Abstract
The process through which multipotential hematopoietic cells commit to distinct lineages involves the induction of specific transcription factors. PU.1 (also known as Spi-1) and GATA-1 are transcription factors essential for the development of myeloid and erythroid lineages, respectively. Overexpression of PU.1 and GATA-1 can block differentiation in lineages in which they normally are down-regulated, indicating that not only positive but negative regulation of these factors plays a role in normal hematopoietic lineage development. Here we demonstrate that a region of the PU.1 Ets domain (the winged helix–turn–helix wing) interacts with the conserved carboxyl-terminal zinc finger of GATA-1 and GATA-2 and that GATA proteins inhibit PU.1 transactivation of critical myeloid target genes. We demonstrate further that GATA inhibits binding of PU.1 to c-Jun, a critical coactivator of PU.1 transactivation of myeloid promoters. Finally, PU.1 protein can inhibit both GATA-1 and GATA-2 transactivation function. Our results suggest that interactions between PU.1 and GATA proteins play a critical role in the decision of stem cells to commit to erythroid vs. myeloid lineages.
The development of specific lineages of peripheral blood cells from hematopoietic stem cells involves the expression of lineage-specific transcription factors. Multiple transcription factors such as CCAAT enhancer-binding protein (C/EBPα), AML-1B, Sp1, and PU.1 contribute to the control of myeloid cell development. PU.1, a member of the Ets transcription factor family, is autoregulated specifically in myeloid cells (1, 2). PU.1 also binds to many myeloid gene promoters, including the granulocyte colony-stimulating factor (G-CSF) receptor, granulocyte/macrophage CSF receptor, macrophage CSF (M-CSF) receptor, CD11b, and myeloperoxidase to regulate their expression (3). Inhibition of PU.1 function blocks myelopoiesis both in vitro (1) and in vivo (4, 5). The amino terminus of PU.1 serves as an activation domain (6). In B cells, the PEST [region rich in proline (P), glutamate (E), serine (S), and threonine (T)] domain of PU.1 recruits a B cell-specific DNA-binding factor, Pip, to a site that is important for immunoglobin κ 3′ enhancer function (7). The C terminus of the PU.1 Ets-homology domain is a winged helix–turn–helix motif (8) that serves as a DNA binding domain.
GATA-1 is a critical factor for erythroid cells development. Its N- and C-terminal zinc finger region is conserved among GATA family members. The C finger itself is capable of binding to DNA (9). The N terminus of GATA-1 serves as a transactivation domain. GATA-1 regulates many erythroid genes, including itself, the erythropoietin receptor, SCL, and the β-globin promoter (10). Disruption of GATA-1 function in mice leads to a loss of erythroid differentiation (11, 12).
PU.1 and GATA-1 are expressed in both early progenitor cells (reviewed in refs. 3 and 13). During erythroid development from multipotential progenitors, GATA-1 is activated and monocytic genes like PU.1 and its target, the M-CSF receptor, are repressed (1). Overexpression of PU.1 blocks erythroid cell differentiation and results in erythroleukemia in mice (14). Conversely, in parallel with the activation of PU.1 expression during monocytic differentiation, GATA-1 (1) and GATA-2 (15) are down-regulated. Enforced expression of GATA-1 or GATA-2 in an early myeloid cell line, 416B, blocked differentiation to myeloid cells and decreased the expression of the myeloid cell surface marker Mac-1 and induced differentiation to megakaryocytic cells (16, 17). These studies indicate that not only positive but also negative regulation of these factors plays a role in normal hematopoietic lineage development (14, 17, 18). However, the mechanism of inactivation of transcription factor function to accomplish this negative regulation still remains unclear. One way to accomplish this inactivation is to represses the expression of those genes that should not function in a particular lineage. An example is that GATA-1 represses PU.1 promoter function 2-fold (2). But to inactivate factors that are already at significant levels may require a different mechanism. One possible mechanism is to block function through physical interactions, because these lineage-specific factors are expressed in the same precursors.
PU.1 acts as a weak transactivator (3), suggesting that it requires coactivators to achieve activation function through physical interactions. The DNA-binding domain of PU.1 has been found to interact physically with many proteins, including myeloid regulators such as AML-1B and C/EBPα (3, 19). We recently have demonstrated that the β3/β4 region (amino acids 243–254) of PU.1 in the Ets domain interacts with a number of proteins (N.W.-a., Y. Koyama, G.B., S. Tetradis, J. Tsukada, Y.-T. Ro, D.G.T., and P.E.A., unpublished results), including c-Jun, which acts as a critical coactivator of PU.1 transactivation of myeloid promoters (20). GATA-1 also has been reported to interact with Sp1 (21), EKLF (21), and RBTN2 (22). In addition, the N finger of GATA-1 interacts with FOG (Friend of GATA-1) (23) and the C finger interacts with CREB-binding protein (CBP)/p300 (24). These physical interactions lead to synergistic activation. Obstructing these physical interactions results in inhibition of the synergistic transactivation function of PU.1 and GATA-1 with their respective coactivators. For example, E1A blocks GATA-1 synergy with CBP (24).
Here, we demonstrate by using a yeast two-hybrid screen and in vitro glutathione S-transferase (GST) pull-down and in vivo immunoprecipitation assays that the DNA-binding domain of PU.1 interacts with the zinc finger region of GATA-2 and the same highly conserved region of GATA-1 (15). These interactions result in an inhibition of PU.1 transactivation function as a result of blocking c-Jun binding to PU.1, as both c-Jun and GATA proteins interact with the same region of PU.1, the β3/β4 region.
MATERIALS AND METHODS
Yeast Two-Hybrid System.
The PEST and DNA-binding domains of human PU.1 (amino acids 100–272) were fused in-frame to the GAL4 DNA-binding domain in the pGBT9 vector (CLONTECH). An EML [multipotential cell line (25)] cDNA library with a VP16 activation domain was used for screening as described (26). Plasmid DNA from potential PU.1-binding clones was analyzed by DNA sequencing.
In Vitro Protein Interaction Assays.
GST-PU.1 fusion proteins were constructed as described (N.W.-a. et al., unpublished results). Both the full-length human GATA-2 cDNA and portions containing either the N finger (amino acids 265–327) or the C finger (amino acids 330–407) were subcloned into pcDNA3 (Invitrogen). Proteins were generated and labeled in vitro by using a coupled transcription and translation kit (Promega). Expression and purification of GST fusion proteins were performed as described (27). To test binding, 2 μg of agarose-bound GST fusion proteins was incubated with 35S-methionine-labeled full-length or truncated GATA-1 or GATA-2 proteins for 1 hr at 4°C in NETN buffer (20 mM Tris⋅HCl, pH 7.8/1 mM EDTA/1% NP-40/150 mM NaCl/0.5% glycerol/protein inhibitors). Beads were washed four times with NETN buffer and then analyzed on SDS/PAGE gels and detected by fluorography.
Coimmunoprecipitation.
PU.1 and either GATA-1 or GATA-2 expression constructs were cotransfected into CV1 cells by using Lipofectamine (GIBCO/BRL). Forty-eight hours after transfection, whole-cell lysates or K562 cell nuclear extracts (28) were incubated with primary antibody diluted 1:500 to 1:1,000 and bound to protein A-Sepharose beads for 90–120 min on ice in 2% glycerol/0.5% Nonidet P-40/1 mM EDTA/20 mM Tris⋅HCl, pH 8/100 mM NaCl/10 mM MgCl2/0.1 mM ZnSO4. Beads then were washed with prechilled NETN three times, and bound proteins were separated on SDS/PAGE gels and transferred to nitrocellulose membranes for Western blotting. For coimmunoprecipitation of PU.1 and GATA-2, 10 μg of both PU.1 and GATA-2 polyclonal antibodies first were coupled to protein A beads. Proteins were detected by enhanced chemiluminescence (Amersham Pharmacia). Primary antibodies used were rabbit anti-PU.1 (sc-352; Santa Cruz Biotechnology); rat anti-murine anti-GATA-1 monoclonal (sc-265; Santa Cruz Biotechnology); and rabbit anti-GATA-2 (29).
Transfection Assays.
Transient transfections were carried out with the Lipofectamine transfection kit (GIBCO/BRL). Approximately 4 hr after the transfection was initiated, cells were placed in 10% FCS and incubated for another 40 hr, and firefly luciferase activity was measured as relative light units (RLU). The RLU from individual transfections were normalized by measurement of Renilla luciferase activity expressed from a cytomegalovirus promoter-driven vector in the same samples (30). Individual transfection experiments were done in triplicate, and the results are reported as mean firefly RLU/Renilla (±SD) from representative experiments. HeLa cell and NIH 3T3 cell transfections were done by calcium phosphate precipitation (19). For detection of PU.1, GATA-1, and GATA-2 protein levels after transfection of CV-1 cells, FLAG-tagged PU.1 in pECE (31) and FLAG-tagged GATA-1 and GATA-2 in pcDNA3 were transfected into CV-1 cells by Lipofectamine. Forty hours after transfection, cells were lysed in 60 μl of 1× passive lysis buffer (Promega). Twenty microliters of the lysate was used for measurement of luciferase activity, and 40 μl was used for Western blot analysis by using M2 anti-FLAG antibody (F3165; Sigma). The relative amounts of FLAG proteins were quantitated by using imagequant software (Molecular Dynamics).
Electrophoretic Mobility-Shift Assays (EMSAs).
EMSAs were performed as described by using double-stranded oligonucleotides containing the PU.1-binding site from CD11b promoter (3′-GCTCAAAGAAGGGCAGAAAAGGAGAAGTAG-5′) (32).
RESULTS
GATA-1 and GATA-2 Interact with PU.1 in Vitro and in Vivo.
PU.1 contains three functional domains: an activation domain, a PEST domain, and an Ets DNA-binding domain (Fig. 1a). Using PU.1 amino acids 100–272 as “bait” in a yeast two-hybrid screening, we isolated 26 putative PU.1-binding cDNA clones from a myeloid cDNA library (33). Two of these clones perfectly matched the sequence encoding amino acids 230–400 of GATA-2, including both the N and C zinc fingers (17). Cotransformant yeast containing both the GAL4-PU.1 DNA-binding domain and a plasmid in which the VP16 activation domain was fused to GATA-2 showed a strong interaction as evidenced by high expression of both histidine and β-galactosidase reporter genes (data not shown). These results verify that PU.1 specifically interacts with GATA-2 in yeast cells.
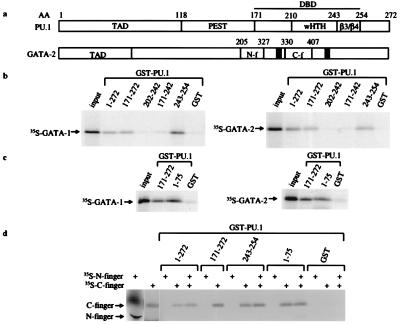
Figure 1.
PU.1 specifically interacts with GATA-1 and GATA-2. (a) Schematic of PU.1 and GATA-2 protein structures is shown. Functional domains are designated according to amino acid residue. (b–d) GST fusion protein interaction studies. 35S-methionine-labeled GATA-1 or GATA-2 was incubated with equal amounts of GST or GST-PU.1 fusion proteins. Truncated GST-PU.1 fusion proteins are marked according to amino acid residue. TAD, transactivation domain; PEST, PEST domain; wHTH, winged helix–turn–helix motif; N-f, N finger; C-f, C finger.
Because the N and C fingers of both GATA-1 and GATA-2 are highly conserved (15), we tested whether GATA-1 as well as GATA-2 interacts with PU.1. As seen in Fig. 1b, GST-PU.1 fusion proteins containing the PU.1 β3/β4 region (amino acids 243–254) precipitated both in vitro-translated GATA-1 and GATA-2. In contrast, GST alone and GST-PU.1 fusion protein lacking the β3/β4 region failed to retain either GATA protein above background levels. The β3/β4 region of PU.1 is an interaction domain that binds to a number of other important transcription factors, including c-Jun, C/EBP, and AML1 (N.W.-a. et al., unpublished results). GATA-1 and GATA-2 also interact with GST-PU activation domain (Fig. 1c), although we observe no functional effects of this interaction (see below). The binding of GATA-1 and GATA-2 to the PU.1 activation domain and β3/β4 region is mediated by the C finger but not the N finger (Fig. 1d).
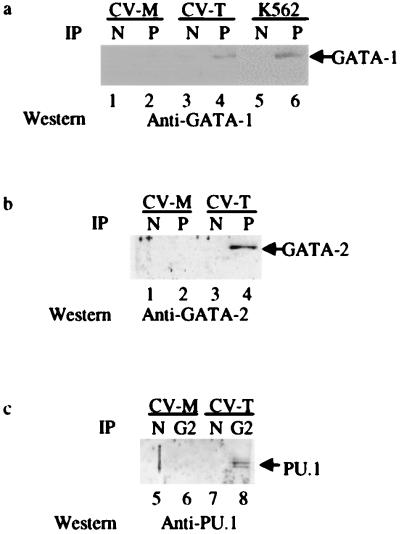
We also could demonstrate that the physical interaction between PU.1 and GATA-1 occurs in vivo in cells in which both are endogenously expressed by using the human cell line K562, which has both early myeloid and erythroblastic characteristics (34). K562 nuclear extracts were immunoprecipitated with either normal rabbit serum or a rabbit anti-PU.1 polyclonal antibody. GATA-1 protein was detected by Western blotting with a GATA-1 murine mAb only in PU.1 immunoprecipitates (Fig. 2a). Similar interactions could be demonstrated in CV1 cells cotransfected with PU.1 and GATA-1 or GATA-2 expression constructs (Fig. 2 a and b). To confirm these in vivo PU.1-GATA interactions, cell extracts from PU.1 + GATA-2 and mock-transfected CV1 cells were immunoprecipitated with the anti-GATA-2 antibody and then Western-blotted with anti-PU.1 antibody. PU.1 protein was detected only in GATA-2 immunoprecipitates from the PU.1 + GATA-2 transfected cells, but not in mock-transfected cells or in control immunoprecipitates by using nonimmune rabbit IgG (Fig. 2c). These results confirmed that interaction between PU.1 and GATA-1 or GATA-2 also occurs in vivo.
Figure 2.
Coimmunoprecipitation of PU.1 and GATA proteins. (a) Coimmunoprecipitation of PU.1 and GATA-1 from whole-cell lysates of transfected CV-1 cells and endogenous PU.1 and GATA-1 from K562 cells with anti-PU.1 antibody (P) or with normal IgG (N). Western blot analysis was performed by using anti-GATA-1 antibody. (b) Coimmunoprecipitation of PU.1 and GATA-2 from whole-cell lysates of transfected CV-1 cells. Western blot analysis was performed by using anti-GATA-2 antibody. (c) Coimmunoprecipitation of PU.1 and GATA-2 with anti-GATA-2 antibody (G2) or with normal IgG in transfected CV-1 cells. Western blot analysis was performed with anti-PU.1 antibody. CV-M, mock-transfected CV-1 cells; CV-T, CV-1 cells transfected with PU.1 and GATA-1 (a) or PU.1 and GATA-2 (b and c).
GATA-1 and GATA-2 Inhibit the Ability of PU.1 to Transactivate Myeloid Target Promoters.
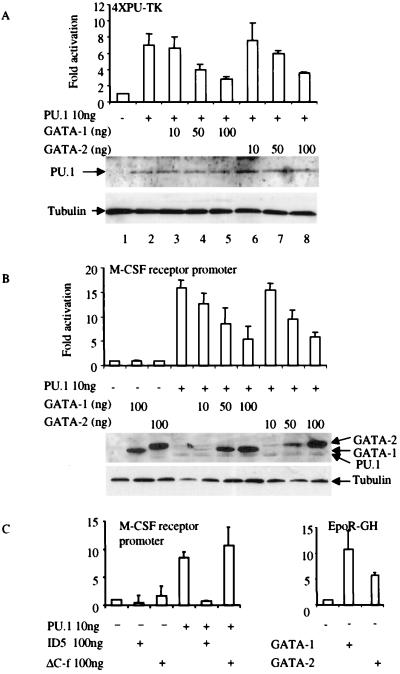
Transient transfections were performed by using a luciferase construct driven by four PU.1 DNA-binding sites in front of a minimal thymidine kinase (TK) promoter (4XPU-TK) (35). In PU.1 transfectants, approximately 7-fold induction of PU.1 transcriptional activity was obtained (Fig. 3a). PU.1 transactivation was repressed by cotransfection of either GATA-1 or GATA-2. The extent of repression depended on the ratio of GATA to PU.1 expression plasmid; >50% inhibition was observed at GATA/PU.1 plasmid ratios of 10:1 (Fig. 3a Upper). Suppression of PU.1-mediated reporter gene expression by GATA-1 or GATA-2 was not associated with any change in the intracellular steady-state level of PU.1 protein (Fig. 3a Lower), suggesting that repression resulted from direct effects on PU.1 transactivation of its target promoter and not on down-regulation of PU.1 expression. GATA-1 and GATA-2 protein levels in transfected CV-1 cells are equal to the level of PU.1 protein when equal amounts of FLAG-tagged PU.1, GATA-1, and GATA-2 expression vectors are used (Fig. 3b Lower). When FLAG-GATA-1 and FLAG-GATA-2 expression vectors are increased to a ratio of 5- and 10-fold above that of PU.1, GATA-1 and GATA-2 proteins levels also are increased 3- and 5-fold, respectively, more than PU.1 (Fig. 3b). Therefore, excess GATA-1 or GATA-2 proteins inhibit PU.1-mediated gene activation, as is likely to be the case in primary erythroid precursors in which GATA-1 levels are higher than PU.1 (1, 36). Suppression of PU.1-induced activation also was observed with the M-CSF receptor promoter (−416 to +124 bp) (Fig. 3b), which has been shown previously to be regulated by PU.1 (37). The entire GATA protein was necessary for PU.1 suppression, because the N or C finger alone was not sufficient to repress PU.1 activation of the PU.1 reporter constructs (data not shown). However, GATA mutant ID5, which consists of GATA-1 with a deletion of the first 63 aa, still could inhibit PU.1 function, but a C finger GATA-1 deletion of amino acids 248–290 did not retain the inhibition function (Fig. 3c Left). To ensure that the observed GATA-1- or GATA-2-mediated inhibition did not reflect toxic or other nonspecific effects of the cotransfected GATA plasmids, luciferase expression from the PU.1-inducible promoters was normalized to that of a cotransfected plasmid expressing Renilla luciferase (Promega) from a cytomegalovirus promoter (30). To confirm that the GATA proteins were functionally active, transient transfections were performed by using an erythropoietin (Epo) receptor promoter expressing human growth hormone (GH) as a reporter (38). The GATA-1 and GATA-2 expression plasmids activated the Epo receptor promoter 10- and 6-fold, respectively (Fig. 3C Right).
Figure 3.
GATA-1 and GATA-2 repress PU.1 transactivation. (a) CV-1 cells were transfected with 100 ng of the multimerized PU.1-binding-site reporter (4XPU-TK), 10 ng of PU.1 expression vector PU.PECE (31), and GATA-1 or GATA-2 expression vectors as indicated. Luciferase activity is shown as fold activation above the level of activity seen with the luciferase reporter in the absence of any added PU.1 expression vector (±SD; n = 4). Western blot analysis was performed by using anti-PU.1 antibody to detect the expression level of PU.1 protein in transfected CV-1 cells as shown, with antitubulin antibody as a loading control. (b) GATA-1 and GATA-2 inhibit PU.1 transactivation of the M-CSF receptor promoter. CV-1 cells were transfected with 100 ng of M-CSF receptor promoter-luciferase, 10 ng of PU.PECE, and GATA-1 or GATA-2 expression vectors as indicated. Luciferase activity is shown as fold activation (±SD; n = 4). GATA-1 or GATA-2 alone did not repress the basal activity of either the 4XPU-TK (a) or the M-CSF receptor promoter (b). Western blot analysis was performed by using anti-FLAG antibody to detect the expression levels of FLAG-tagged PU.1, GATA-1, and GATA-2 proteins in transfected CV-1 cells as shown, with antitubulin antibody as a loading control. (c) Deletion mutation of the C finger of GATA-1 loses the ability to inhibit PU.1 function. (Left) CV-1 cells were transfected with the M-CSF receptor promoter and PU.1, ID5, and ΔC-f expression vectors as indicated. Luciferase activity is shown as fold activation (±SD; n = 3). (Right) The reporter is the erythropoietin receptor promoter-driving human growth hormone (EpoR-GH). Four micrograms of EpoR-GH and 9 μg of GATA-1 and GATA-2 were used in these transfection experiments. ID5, N-terminal deletion (amino acids 1–63 were deleted) of GATA-1; ΔC-f, C finger deletion mutation of GATA-1.
GATA-1 and GATA-2 Do Not Inhibit PU.1 Transactivation Through Inhibition of DNA Binding or the PU.1 Activation Domain.
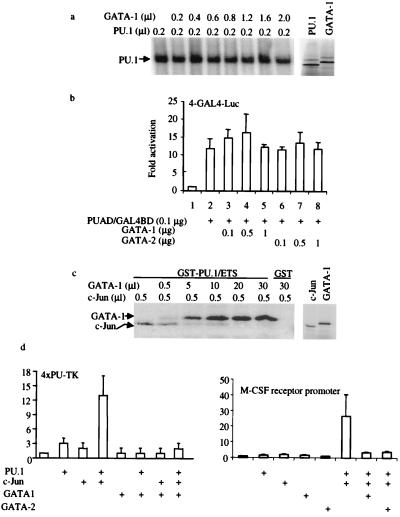
We investigated further the mechanism by which GATA proteins repress PU.1 activity. EMSAs demonstrated that neither GATA-1 (Fig. 4a) nor GATA-2 (data not shown) interferes with PU.1 DNA-binding activity (Fig. 4a). We also investigated whether GATA proteins repress the PU.1 activity through inhibition of its activation domain. As shown in Fig. 4b, neither GATA-1 nor GATA-2 represses the ability of a fusion protein containing the GAL4 DNA-binding domain and the PU.1 activation domain to transactivate a luciferase reporter containing multiple GAL4 DNA-binding sites (39). Therefore, we do not believe that the inhibition of PU.1 function is mediated by GATA binding to the PU.1 activation domain.
Figure 4.
GATA proteins inhibit PU.1 transactivation by displacing the PU.1 coactivator, c-Jun. (a) GATA proteins do not inhibit PU.1 binding to DNA. (Left) EMSA using a α-32P-ATP-labeled PU.1-binding-site probe (32) and in vitro transcribed and translated PU.1 and GATA-1 proteins. PU.1 protein (0.2 μl) was used for each reaction, along with increasing amounts of GATA-1 protein as indicated. (Right) SDS/PAGE gel of in vitro transcribed and translated 2 μl of PU.1 and 2 μl of GATA used in EMSA. (b) GATA-1 and GATA-2 do not inhibit PU.1 activation domain function. CV-1 cells were transfected with 4 μg of multimerized GAL4 DNA-binding sites in front of a minimal TK promoter used as a reporter. One hundred nanograms of PU.1 activation domain/GAL4 DNA-binding domain fusion protein (PUAD/GAL4BD) in pcDNA3 was used as a transactivator, and GATA-1 or GATA-2 was added as indicated. Luciferase activities were corrected by an internal-control Renilla luciferase vector and shown as fold activation (±SD; n = 3). (c) GATA-1 inhibits c-Jun binding to PU.1. (Left) In vitro 35S-methionine-labeled c-Jun or GATA-1 was incubated with GST-PU.1 Ets domain (GST-PU.1/ETS) or GST alone (GST). The amounts of GATA-1 and c-Jun added are indicated at the top. (Right) SDS/PAGE gel of c-Jun (1 μl) and GATA-1 (2 μl) used in the GST pull-down experiment. (d) GATA-1 inhibits c-Jun coactivation of PU.1 function. F9 cells were transfected by using Lipofectamine Plus with 0.3 μg of 4XPU-TK, 0.1 μg of PU.PECE, 0.05 μg of pSV-Sport1-c-Jun [a c-Jun expression vector (20)], and 1.3 μg of pcDNA3-GATA-1. Luciferase activates were determined 24 hr after transfection and shown as fold activation (±SD; n = 3). The same experiments were done by using the M-CSF receptor promoter-luciferase reporter as shown (Right) using GATA-1 or GATA-2.
GATA-1 and GATA-2 Inhibit c-Jun Coactivation of PU.1 Transcriptional Activation Function by Competing for Binding to a Common Interaction Region in the PU.1 Ets Domain.
We have shown recently that the human protooncogene, c-Jun, interacts with the β3/β4 region of PU.1 and enhances the ability of PU.1 to transactivate myeloid target promoters such as the M-CSF receptor (20). PU.1 cannot transactivate the M-CSF receptor in F9 cells, which do not express endogenous c-Jun, unless c-Jun is cotransfected into these cells (20). Because c-Jun and GATA proteins both bind to the β3/β4 region of PU.1, the repression of PU.1 transactivation by GATA proteins might be a result of disruption of the interaction of PU.1 with c-Jun. Therefore, we investigated whether GATA proteins repress PU.1 transactivation by directly interrupting the PU.1/c-Jun interaction. Both c-Jun and GATA-1 can bind specifically to the PU.1 DNA-binding domain individually (Figs. 1 and 2; ref. 20). Addition of excess GATA-1 abrogated c-Jun interaction with the PU.1 DNA-binding domain. Importantly, transient transfection experiments demonstrated that the coactivation of PU.1 function by c-Jun in F9 cells could be reversed by cotransfection of the GATA-1 expression vector on the multimerized PU.1-binding-site promoter or both GATA-1 or GATA-2 on the M-CSF receptor promoter (Fig. 4d). These results indicated that repression of PU.1 activity by GATA proteins is mediated by displacement of the PU.1 coactivator, c-Jun.
PU.1 Inhibits Both GATA-1 and GATA-2 Activation.
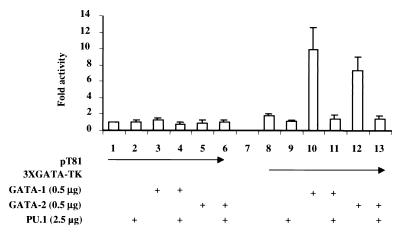
To investigate whether the interaction between PU.1 and GATA affects GATA function, transient transfections were performed by using a multimerized GATA-binding site in front of a minimal TK promoter-luciferase reporter. Cotransfection of GATA-1 and GATA-2 activated the reporter 12- and 9-fold, respectively (Fig. 5, columns 10 and 12), but not the minimal TK-luciferase reporter. A 5-fold excess of PU.1 plasmid fully inhibited both GATA-1 and GATA-2 function (Fig. 5, columns 11 and 13).
Figure 5.
PU.1 inhibits GATA transactivation function. CV-1 cells were transfected with 4 μg of pT81 (minimal TK promoter-driving luciferase gene expression) or multimerized GATA-binding-site reporter (3XGATA-TK), 0.5 μg of GATA-1 or GATA-2 expression vector, and 2.5 μg of PU.1 expression vector PU.pECE as indicated by the CaPO4 method. Luciferase activity is shown as fold activation above the level of activity seen with pT81 in the presence of the expression vector backbone (pECE for PU.1 and pcDNA3 for GATA-1 and GATA-2) (±SD; n = 4).
DISCUSSION
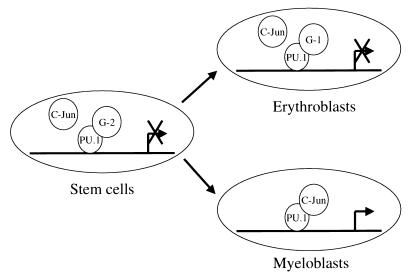
PU.1, GATA-1, and GATA-2 are lineage-specific transcription factors that are expressed in multipotential progenitor cells (40). During lineage commitment, activation of specific transcription factors has been shown to direct precursor cells to a particular differentiation pathway (3, 41). Other lineage regulators must be inactivated to ensure normal lineage differentiation. PU.1 transgenic mice induce erythroleukemia, demonstrating that it is essential to block PU.1 function for normal erythroid cell development (42). GATA-1, a critical erythroid transcription factor, is likely to play a role in inhibition of PU.1 function during erythroid cell differentiation. We postulate that the activation of GATA proteins and subsequent suppression of PU.1 function is a critical step in the commitment of stem cells to the erythroid lineage and erythroid cell differentiation. As shown in the model in Fig. 6, our results suggest that in hematopoietic stem cells and/or multipotential progenitors, GATA-2 occupies the β3/β4 region of PU.1 to block PU.1 interaction with its coactivator, c-Jun. As the result of this inhibition, stem cells either remain undifferentiated or commit to the erythroid lineage. In erythroblasts, GATA-1 blocks PU.1 function to ensure fully the maturation of erythroid cells.
Figure 6.
GATA-1 and GATA-2 inhibit PU.1 function at different stages during hematopoietic cell differentiation to specific lineages through protein—protein interactions. The model hypothesizes that in stem cells, GATA-2 blocks PU.1 and c-Jun interaction and, therefore, inhibits PU.1 activation of its downstream target genes. In erythroblasts, up-regulation of GATA-1 blocks coactivation of PU.1 by c-Jun. But in developing myeloid progenitors, with decreased expression of GATA-1 and GATA-2, PU.1 and c-Jun synergistically activate PU.1 target genes such as the M-CSF receptor (20). G-1, GATA-1; G-2, GATA-2.
Here we have shown that a specific effect of both GATA-1 and GATA-2 is to functionally suppress PU.1 activity. Our data suggest a mechanism in which negative regulation of one lineage-specific regulator by another modulates function through critical protein–protein interactions (N.W.-a. et al., unpublished results; refs. 23, 43, and 44). This supports a model in which the timing, levels of expression (1), and combinations of different regulators (3, 44) play a critical role in hematopoietic differentiation.
The PU.1 and GATA protein interactions were found by a yeast two-hybrid screen method using the PU.1 DNA-binding domain as a “bait” to screen a cDNA library derived from an early myeloid cell line, EML. This screen detected GATA-2 but not GATA-1 because GATA-2 is highly expressed in EML cells compared with GATA-1 (data not shown). Both yeast two-hybrid and GST pull-down experiments demonstrated that the C finger of GATA proteins interacts with the β3/β4 region of PU.1, which is located in the winged helix–turn–helix PU.1 DNA-binding motif. It has been demonstrated that the β3/β4 region interacts with a number of proteins (N.W.-a., unpublished results). Interestingly, the C finger of GATA proteins also interacts with the PU.1 amino-terminal activation domain. The GATA N finger binds neither the PU.1 activation domain nor the β3/β4 region. Deletion mutations of the PU.1 activation domain and the β3/β4 region result in a loss of the ability of GATA to bind to PU.1. These data demonstrate that the C finger of GATA proteins not only possesses DNA-binding function but also serves as a protein—protein interaction motif. Coimmunoprecipitation experiments were performed to confirm that GATA proteins interact with PU.1 in vivo.
Repression of PU.1 function is a critical step for erythroid cell differentiation, because overexpression of PU.1 in erythroblast blocks erythroid cell differentiation (42). We previously have reported that GATA-1 represses the PU.1 promoter 2-fold (2), and changes of transcription factor levels of this magnitude can effect major changes in myeloid differentiation (45). But PU.1 is still expressed in erythroblasts, albeit at low levels compared with myeloid cells. Therefore, GATA-1 may serve to inactivate those levels of PU.1 by interacting directly with PU.1. AML-1B (19), C/EBPα (46), and c-Jun (20) all interact with the β3/β4 region of PU.1 and synergize with PU.1 to activate downstream myeloid genes. Although GATA proteins bind to the same region of PU.1 as these activators of PU.1 function, they repress the ability of PU.1 to activate both artificial and natural target promoters. The C finger portion of GATA proteins interacts with PU.1. A deletion mutation of the C finger of GATA-1 lost the ability to inhibit PU.1 function. However, an N terminal deletion of GATA-1 still inhibited PU.1 function. These results suggest that one mechanism of GATA-1 inactivation of PU.1 is direct interaction with PU.1 in committed erythroblasts, enabling full maturation of erythrocytes, although it is still possible that GATA proteins could work through indirect mechanisms such as alteration of chromatin structure.
The PU.1 activation domain is known to interact with the basal transcription factor TATAA-binding protein (47). Because both GATA-1 and GATA-2 also interact with this same region of PU.1, they might block PU.1 function by blocking TATAA-binding protein binding to PU.1. This hypothesis was tested by using a system with multiple GAL4-binding sites to drive luciferase gene expression as a reporter and using a PU.1 activation domain–GAL4 DNA-binding domain fusion protein as an activator. Cotransfection of neither GATA-1 nor GATA-2 affected the transactivation function of the fusion protein. Therefore, we investigated whether GATA inhibited PU.1 function through other mechanisms.
Although GATA-1 and GATA-2 bind to the β3/β4 region of the PU.1 DNA-binding domain, they do not block PU.1 binding to DNA. GST pull-down experiments demonstrate that GATA-1 could compete with PU.1 interaction with its coactivator c-Jun. Transient transfection assays demonstrated that PU.1 itself could not activate myeloid target promoters in c-Jun-deficient F9 cells (20). Cotransfected c-Jun acts as a coactivator of PU.1 function in F9 cells, and expression of GATA-1 reversed coactivation of PU.1 by c-Jun. Inactivation of PU.1 in erythroblasts by GATA-1 may be due to disruption of interaction of PU.1 with its coactivators, including c-Jun.
Low levels of GATA-1 and GATA-2 are detectable in early myeloid progenitor cells (reviewed in refs. 3 and 13). Blocking the function of GATA-1 and GATA-2 is a critical step for myeloid differentiation (17, 18). As shown in Fig. 5, PU.1 inhibits both GATA-1 and GATA-2 activation in transient transfections. Both the N and C fingers of GATA-1 interact with other activators and coactivators, including Sp1, RBTN2, FOG, and CBP. The GATA C finger is important for binding to DNA, self-association, and protein—protein interactions (48). PU.1 potentially could block these functions of GATA-1 as well as compete for factors such as Sp1 and CBP (49). Studies of the mechanism of how PU.1 inhibits GATA function will add to our understanding of how positive and negative regulation of transcription factors during stem cell commitment leads to lineage differentiation.
Acknowledgments
We thank Shickwann Tsai for the yeast EML library, Stuart Orkin for GATA-1 and GATA-2 expression vectors, Thomas Quertermous for GATA-2 antibody, Rich Maki and Tony Kouzarides for PU.1 plasmids, and Donald E. Ayer for pVP16 and GAL4 plasmids. We also thank Stuart Orkin for helpful discussions and suggestions. This work was supported by National Institutes of Health Grants CA41456, CA72009, and CA70297 and American Cancer Society Grant RPG98213. A.I. is supported by a long-term fellowship from the Human Frontier Science Program.
ABBREVIATIONS
- CBP
CREB-binding protein
- C/EBPα
CCAAT enhancer-binding protein
- GST
glutathione S-transferase
- M-CSF
macrophage-granulocyte stimulating factor
- PEST
region rich in proline (P), glutamate (E), serine (S), and threonine (T)
- EMSA
electrophoretic mobility-shift assay
- TK
thymidine kinase
Note Added in Proof:
A recent report described similar physical interactions between GATA proteins and PU.1 and inhibition by PU.1 of GATA function in erythroid cells (50).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Voso M T, Burn T C, Wulf G, Lim B, Leone G, Tenen D G. Proc Natl Acad Sci USA. 1994;91:7932–7936. doi: 10.1073/pnas.91.17.7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H M, Ray-Gallet D, Zhang P, Hetherington C J, Gonzalez D A, Zhang D-E, Moreau-Gachelin F, Tenen D G. Oncogene. 1995;11:1549–1560. [PubMed] [Google Scholar]
- 3.Tenen D G, Hromas R, Licht J D, Zhang D-E. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- 4.Scott E W, Simon M C, Anastai J, Singh H. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 5.McKercher S R, Torbett B E, Anderson K L, Henkel G W, Vestal D J, Baribault H, Klemsz M, Feeney A J, Wu G E, Paige C J, et al. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 6.Klemsz M J, Maki R A. Mol Cell Biol. 1996;16:390–397. doi: 10.1128/mcb.16.1.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pongubala J M, Nagulapalli S, Klemsz M J, McKercher S R, Maki R A, Atchison M L. Mol Cell Biol. 1992;12:368–378. doi: 10.1128/mcb.12.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kodandapani R, Pio F, Ni C Z, Piccialli G, Klemsz M, McKercher S, Maki R A, Ely K R. Nature (London) 1996;380:456–460. doi: 10.1038/380456a0. [DOI] [PubMed] [Google Scholar]
- 9.Martin D I, Orkin S H. Genes Dev. 1990;4:1886–1898. doi: 10.1101/gad.4.11.1886. [DOI] [PubMed] [Google Scholar]
- 10.Orkin S H. Blood. 1992;80:575–581. [PubMed] [Google Scholar]
- 11.Pevny L, Simon M C, Robertson E, Klein W H, Tsai S F, D’Agati V, Orkin S H, Costantini F. Nature (London) 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 12.Fujiwara Y, Browne C P, Cunniff K, Goff S C, Orkin S H. Proc Natl Acad Sci USA. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shivdasani R A, Orkin S H. Blood. 1996;87:4025–4039. [PubMed] [Google Scholar]
- 14.Moreau-Gachelin F, Tavitian A, Tambourin P. Nature (London) 1988;331:277–280. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- 15.Lee M E, Temizer D H, Clifford J A, Quertermous T. J Biol Chem. 1991;266:16188–16192. [PubMed] [Google Scholar]
- 16.Visvader J E, Elefanty A G, Strasser A, Adams J M. EMBO J. 1992;11:4557–4564. doi: 10.1002/j.1460-2075.1992.tb05557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Visvader J E, Crossley M, Hill J, Orkin S H, Adams J M. Mol Cell Biol. 1995;15:634–641. doi: 10.1128/mcb.15.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulessa H, Frampton J, Graf T. Genes Dev. 1995;9:1250–1262. doi: 10.1101/gad.9.10.1250. [DOI] [PubMed] [Google Scholar]
- 19.Petrovick M S, Hiebert S W, Friedman A D, Hetherington C J, Tenen D G, Zhang D-E. Mol Cell Biol. 1998;18:3915–3925. doi: 10.1128/mcb.18.7.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Behre G, Whitmarsh A J, Coghlan M P, Hoang T, Carpenter C L, Zhang D-E, Davis R J, Tenen D G. J Biol Chem. 1999;274:4939–4946. doi: 10.1074/jbc.274.8.4939. [DOI] [PubMed] [Google Scholar]
- 21.Merika M, Orkin S H. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osada H, Grutz G, Axelson H, Forster A, Rabbitts T H. Proc Natl Acad Sci USA. 1995;92:9585–9589. doi: 10.1073/pnas.92.21.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsang A P, Visvader J E, Turner C A, Fujiwara Y, Yu C, Weiss M J, Crossley M, Orkin S H. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 24.Blobel G A, Nakajima T, Eckner R, Montminy M, Orkin S H. Proc Natl Acad Sci USA. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai S, Bartelmez S, Sitnicka E, Collins S. Genes Dev. 1994;8:2831–2841. doi: 10.1101/gad.8.23.2831. [DOI] [PubMed] [Google Scholar]
- 26.Fields S, Song O. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 27.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 28.Andrews N C, Faller D V. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawana M, Lee M E, Quertermous E E, Quertermous T. Mol Cell Biol. 1995;15:4225–4231. doi: 10.1128/mcb.15.8.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Behre G, Smith L T, Tenen D G. BioTechniques. 1999;26:24–28. doi: 10.2144/99261bm03. [DOI] [PubMed] [Google Scholar]
- 31.Klemsz M J, McKercher S R, Celada A, Van Beveren C, Maki R A. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 32.Pahl H L, Scheibe R J, Zhang D-E, Chen H M, Galson D L, Maki R A, Tenen D G. J Biol Chem. 1993;268:5014–5020. [PubMed] [Google Scholar]
- 33.Lioubin M N, Algate P A, Tsai S, Carlberg K, Aebersold A, Rohrschneider L R. Genes Dev. 1996;10:1084–1095. doi: 10.1101/gad.10.9.1084. [DOI] [PubMed] [Google Scholar]
- 34.Koeffler H P, Golde D W. Blood. 1980;56:344–350. [PubMed] [Google Scholar]
- 35.Smith L T, Hohaus S, Gonzalez D A, Dziennis S E, Tenen D G. Blood. 1996;88:1234–1247. [PubMed] [Google Scholar]
- 36.Galson D L, Hensold J O, Bishop T R, Schalling M, D’Andrea A D, Jones C, Auron P E, Housman D E. Mol Cell Biol. 1993;13:2929–2941. doi: 10.1128/mcb.13.5.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang D-E, Hetherington C J, Chen H M, Tenen D G. Mol Cell Biol. 1994;14:373–381. doi: 10.1128/mcb.14.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zon L I, Youssoufian H, Mather C, Lodish H F, Orkin S H. Proc Natl Acad Sci USA. 1991;88:10638–10641. doi: 10.1073/pnas.88.23.10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ayer D E, Laherty C D, Lawrence Q A, Armstrong A P, Eisenman R N. Mol Cell Biol. 1996;16:5772–5781. doi: 10.1128/mcb.16.10.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng T, Shen H, Giokas D, Gere J, Tenen D G, Scadden D T. Proc Natl Acad Sci USA. 1996;93:13158–13163. doi: 10.1073/pnas.93.23.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nerlov C, Graf T. Genes Dev. 1998;12:2403–2412. doi: 10.1101/gad.12.15.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreau-Gachelin F, Wendling F, Molina T, Denis N, Titeux M, Grimber G, Briand P, Vainchenker W, Tavitian A. Mol Cell Biol. 1996;16:2453–2463. doi: 10.1128/mcb.16.5.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sieweke M H, Tekotte H, Frampton J, Graf T. Cell. 1996;85:49–60. doi: 10.1016/s0092-8674(00)81081-8. [DOI] [PubMed] [Google Scholar]
- 44.Sieweke M H, Graf T. Curr Opin Genet Dev. 1998;8:545–551. doi: 10.1016/s0959-437x(98)80009-9. [DOI] [PubMed] [Google Scholar]
- 45.Radomska H S, Huettner C S, Zhang P, Cheng T, Scadden D T, Tenen D G. Mol Cell Biol. 1998;18:4301–4314. doi: 10.1128/mcb.18.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang D-E, Hetherington C J, Meyers S, Rhoades K L, Larson C J, Chen H M, Hiebert S W, Tenen D G. Mol Cell Biol. 1996;16:1231–1240. doi: 10.1128/mcb.16.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hagemeier C, Bannister A J, Cook A, Kouzarides T. Proc Natl Acad Sci USA. 1993;90:1580–1584. doi: 10.1073/pnas.90.4.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crossley M, Merika M, Orkin S H. Mol Cell Biol. 1995;15:2448–2456. doi: 10.1128/mcb.15.5.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen H M, Pahl H L, Scheibe R J, Zhang D-E, Tenen D G. J Biol Chem. 1993;268:8230–8239. [PubMed] [Google Scholar]
- 50.Rekhtman N, Radparvar F, Evans T, Skoultchi A I. Genes Dev. 1999;13:1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]