Abstract
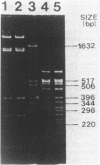
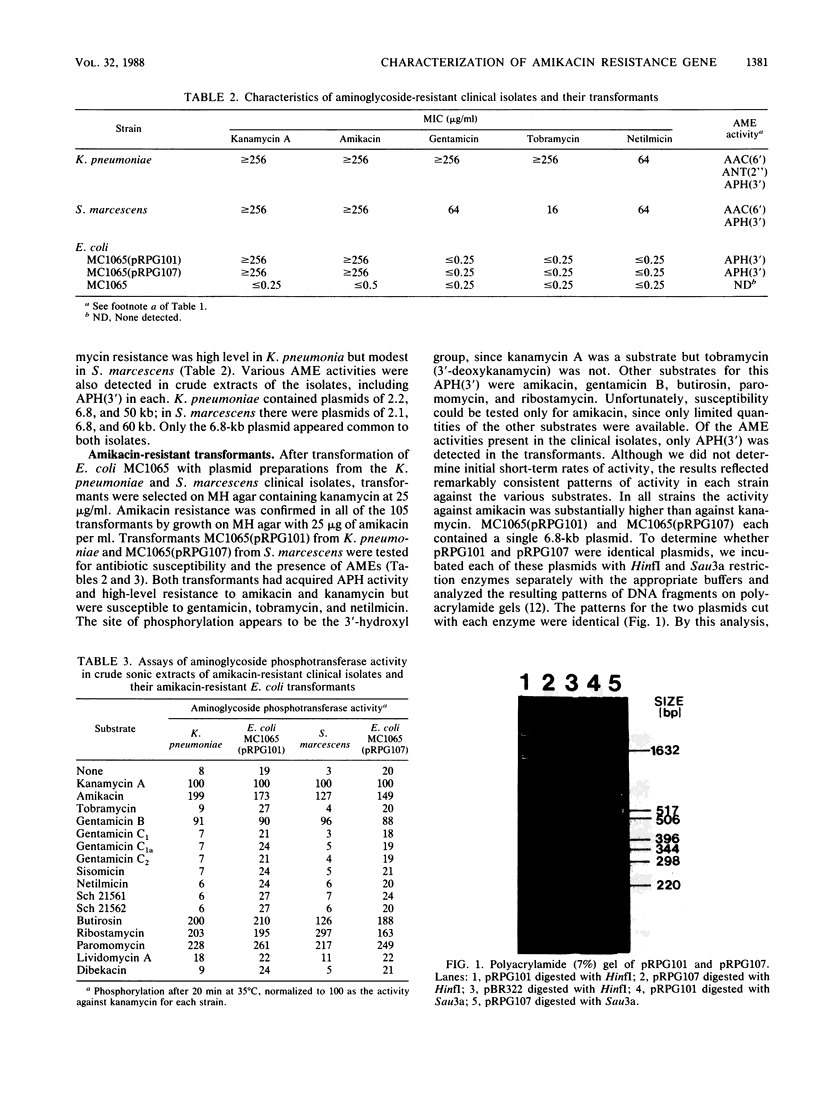
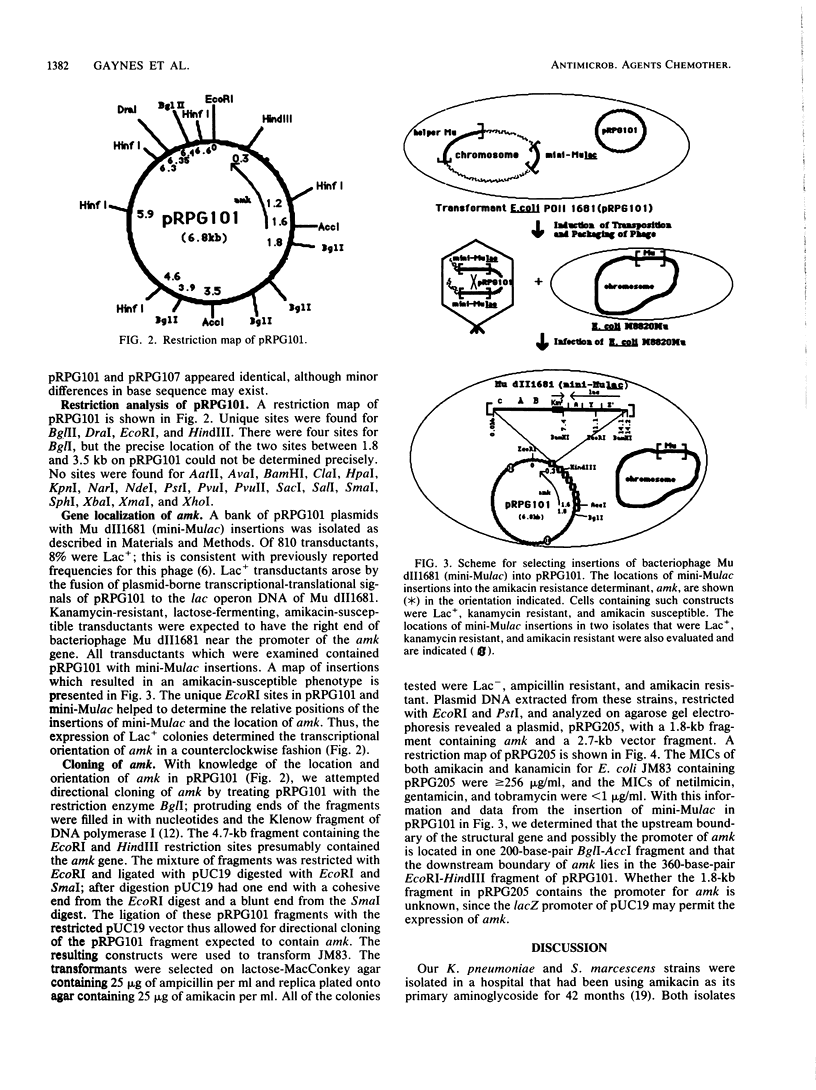
Clinical isolates of Klebsiella pneumoniae and Serratia marcescens at a hospital that had used amikacin as its principal aminoglycoside for the preceding 42 months demonstrated high-level resistance to amikacin (greater than or equal to 256 micrograms/ml), kanamycin (greater than or equal to 256 micrograms/ml), gentamicin (greater than or equal to 64 micrograms/ml), netilmicin (64 micrograms/ml), and tobramycin (greater than or equal to 16 micrograms/ml). The resistant strains contained an identical 6.8-kilobase plasmid, pRPG101. Transformation of pRPG101 into Escherichia coli produced high-level resistance to amikacin (greater than or equal to 256 micrograms/ml) and kanamycin (greater than or equal to 256 micrograms/ml) but unchanged susceptibilities to gentamicin, netilmicin, and tobramycin. The clinical isolates and transformants produced a novel 3'-phosphotransferase, APH(3'), that modified amikacin and kanamycin in vitro. The location and orientation of the amk gene encoding this APH(3') were determined by analysis of insertions in pRPG101 of the defective gene fusion phage Mu dII1681 (mini-Mulac). Cells containing plasmids with insertions into amk that had the lac operon fused to the amk promoter were selected as Lac+ and amikacin susceptible. A collection of these mini-Mulac insertions was mapped by restriction enzyme analysis. This characterization of amk facilitated its cloning as a 1.8-kilobase EcoRI-Bg/I fragment of pRPG101 into the pUC19 vector. E. coli strains containing this recombinant plasmid had APH(3') activity and demonstrated high-level resistance to amikacin and kanamycin (greater than or equal to 256 micrograms/ml) but were as susceptible to gentamicin, tobramycin, and netilmicin (less than or equal to 1.0 microgram/ml) as the strains harboring the original pRPG101 plasmid.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi T., Mizuuchi M., Robinson E. A., Appella E., O'Dea M. H., Gellert M., Mizuuchi K. DNA sequence of the E. coli gyrB gene: application of a new sequencing strategy. Nucleic Acids Res. 1987 Jan 26;15(2):771–784. doi: 10.1093/nar/15.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belas R., Mileham A., Simon M., Silverman M. Transposon mutagenesis of marine Vibrio spp. J Bacteriol. 1984 Jun;158(3):890–896. doi: 10.1128/jb.158.3.890-896.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Chou J. In vivo formation of gene fusions encoding hybrid beta-galactosidase proteins in one step with a transposable Mu-lac transducing phage. Proc Natl Acad Sci U S A. 1984 Jan;81(2):535–539. doi: 10.1073/pnas.81.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho B. A., Olfson P., Casadaban M. J. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J Bacteriol. 1984 May;158(2):488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeHertogh D. A., Lerner S. A. Correlation of aminoglycoside resistance with the KmS and Vmax/Km ratios of enzymatic modification of aminoglycosides by 2''-O-nucleotidyltransferase. Antimicrob Agents Chemother. 1985 Apr;27(4):670–671. doi: 10.1128/aac.27.4.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas M. J., Dowding J. E. Aminoglycoside-modifying enzymes. Methods Enzymol. 1975;43:611–628. doi: 10.1016/0076-6879(75)43124-x. [DOI] [PubMed] [Google Scholar]
- Lambert T., Gerbaud G., Courvalin P. Transferable amikacin resistance in Acinetobacter spp. due to a new type of 3'-aminoglycoside phosphotransferase. Antimicrob Agents Chemother. 1988 Jan;32(1):15–19. doi: 10.1128/aac.32.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine J. F., Maslow M. J., Leibowitz R. E., Pollock A. A., Hanna B. A., Schaefler S., Simberkoff M. S., Rahal J. J., Jr Amikacin-resistant gram-negative bacilli: correlation of occurrence with amikacin use. J Infect Dis. 1985 Feb;151(2):295–300. doi: 10.1093/infdis/151.2.295. [DOI] [PubMed] [Google Scholar]
- Meyer R. D. Patterns and mechanisms of emergence of resistance to amikacin. J Infect Dis. 1977 Sep;136(3):449–452. doi: 10.1093/infdis/136.3.449. [DOI] [PubMed] [Google Scholar]
- Moody M. M., de Jongh C. A., Schimpff S. C., Tillman G. L. Long-term amikacin use. Effects on aminoglycoside susceptibility patterns of gram-negative bacilli. JAMA. 1982 Sep 10;248(10):1199–1202. doi: 10.1001/jama.248.10.1199. [DOI] [PubMed] [Google Scholar]
- Oka A., Sugisaki H., Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981 Apr 5;147(2):217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- Perlin M. H., Lerner S. A. High-level amikacin resistance in Escherichia coli due to phosphorylation and impaired aminoglycoside uptake. Antimicrob Agents Chemother. 1986 Feb;29(2):216–224. doi: 10.1128/aac.29.2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price K. E., Kresel P. A., Farchione L. A., Siskin S. B., Karpow S. A. Epidemiological studies of aminoglycoside resistance in the U.S.A. J Antimicrob Chemother. 1981 Jul;8 (Suppl A):89–105. doi: 10.1093/jac/8.suppl_a.89. [DOI] [PubMed] [Google Scholar]
- Saavedra S., Vera D., Ramírez-Ronda C. H. Susceptibility of aerobic gram-negative bacilli to aminoglycosides. Effects of 45 months of amikacin as first-line aminoglycoside therapy. Am J Med. 1986 Jun 30;80(6B):65–70. doi: 10.1016/0002-9343(86)90481-x. [DOI] [PubMed] [Google Scholar]
- Thompson C. J., Gray G. S. Nucleotide sequence of a streptomycete aminoglycoside phosphotransferase gene and its relationship to phosphotransferases encoded by resistance plasmids. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5190–5194. doi: 10.1073/pnas.80.17.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Courvalin P. Evolution and transfer of aminoglycoside resistance genes under natural conditions. J Antimicrob Chemother. 1986 Oct;18 (Suppl 100):93–102. doi: 10.1093/jac/18.supplement_c.93. [DOI] [PubMed] [Google Scholar]
- Trieu-Cuot P., Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3'5"-aminoglycoside phosphotransferase type III. Gene. 1983 Sep;23(3):331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- Umezawa Y., Yagisawa M., Sawa T., Takeuchi T., Umezawa H. Aminoglycoside 3'-phosphotransferase III, a new phosphotransferase. Resistance mechanism. J Antibiot (Tokyo) 1975 Nov;28(11):845–853. doi: 10.7164/antibiotics.28.845. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Young S. A., Tenover F. C., Gootz T. D., Gordon K. P., Plorde J. J. Development of two DNA probes for differentiating the structural genes of subclasses I and II of the aminoglycoside-modifying enzyme 3'-aminoglycoside phosphotransferase. Antimicrob Agents Chemother. 1985 May;27(5):739–744. doi: 10.1128/aac.27.5.739. [DOI] [PMC free article] [PubMed] [Google Scholar]