, 3725–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea D.E. and Coulson,C.A. (1949) The distribution of the numbers of mutants in bacterial populations. J. Genet., 49, 264–285. [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie,A.,3rd, Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Lopes M., Cotta-Ramusino,C., Pellicioli,A., Liberi,G., Plevani,P., Muzi-Falconi,M., Newlon,C.S. and Foiani,M. (2001) The DNA replication checkpoint response stabilizes stalled replication forks. Nature, 412, 557–561. [DOI] [PubMed] [Google Scholar]
- Maga G., Villani,G., Tillement,V., Stucki,M., Locatelli,G.A., Frouin,I., Spadari,S. and Hubscher,U. (2001) Okazaki fragment processing: modulation of the strand displacement activity of DNA polymerase δ by the concerted action of replication protein A, proliferating cell nuclear antigen and flap endonuclease-1. Proc. Natl Acad. Sci. USA, 98, 14298–14303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka J. and Burgers,P.M. (2003) Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc. Natl Acad. Sci. USA, 100, 2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M.L., Gygi,S.P., Aebersold,R. and Hieter,P. (2001) Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S.cerevisiae. Mol. Cell, 7, 959–970. [DOI] [PubMed] [Google Scholar]
- Merrill B.J. and Holm,C. (1998) The RAD52 recombinational repair pathway is essential in pol30 (PCNA) mutants that accumulate small single-stranded DNA fragments during DNA synthesis. Genetics, 148, 611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B. (2000) Replication fork arrest and DNA recombination. Trends Biochem. Sci., 25, 173–178. [DOI] [PubMed] [Google Scholar]
- Michel B., Flores,M.J., Viguera,E., Grompone,G., Seigneur,M. and Bidnenko,V. (2001) Rescue of arrested replication forks by homologous recombination. Proc. Natl Acad. Sci. USA, 98, 8181–8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossi R. and Hubscher,U. (1998) Clamping down on clamps and clamp loaders—the eukaryotic replication factor C. Eur. J. Biochem., 254, 209–216. [PubMed] [Google Scholar]
- Mullen J.R., Kaliraman,V., Ibrahim,S.S. and Brill,S.J. (2001) Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics, 157, 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiki T., Shimomura,T., Kondo,T., Matsumoto,K. and Sugimoto,K. (2000) Rfc5, in cooperation with rad24, controls DNA damage checkpoints throughout the cell cycle in Saccharomyces cerevisiae. Mol. Cell. Biol., 20, 5888–5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiki T., Kondo,T., Nakada,D., Matsumoto,K. and Sugimoto,K. (2001) Chl12 (Ctf18) forms a novel replication factor C-related complex and functions redundantly with Rad24 in the DNA replication checkpoint pathway. Mol. Cell. Biol., 21, 5838–5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov V.N., Araki,H. and Sugino,A. (1998) The RFC2 gene, encoding the third-largest subunit of the replication factor C complex, is required for an S-phase checkpoint in Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 4914–4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S., Shiomi,Y., Sugimoto,K., Obuse,C. and Tsurimoto,T. (2002) A proteomics approach to identify proliferating cell nuclear antigen (PCNA)-binding proteins in human cell lysates. Identification of the human CHL12/RFCs2–5 complex as a novel PCNA-binding protein. J. Biol. Chem., 277, 40362–40367. [DOI] [PubMed] [Google Scholar]
- Osborn A.J., Elledge,S.J. and Zou,L. (2002) Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol., 12, 509–516. [DOI] [PubMed] [Google Scholar]
- Parenteau J. and Wellinger,R.J. (1999) Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell. Biol., 19, 4143–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich A.G. and Hartwell,L.H. (1995) A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell, 82, 841–847. [DOI] [PubMed] [Google Scholar]
- Paulovich A.G., Margulies,R.U., Garvik,B.M. and Hartwell,L.H. (1997) RAD9, RAD17 and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics, 145, 45–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A., Lucca,C., Liberi,G., Marini,F., Lopes,M., Plevani,P., Romano,A., Di Fiore,P.P. and Foiani,M. (1999) Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J., 18, 6561–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R., Michel,B. and Gangloff,S. (2000) Replication fork pausing and recombination or ‘gimme a break’. Genes Dev., 14, 1–10. [PubMed] [Google Scholar]
- Scholes D.T., Banerjee,M., Bowen,B. and Curcio,M.J. (2001) Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics, 159, 1449–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Shimomura T., Ando,S., Matsumoto,K. and Sugimoto,K. (1998) Functional and physical interaction between Rad24 and Rfc5 in the yeast checkpoint pathways. Mol. Cell. Biol., 18, 5485–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siede W., Friedberg,A.S., Dianova,I. and Friedberg,E.C. (1994) Characterization of G1 checkpoint control in the yeast Saccharomyces cerevisiae following exposure to DNA-damaging agents. Genetics, 138, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. and Rothstein,R. (1999) An allele of RFA1 suppresses RAD52-dependent double-strand break repair in Saccharomyces cerevisiae. Genetics, 151, 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo J.M., Lopes,M. and Foiani,M. (2002) Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science, 297, 599–602. [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Shimomura,T., Hashimoto,K., Araki,H., Sugino,A. and Matsumoto,K. (1996) Rfc5, a small subunit of replication factor C complex, couples DNA replication and mitosis in budding yeast. Proc. Natl Acad. Sci. USA, 93, 7048–7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Ando,S., Shimomura,T. and Matsumoto,K. (1997) Rfc5, a replication factor C component, is required for regulation of Rad53 protein kinase in the yeast checkpoint pathway. Mol. Cell. Biol., 17, 5905–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K. and Russell,P. (2001) Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat. Cell Biol., 3, 966–972. [DOI] [PubMed] [Google Scholar]
- Tercero J.A. and Diffley,J.F. (2001) Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature, 412, 553–557. [DOI] [PubMed] [Google Scholar]
- Tong A.H. et al. (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science, 294, 2364–2368. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T. and Stillman,B. (1989) Purification of a cellular replication factor, RF-C, that is required for coordinated synthesis of leading and lagging strands during simian virus 40 DNA replication in vitro. Mol. Cell. Biol., 9, 609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye B.K. (1999) Minichromosome maintenance as a genetic assay for defects in DNA replication. Methods, 18, 329–334. [DOI] [PubMed] [Google Scholar]
- Virshup D.M. and Kelly,T.J. (1989) Purification of replication protein C, a cellular protein involved in the initial stages of simian virus 40 DNA replication in vitro. Proc. Natl Acad. Sci. USA, 86, 3584–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T.A., Kiser,G.L. and Hartwell,L.H. (1994) Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev., 8, 652–665. [DOI] [PubMed] [Google Scholar]
- Winzeler E.A. et al. (1999) Functional characterization of the S.cerevisiae genome by gene deletion and parallel analysis. Science, 285, 901–906. [DOI] [PubMed] [Google Scholar]
- Xie Y., Counter,C. and Alani,E. (1999) Characterization of the repeat-tract instability and mutator phenotypes conferred by a Tn3 insertion in RFC1, the large subunit of the yeast clamp loader. Genetics, 151, 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Liu,Y., Argueso,J.L., Henricksen,L.A., Kao,H.I., Bambara,R.A. and Alani,E. (2001) Identification of rad27 mutations that confer differential defects in mutation avoidance, repeat tract instability and flap cleavage. Mol. Cell. Biol., 21, 4889–4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao N., Turner,J., Kelman,Z., Stukenberg,P.T., Dean,F., Shechter,D., Pan,Z.Q., Hurwitz,J. and O’Donnell,M. (1996) Clamp loading, unloading and intrinsic stability of the PCNA, β and gp45 sliding clamps of human,E.coli and T4 replicases. Genes Cells, 1, 101–113. [DOI] [PubMed] [Google Scholar]