Abstract
The p21WAF1 (p21) cyclin-dependent kinase inhibitor plays a major role in regulating cell cycle arrest. It was recently reported that the p53-independent elevation of p21 protein levels is essential in mediating the G1 arrest resulting from signal transduction events initiated by the crosslinking of membrane IgM on Daudi Burkitt lymphoma cells. Although the role of p21 in cell cycle regulation is well documented, there is little information concerning its role in antibody-mediated apoptosis. In the present study, we examined the involvement of p21 in the regulation of apoptosis by suppressing its induction in anti-IgM-treated Daudi cells through a p21 antisense expression construct approach. Reduction in induced p21 protein levels resulted in diminished G1 arrest and increased apoptosis. The increased susceptibility to anti-IgM-mediated apoptosis was associated with increased caspase-3-like activity and poly-(ADP)ribose polymerase cleavage. These data suggest that p21 may directly interfere with the caspase cascade, thus playing a dual role in regulating both cell cycle progression and apoptosis.
Keywords: apoptosis, B cell lymphoma, caspase
There is accumulating evidence that, if tumor-reactive Abs are selected for their direct antiproliferative activity, they make highly effective antitumor agents. Thus, in an experimental model of murine B lymphoma, the induction of a dormant state by antiidiotype Abs indicates a predominant role for “negative signaling” of tumor cells (1–3). Indeed, in vitro studies (4–8) indicate that antiidiotype or anti-IgM Abs can induce cell cycle arrest and/or apoptosis in both murine and human lymphoma cell lines. At the clinical level, promising results have been obtained in non-Hodgkin’s lymphoma by using antiidiotype (9, 10) or anti-CD20 mAbs (11–14). After relapse, the latter appears as effective as chemotherapy for the treatment of patients with low-grade non-Hodgkin’s lymphoma (15). In addition to the contribution of effector mechanisms to the observed clinical activity, anti-CD20 mAbs have direct antiproliferative activity, including induction of growth arrest and apoptosis in B lymphoma cell lines (16–19). Also, an anti-Her2 mAb (Herceptin), which recently has been approved for treatment of advanced breast cancer, has the capacity to negatively signal the Her2-overexpressing tumor cells (20, 21).
The common mechanism(s) by which stressful stimuli, including signaling Abs, chemotherapeutic agents, irradiation, etc., exert their inhibitory effect is by blocking cell progression at key cell cycle checkpoints and/or activation of apoptotic pathway(s). These checkpoints are mainly controlled by negative regulation of cyclin/cyclin-dependent kinase (CDK) activity through the CDK inhibitory proteins. p21WAF1 (p21) is one of the several key CDK inhibitors that act ubiquitously on CDKs and is responsible for the induction of cell cycle arrest (22).
Previous work from our laboratory aimed at understanding the major components of the signaling cascade initiated by hypercrosslinking of membrane IgM (mIgM) on Burkitts B lymphoma (Daudi) cells has shown that induction of p21 protein and subsequent inhibition of the retinoblastoma kinase activity associated with CDK2 are the central events responsible for the late G1 arrest. Changes induced by anti-IgM also include loss of the hyperphosphorylated form of retinoblastoma and down-regulation of cyclin A (23). The lack of functional p53 in Daudi cells indicates that the G1 arrest is p53-independent. Recent reports of others have shown that the G0/G1 growth arrest of Daudi cells treated with IFNα is also caused by the induction of p21 expression, the resultant inhibition of CDK2 kinase activity, and the inhibition of retinoblastoma phosphorylation (24–26).
It has been suggested that cell cycle arrest mediated by expression of p21 is an early event in the sequence by which activation of p53 leads to apoptosis (27). In contrast, recent evidence obtained with antisense strategies suggests that p21 protects cells against apoptosis (28, 29). Also, the overexpression of p21 transcripts protects against p53-mediated apoptosis in human melanoma cells (30). Thus, there is uncertainty regarding the role(s) of p21 in programmed cell death.
The purpose of the present study was to define the role of p21 CDK inhibitor in the cell cycle arrest and apoptosis induced by hypercrosslinking mIgM on Daudi cells. We show that the reduction of the inducible endogenous p21 protein levels in anti-IgM-treated Daudi cells through the use of antisense p21 expression vectors decreases their cell cycle arrest response and increases their susceptibility to caspase-mediated apoptosis. Therefore, we conclude that the nature of the growth inhibitory signal triggered by anti-IgM depends on the level of inducible p21.§
METHODS
Cell Lines.
The human Burkitt’s lymphoma cell line “Daudi” was maintained in culture by serial passage in RPMI medium 1640 containing 25 mM Hepes, 10% heat-inactivated fetal bovine serum, and 100 mM l-glutamine (complete medium). The cells were grown in a humidified atmosphere of 5% CO2 and air. Daudi cells transfected with pCEP4 or pCEP-WAF1-AS plasmid constructs were cultured as above except that the culture medium also contained 100 μg/ml hygromycin B (Boehringer Mannheim).
Antibodies.
Purified goat IgG specific for human IgM was purchased from Chemicon. Purified mAb against human p21 was purchased from PharMingen. Rabbit polyclonal serum against poly-(ADP ribose) polymerase (PARP) was purchased from Boehringer Mannheim. mAbs against human μ domain (HB57) and Her2 (Her81) were prepared and purified in our laboratory.
Analysis by Flow Cytometry of the Cell Cycle Status and Apoptosis.
Cells were examined simultaneously for viability and cell cycle status by flow cytometry using a FACStar Plus (Becton Dickinson). Daudi cells (5 × 105) were plated in flat-bottom 12-well plates and were treated with Abs (5), and, after 24 hours, cells were harvested and washed twice with PBS containing 1% fetal bovine serum. The pellet was incubated for 30 minutes on ice with 50 μl of 400 μM 7-aminoactinomycin D (7-AAD) (Molecular Probes), a vital dye that stains the DNA of permeable cells. Cells then were fixed with 1.0 ml of 0.5% paraformaldehyde in PBS and simultaneously were permeabilized and stained for 16 hours at 4°C with 220 μl of 10 μg/ml Hoechst 33342 dye (also a DNA-binding dye) containing 5% Tween 20 (31). The data from 104 cells were collected and stored by using lysys ii software (Becton Dickinson) and were analyzed by using paint-a-gate software (Becton Dickinson). The viability and percentage of cells in each stage of the cell cycle were determined on gated single cells (cell doublets were excluded by pulse analysis of width vs. area of the Hoechst signal).
DNA Synthesis.
Triplicate cultures of 5 × 104 cells/well were plated in 96-well flat-bottom plates, Abs were added, and plates were incubated for 24 hours. During the last 8 hours of culture, cells were pulsed with 1 μCi of 3H-thymidine (Amersham Pharmacia) and were harvested.
Western Blotting.
Cells (5 × 106–107) were lysed in ice-cold Nonidet P-40 lysis buffer (0.1% NP40/50 mM Tris, pH 7.5/120 mM NaCl/1 mM EDTA/0.1 mM sodium orthovanadate/50 mM sodium fluoride/1 mM PMSF/10 μg/ml each of aprotinin and leupeptin). Lysates were sonicated and clarified by centrifugation. Total protein content was quantitated with a microbicinchoninic acid kit (Pierce). Equal amounts of protein were resolved by SDS/PAGE and then were transferred to a poly(vinylidene difluoride) membrane (NEN). The membranes were blocked in Tris-buffered saline containing 5% milk and 0.1% Tween 20 and were incubated overnight at 4°C with specific Abs. The levels of protein were analyzed by using the enhanced chemiluminescence (ECL) system (Amersham Pharmacia).
DNA Transfection.
The pCEP-WAF1-AS antisense p21 expression plasmid construct was kindly provided by Bert Vogelstein (Johns Hopkins Oncology Center, Baltimore). The antisense p21 expression vector consists of the full length cDNA of the entire coding region of the human WAF-1 gene cloned in the antisense orientation into the NotI cassette under the control of the CMV promoter of the pCEP4 mammalian expression vector (Invitrogen). This expression vector carries a hygromycin B resistance gene. Daudi cells transfected with empty vector pCEP4 were used as “controls.” For transfection, cells were suspended in ice-cold PBS at 107 cells/0.8 ml; then, 20 μg of linearized DNA from antisense or empty vector was added, and cells were kept on ice for 10 minutes, were electroporated at 960 μF, 250 V with a Gene pulser (Bio-Rad), and were maintained on ice for another 10 minutes. After 48 hours of incubation in culture medium, the cells were selected for 2 weeks in culture medium containing 100 μg/ml hygromycin B (selection medium). The antisense clones were obtained from the stable transfectant pool by limiting dilution. Hygromycin B-resistant clones were isolated and tested by PCR for the presence of the hygromycin B resistance gene in the genomic DNA by using a set of primers: 5′-AGAAGATGTTGGCGACCTCG-3′ and 5′-TACACTACATGGCGTGATTTC-3′.
Caspase Activity.
Caspase-3 activity was analyzed by using an ApoAlert CPP32 kit (CLONTECH). Whole cell extracts prepared from 5 × 106 cells were incubated for 1 hour at 37°C with an Asp-Glu-Val-Asp-p-nitroanilide (DEVD-pNA) substrate. Caspase activity was determined spectrophotometrically at 405 nm by measurement of the free pNA produced on cleavage of the substrate. Quantitation of pNA was determined by comparison to a pNA standard curve.
RESULTS
Transfection of Daudi Cells with p21 Antisense Plasmid Constructs Decreases the Level of Inducible p21.
To suppress the level of inducible p21, Daudi cells were stably transfected with the pCEP-WAF1-AS antisense p21 construct. As a negative control, the pCEP4 vector without the cDNA insert was used. Selection with hygromycin B yielded a stable transfectant pool that, after limiting dilution, yielded several clonal antisense lines.
The effect of the antisense construct on inducible p21 protein level was investigated by Western blot analysis of the lysates from transfected cells grown for 24 hours in the presence or absence of 20 μg/ml anti-IgM. The seven antisense clones tested had inducible p21 protein levels that were 1.5- to 4-fold lower than those in parental untransfected [wild-type (WT)] cells. The one with the lowest inducible p21 protein level was selected for subsequent studies. Treatment of WT cells with anti-IgM or of cells transfected with the empty pCEP4 vector (control) led to the induction of the same protein levels of p21 (Fig. 1). In contrast, Daudi cells transfected with the antisense p21-expression vector (AS) showed ≈4-fold less inducible p21.
Figure 1.
The level of p21 protein in antisense p21-transfected Daudi cells after treatment with anti-IgM. Equal amounts (50 μg of total protein) of cell lysates from WT, control, and AS cells treated or untreated for 24 hours with 20 μg/ml anti-IgM were electrophoresed on a 12% SDS/PAGE gel and were transferred to poly(vinylidene difluoride) membranes. The p21 protein in each sample was detected with 1 μg/ml anti-p21 mAb followed by enhanced chemiluminescence detection. Values represent the relative induction of p21 as normalized to p21 protein levels in untreated WT cells as detected by densitometric scanning of the bands. A representative experiment of three is shown.
Daudi Antisense-p21WAF1 Transfectants Are More Susceptible to Inhibition by anti-IgM Than Control Cells.
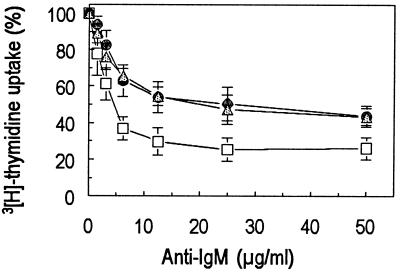
The functional consequence of lower levels of inducible p21 in AS cells was investigated by analyzing the in vitro growth inhibition after incubation with anti-IgM. The level of thymidine incorporation by cells incubated for 24 hours with various concentrations of anti-IgM indicated that the inhibition of proliferation of the AS clone (IC50 = 4 μg/ml) was achieved at concentrations ≈5-fold lower as compared with that of the WT (IC50 = 25 μg/ml) or control (IC50 = 20 μg/ml) (Fig. 2).
Figure 2.
Dose-dependent inhibition of DNA synthesis in Daudi cells by anti-IgM. AS (-□-), WT (-●-), or control transfected (-▵-) Daudi cells (5 × 104) were incubated at 37°C for 24 hours with the indicated concentrations of anti-IgM. DNA synthesis was assessed by measuring the level of 3H-thymidine incorporation. Values represent the average of four independent experiments.
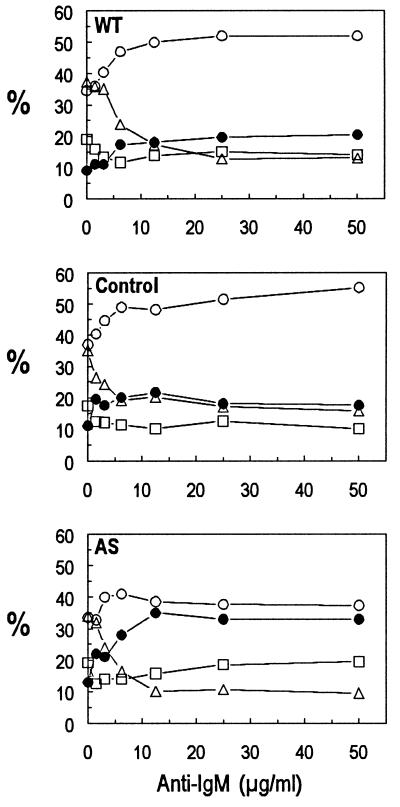
The nature of this inhibitory effect generated by anti-IgM on AS cells was determined by measuring both changes in DNA content and viability by using flow cytometry (Fig. 3). Incubation for 24 hours with the same range of anti-IgM concentrations as those used for DNA replication assays led to the induction of a G1 arrest, with a concurrent decline of cells in S phase. Cell death was ≈25% in both WT cells and control transfectants. In contrast, the inhibitory effect induced in AS cells was caused mainly by a marked increase in cell death with a concurrent decrease in G1 arrest (Fig. 3). Therefore, we conclude that the nature of the growth inhibitory signal triggered by anti-IgM depends on the level of inducible p21.
Figure 3.
Effect of anti-IgM on cell cycle status and viability in antisense p21-transfected Daudi cells. Daudi cells (1 × 106/ml) were incubated for 24 hours with the indicated concentrations of anti-IgM, after which the cells were stained with 7-AAD and Hoechst. The percentages of apoptotic (dead) (-●-) and viable cells in G1 (-○-), S-phase (-▵-), and G2-M (-□-) were determined by flow cytometric analysis of the DNA content and membrane permeability. A representative experiment of three is shown.
Analysis of mIgM Expression in Antisense p21 Transfectants.
The possibility was considered that the increased susceptibility of AS cells could be caused by an up-regulation of mIgM expression. Flow cytometric analysis of cells stained with an anti-human IgM mAb conjugated to FITC indicated that all of the above cell lines expressed an equal intensity of mIgM (Fig. 4).
Figure 4.
Expression of mIgM on transfected Daudi cells. Cell surface expression of IgM was analyzed by flow cytometry of cells that were stained directly with FITC-conjugated IgG1 anti-human μ mAb (HB57). A total of 5 × 105 cells were stained for each sample with a preliminary-determined optimal concentration of mAb. The thin line indicates the background staining with a FITC-conjugated IgG1 anti-HER2 control mAb. A representative experiment of three is shown.
Apoptotic Response Correlates with Caspase-3 Activation.
To assess the molecular substrate of the increased apoptosis in antisense transfectants, we examined caspase activity in treated cells. Current evidence indicates that caspases play a pivotal role in the initiation and execution of apoptosis in response to a number of cell death stimuli. To date, at least 10 different members of the caspase family have been identified in mammalian cells. Because caspase-3 has been reported to play a critical role in the execution machinery of mammalian apoptosis, we determined whether the apoptotic response in these cells was related to caspase-3 activation. This was monitored by measuring the proteolytic activity of the cell lysates from cells treated for 24 hours with anti-IgM on a pNA-derived tetrapeptide substrate. The basal levels of caspase-3 activity were similar in all of these cell lines. As shown in Fig. 5, the caspase activity after stimulation for 24 hours with anti-IgM did not exceed 1 unit in WT and control cells and was ≈5-fold higher in AS cells. We, therefore, conclude that caspase-mediated apoptosis in anti-IgM treated cells depends on the level of p21.
Figure 5.
Proteolytic cleavage of PARP and caspase activity in Daudi cells after treatment for 24 h with 20 μg/ml anti-IgM. Fifty micrograms of protein of total cell lysate per lane were separated by 10% SDS/PAGE. Immunodetection of PARP was carried out by using a 1:2,000 dilution of a Rb anti-PARP serum (Upper). Caspase-3-like protease activity was detected with ApoAlert CPP32 protease Assay kit (Lower). A representative experiment of three is shown.
Proteolytic Cleavage of PARP Occurs During Anti-IgM-Induced Apoptosis.
One of the immediate effects of caspase-3 activation is the cleavage of PARP. Western blot analysis of lysates of Daudi cells incubated for 24 hours with anti-IgM demonstrated cleavage of the 116-kDa PARP protein into its characteristic 85-kDa fragment. The extent of PARP cleavage paralleled the increase in caspase activity in antisense transfectants. Thus, in the AS transfectant, the 85-KDa cleavage product was 3.5-fold higher than in WT and control transfectants (Fig. 5). Taken together, these results suggest that induction of apoptosis in Daudi cells is linked to the pathway leading to cleavage of PARP and that p21 blocks or interferes at a point upstream of PARP cleavage. This suggests that p21 regulates both apoptosis and cell cycle progression in anti-IgM-treated Daudi cells. This anti-apoptotic role of p21 is consistent with that described by others with prostaglandin A2 in human breast carcinoma cells through expression of antisense p21 transcripts (28).
DISCUSSION
We have studied a murine B lymphoma model of dormancy in which anti-idiotype (or anti-IgM) antibody plays a major role in inducing and maintaining the dormant state (2, 3). Thus antiidiotype induces cell cycle arrest and apoptosis in these cells both in vivo and in vitro (4). One of the major questions is how the signal transduction pathways originating from the mIgM antigen receptor results in both cell death and cell cycle arrest. Understanding of this issue could be exploited therapeutically to shift the balance from one outcome to another, depending on the particular goals of the therapy: namely, killing tumor cells vs. maintaining a dormant state. To this end, we have been studying the role of CDK inhibitors in inducing cell cycle arrest after hypercrosslinking mIgM on a human lymphoma cell line (Daudi). We have found that hypercrosslinking of mIgM results in the induction of p21, which stoichiometrically inhibits the kinase activity of the cyclin E-CDK2 complex and consequently activates a late-G1 checkpoint. However, as mentioned above, anti-IgM treatment also results in the induction of apoptosis. The possible explanations to account for the observed anti-IgM-induced cell cycle arrest and apoptosis include (i) independent outcomes signaled through independent pathways, (ii) dependent outcomes initially signaled through a common pathway, in which arrest via the checkpoint regulator may be a necessary prelude to induction of apoptosis, or (iii) independent outcomes signaled through a common pathway, in which arrest initiated via the checkpoint regulator prevents rather than promotes apoptosis. The precise contribution of p21 to these different outcomes is not yet clear. Previous studies of others have demonstrated that p21 expression in response to exogenous stress factors results in cell cycle arrest (32, 33) and either stimulates (34) or inhibits (28, 35, 36) apoptosis, depending on the cell type and experimental conditions.
The major finding to emerge from the present study is that, in Daudi cells, p21 has a second function in addition to its role in the induction of cell cycle arrest: namely, direct interference with apoptosis. In this study, we analyzed the outcome of mIgM hypercrosslinking on Daudi cells in which the level of inducible p21 had been lowered by transfection with antisense p21 constructs. We found that a 4-fold reduction of the level of inducible p21 in anti-IgM-treated Daudi cells renders these cells more susceptible to apoptosis. We have considered two possibilities for this effect of p21. The simplest explanation is that prevention of replication is responsible for the above effect. Although there is no definitive evidence that progression through the cell cycle is required for apoptosis, the majority of cells that undergo apoptosis frequently depend on their cell cycle status (37). For example, Waldman et al. (38, 39) have demonstrated that the lack of a p21 checkpoint in p21 −/− human colon carcinoma cells increases their sensitivity to chemotherapeutic drugs or x-irradiation. This could be attributable to replication of cells with damaged DNA because of the failure of DNA repair. In a variety of systems in which exposure to stressful stimuli results in p53-independent cell death, p21 has been implicated in survival. Suppression of p21 expression through transfection with antisense p21 oligonucleotides or full transcripts was found to promote apoptosis by either blocking growth factor-induced differentiation of neuroblastoma cells (29) or by attenuating the CDK2-mediated growth arrest induced by prostaglandin A2 in human breast cells (28). Adenoviral overexpression of p21 in human colorectal carcinoma cells conferred protection against prostaglandin A2-mediated cell death (40). The protective role of p21 has also been recently demonstrated in the case of p53-dependent cell death. Thus, adenovirus-driven expression of p21 in human melanoma cells ectopically overexpressing p53 resulted in protection against p53-induced apoptosis (30). Taken together, the above results indicate a protective function of p21 in the decision between cell survival and cell death and suggests that the survival response conferred by p21 might depend, at least in part, on the direct control of the CDK2 activity required for G1/S transition. The importance of this issue is highlighted by the increasing evidence that specific cleavage of p21 by caspases results in the inability of p21 to bind and inactivate either CDK2 or proliferating cell nuclear antigen and that this is the central event during DNA-damage induced apoptosis in endothelial cells (41), myeloblastic leukemia (42), hepatoma cells (43), or lung cancer cells (44) of human origin.
Another possibility, which is not mutually exclusive, is that p21 per se inhibits apoptosis. A recent report indicates that protection from apoptosis by p21 involves not only G1 arrest but also the interruption of the caspase cascade at a point upstream from caspase-3 activation (45). In accordance with these results, we postulate that p21 might act not only as a substrate for caspases but also as an inhibitor of caspase-3 activation in the same manner as p21’s stoichiometric inhibition of cyclin/CDK complexes. This is suggested by the present results and the absence of any proteolytic cleavage of p21 in our previous studies. According to this scenario, the presence or absence of p21 after stimulation with anti-IgM will be reflected in the level of caspase-3 activation and, subsequently, in the level of PARP cleavage. Indeed, while our experiments were in progress, Suzuki et al. (46, 47) reported that regulation of caspase-3 activation by p21 is a key event in the resistance to Fas-mediated apoptosis in human hepatoma cells and that it involves the formation of a procaspase-3/p21 complex. This is accomplished via a caspase-3 binding domain in the N-terminal region of p21, distinct from those for CDK2 and proliferating cell nuclear antigen. Taken together with the aforementioned reports, our data suggest that p21 is a factor that protects cells from caspase-mediated apoptosis. The possibility that p21 interacts with procaspase-3 at different time points during anti-IgM stimulation of WT can be explored by immunoprecipitation. In addition, the distribution of p21 between viable and apoptotic cells using WT and AS cells can be determined. If this “stoichiometric” inhibition of caspase activation by p21 is a general mechanism for prevention of receptor signaled apoptosis, we postulate that the kinetics of p21 induction is a critical event for the final outcome. These observations have implications for our goal of understanding how to tip the balance between cell cycle arrest and apoptosis to optimize a treatment regimen using antitumor Abs and chemotherapy. Thus, pretreatment of cancer cells with a specific Ab that induces a G1 growth arrest mediated by p21 up-regulation could result in increased resistance to killing by chemotherapeutic agents that act downstream of G1. Because most of the chemotherapeutic agents currently used in antitumor therapy are acting on actively proliferating cells, the present results suggest that an antibody that induces cell cycle arrest should be given after chemotherapy because the lack of stimulation of p21 CDK inhibitor expression may markedly increase susceptibility to chemotherapy-induced apoptosis. Indeed, previous data of others (38, 39, 48, 49) indicate that p21-deficient human colorectal cancer cells are more sensitive to killing induced by a number of chemotherapeutic agents or x-irradiation.
Acknowledgments
We are deeply appreciative to Dr. Bert Vogelstein for providing pCEP4-WAF1-AS plasmid. We thank Drs. Richard Scheuermann and John Abrams for their critical reading of the manuscript and helpful suggestions. This research was supported by Leukemia Society of America Grant 6247-98.
ABBREVIATIONS
- AS
antisense
- CDK
cyclin-dependent kinase
- mIgM
membrane IgM
- p21
p21WAF1
- PARP
poly-(ADP ribose) polymerase
- pNA
p-nitroanilide
- WT
wild type
Footnotes
This is paper no. 8 in a series.
References
- 1.Page D, DeFranco A. Mol Cell Biol. 1990;10:3003–3012. doi: 10.1128/mcb.10.6.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uhr J W, Tucker T, May R D, Siu H, Vitetta E S. Cancer Res. 1991;51:5045S–5053S. [PubMed] [Google Scholar]
- 3.Yefenof E, Picker L J, Scheuermann R H, Tucker T F, Vitetta E S, Uhr J W. Proc Natl Acad Sci USA. 1993;90:1829–1833. doi: 10.1073/pnas.90.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Racila E, Scheuermann R H, Picker L J, Yefenof E, Tucker T, Chang W, Marches R, Street N E, Vitetta E S, Uhr J W. J Exp Med. 1995;181:1539–1550. doi: 10.1084/jem.181.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marches R, Racila E, Tucker T F, Picker L, Mongini P, Hsueh R, Vitetta E S, Scheuermann R H, Uhr J W. Ther Immunol. 1996;2:125–136. [PubMed] [Google Scholar]
- 6.Hasbold J, Klaus G G B. Eur J Immunol. 1990;20:1685–1690. doi: 10.1002/eji.1830200810. [DOI] [PubMed] [Google Scholar]
- 7.Beckwith M, Urba W J, Ferris D K, Freter C E, Kuhns D B, Moratz C M, Longo D L. J Immunol. 1991;147:2411–2418. [PubMed] [Google Scholar]
- 8.Joseph L F, Ezhevsky S, Scott D W. Cell Growth Differ. 1995;6:51–57. [PubMed] [Google Scholar]
- 9.Levy R, Miller A R. J Natl Cancer Inst Monogr. 1990;10:61–68. [PubMed] [Google Scholar]
- 10.Hsu F J, Kwak L, Campbell M, Liles T, Czerwinski D, Hart S, Syrengelas A, Miller R, Levy R. Ann NY Acad Sci. 1993;690:385–387. doi: 10.1111/j.1749-6632.1993.tb44039.x. [DOI] [PubMed] [Google Scholar]
- 11.Maloney D G, Grillo-Lopez A J, Bodkin D J, White C A, Liles T M, Royston I, Varns C, Rosenberg J, Levy R. J Clin Oncol. 1997;15:3266–3274. doi: 10.1200/JCO.1997.15.10.3266. [DOI] [PubMed] [Google Scholar]
- 12.Maloney D G, Grillo-Lopez A J, White C A, Bodkin D, Schilder R J, Neidhart J A, Janakiraman N, Foon K A, Liles T M, Dallaire B K, et al. Blood. 1997;90:2188–2195. [PubMed] [Google Scholar]
- 13.McLaughlin P, Grillolopez A J, Link B K, Levy R, Czuczman M S, Williams M E, Heyman M R, Bencebruckler I, White C A, Cabanillas F, et al. J Clin Oncol. 1998;16:2825–2833. doi: 10.1200/JCO.1998.16.8.2825. [DOI] [PubMed] [Google Scholar]
- 14.Tobinai K, Kobayashi Y, Narabayashi M, Ogura M, Kagami Y, Morishima Y, Ohtsu T, Igarashi T, Sasaki Y, Kinoshita T, et al. Ann Oncol. 1998;9:527–534. doi: 10.1023/a:1008265313133. [DOI] [PubMed] [Google Scholar]
- 15.Coiffier C, Haioun N, Ketterer A, Engert H, Tilly H, Ma D, Johnson P, Lister A, Feuring-Buske M, Radford J A, et al. Blood. 1998;92:1927–1932. [PubMed] [Google Scholar]
- 16.Tedder T F, Forsgren A, Boyd A W, Nadler L M, Schlossman S F. Eur J Immunol. 1986;16:881–887. doi: 10.1002/eji.1830160802. [DOI] [PubMed] [Google Scholar]
- 17.Demidem A, Lam T, Alas S, Hariharan K, Hanna N, Bonavida B. Cancer Biother Radiopharmacol. 1997;12:177–186. doi: 10.1089/cbr.1997.12.177. [DOI] [PubMed] [Google Scholar]
- 18.Taji H, Kagami Y, Ogada Y, Andou M, Nishi Y, Saito H, Seto M, Morishima Y. Jpn J Cancer Res. 1998;89:748–756. doi: 10.1111/j.1349-7006.1998.tb03280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shan D, Ledbetter J, Press O W. Blood. 1998;91:1644–1652. [PubMed] [Google Scholar]
- 20.Hudziak R M, Lewis G D, Winget M, Fendly B M, Shepard H M, Ullrich A. Mol Cell Biol. 1989;9:1165–1172. doi: 10.1128/mcb.9.3.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carter P, Presta L, Gorman C M, Ridgway J B, Henner D, Wong W L, Rowland A M, Kotts C, Carver M E, Shepard H M. Proc Natl Acad Sci USA. 1992;89:4285–4289. doi: 10.1073/pnas.89.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. Nature (London) 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 23.Marches R, Scheuermann R H, Uhr J W. Cancer Res. 1998;58:691–697. [PubMed] [Google Scholar]
- 24.Sangfelt O, Erickson S, Einhorn S, Grander D. Oncogene. 1997;14:415–423. doi: 10.1038/sj.onc.1200832. [DOI] [PubMed] [Google Scholar]
- 25.Mandal M, Bandyopadhyay D, Goepfert T M, Kumar R. Oncogene. 1998;16:217–225. doi: 10.1038/sj.onc.1201529. [DOI] [PubMed] [Google Scholar]
- 26.Subramaniam P S, Cruz P E, Hobeika A C, Johnson H M. Oncogene. 1998;16:1885–1890. doi: 10.1038/sj.onc.1201712. [DOI] [PubMed] [Google Scholar]
- 27.Meikrantz W, Schlegel R. J Cell Biochem. 1995;58:160–174. doi: 10.1002/jcb.240580205. [DOI] [PubMed] [Google Scholar]
- 28.Gorospe M, Holbrook N J. Cancer Res. 1996;56:475–479. [PubMed] [Google Scholar]
- 29.Poluha W, Poluha D K, Chang B, Crosbie N E, Schonhoff C M, Kilpatrick D L, Ross A H. Mol Cell Biol. 1996;16:1335–1341. doi: 10.1128/mcb.16.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorospe M, Cirielli C, Wang X, Seth P, Capogrossi M C, Holbrook N J. Oncogene. 1997;14:929–935. doi: 10.1038/sj.onc.1200897. [DOI] [PubMed] [Google Scholar]
- 31.Picker L J, Treer J R, Ferguson-Darnell B, Collins P A, Buck D, Terstappen L W M M. J Immunol. 1993;150:1105–1121. [PubMed] [Google Scholar]
- 32.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 33.Waldman T, Kinzler K W, Vogelstein B. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 34.Duttaroy A, Qian J F, Smith J S, Wang E. J Cell Biochem. 1997;64:434–446. [PubMed] [Google Scholar]
- 35.Canman C E, Gilmer T M, Coutts S B, Kastan M B. Genes Dev. 1995;9:600–611. doi: 10.1101/gad.9.5.600. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, Walsh K. Science. 1996;273:359–361. doi: 10.1126/science.273.5273.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishioka W K, Welsh R M. J Exp Med. 1994;179:769–774. doi: 10.1084/jem.179.2.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waldman T, Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1996;381:713–716. doi: 10.1038/381713a0. [DOI] [PubMed] [Google Scholar]
- 39.Waldman T, Zhang Y, Dillehay L, Yu J, Kinzler K, Vogelstein B, Williams J. Nat Med. 1997;3:1034–1036. doi: 10.1038/nm0997-1034. [DOI] [PubMed] [Google Scholar]
- 40.Gorospe M, Wang X, Guyton K Z, Holbrook N J. Mol Cell Biol. 1996;16:6654–6660. doi: 10.1128/mcb.16.12.6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levkau B, Koyama H, Raines E W, Clurman B E, Herren B, Orth K, Roberts J M, Ross R. Mol Cell. 1998;1:553–563. doi: 10.1016/s1097-2765(00)80055-6. [DOI] [PubMed] [Google Scholar]
- 42.Gervais J L M, Seth P, Zhang H. J Biol Chem. 1998;273:19207–19212. doi: 10.1074/jbc.273.30.19207. [DOI] [PubMed] [Google Scholar]
- 43.Park J A, Kim K-W, Kim S I, Lee S K. Eur J Biochem. 1998;257:242–248. doi: 10.1046/j.1432-1327.1998.2570242.x. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Fujita N, Tsuruo T. Oncogene. 1999;18:1131–1138. doi: 10.1038/sj.onc.1202426. [DOI] [PubMed] [Google Scholar]
- 45.Bissonnette N, Hunting D J. Oncogene. 1998;16:3461–3469. doi: 10.1038/sj.onc.1201899. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki A, Tsutomi Y, Akahane K, Araki T, Miura M. Oncogene. 1998;17:931–939. doi: 10.1038/sj.onc.1202021. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki A, Tsutomi Y, Miura M, Akahane K. Oncogene. 1999;18:1239–1244. doi: 10.1038/sj.onc.1202409. [DOI] [PubMed] [Google Scholar]
- 48.McDonald E R, III, Wu G S, Waldman T, El Deiry W S. Cancer Res. 1996;56:2250–2255. [PubMed] [Google Scholar]
- 49.Wouters B G, Giaccia A J, Denko N C, Brown J M. Cancer Res. 1997;57:4703–4706. [PubMed] [Google Scholar]